Abstract
With the improved secreted protein prediction approach and comprehensive data sources, including Swiss-Prot, TrEMBL, RefSeq, Ensembl and CBI-Gene, we have constructed secretomes of human, mouse and rat, with a total of 18 152 secreted proteins. All the entries are ranked according to the prediction confidence. They were further annotated via a proteome annotation pipeline that we developed. We also set up a secreted protein classification pipeline and classified our predicted secreted proteins into different functional categories. To make the dataset more convincing and comprehensive, nine reference datasets are also integrated, such as the secreted proteins from the Gene Ontology Annotation (GOA) system at the European Bioinformatics Institute, and the vertebrate secreted proteins from Swiss-Prot. All these entries were grouped via a TribeMCL based clustering pipeline. We have constructed a web-based secreted protein database, which has been publicly available at http://spd.cbi.pku.edu.cn. Users can browse the database via a GO assignment or chromosomal-location-based interface. Moreover, text query and sequence similarity search are also provided, and the sequence and annotation data can be downloaded freely from the SPD website.
INTRODUCTION
Secreted proteins such as cytokines, chemokines, hormones, digestive enzymes, antibodies as well as components of the extra-cellular matrix, are secreted from cells into the extra-cellular space. They play pivotal biological regulatory roles and have the potential for protein therapeutics (1). The majority of secreted proteins have a signal peptide according to the signal hypothesis (2). Signal peptides are located at the N-terminal of nascent proteins and their lengths are usually <70 amino acid residues. They are cleaved during the process of entering the endoplasmic reticulum (ER) lumen. The signal peptide is a hallmark of secreted proteins. However, many transmembrane (TM) proteins also have a signal peptide (3–4). Several secreted protein prediction methods have been developed mainly based on the analysis of signal peptides, and genome-wide identification of potential novel secreted proteins has been reported (5–7).
In this study, we implemented an improved secreted protein prediction approach, CJ-SPHMM+TMHMM+PSORT (8–10), to search a comprehensive data source, including Swiss-Prot/TrEMBL (11), RefSeq (12), Ensembl (13) as well as CBI-Gene constructed locally, and constructed the secretomes of human, mouse and rat. We have also set up a complete secreted protein classification pipeline, and classified our predicted secreted proteins into different functional categories. To make our predicted results more comprehensive, we collected nine reference datasets including the Secreted Protein Discovery Initiative (SPDI) (5), the Riken mouse secretomes (6), the secreted proteins from Gene Ontology Annotation (GOA) (14), etc. A TribeMCL (15) based cluster pipeline was implemented, to group our predicted secreted proteins with these reference sequences. All the sequence data and annotation information have been publicly available at http://spd.cbi.pku.edu.cn.
CONSTRUCTION OF THE PIPELINE
Collection and annotation of the core dataset
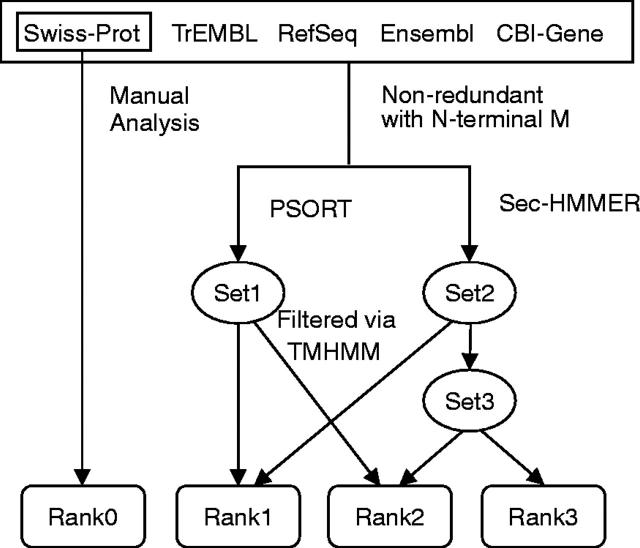
SPD consists of a core dataset and a reference dataset. The core dataset contains 18 152 secreted proteins retrieved from Swiss-Prot/TrEMBL, Ensembl, RefSeq and CBI-Gene. The pipeline of constructing the dataset is shown in Figure 1. A combined strategy was applied to collect as much secreted proteins as possible, using both automatic processing and manual intervening. The dataset Rank0 from Swiss-Prot includes some partial sequences without the N- or C-termini, for they were collected according to database annotation. Given that most of the signal peptides are located at the N-terminal of proteins and fundamental to secreted protein prediction, we eliminated the entries without N-terminal methionine (Met, M) in CBI-Gene, Ensembl, Swiss-Prot/TrEMBL and RefSeq in our prediction results, since some entries from these datasets are hypothetical and truncated. Therefore, proteins in the datasets of Rank1, 2, 3 all have N-terminal Met.
Figure 1.
The pipeline we used to collect secreted proteins. Sequence data were downloaded by December 2003. Non-redundant: identical sequences were excluded via pairwise BLASTP. Sec-HMMER (CJ-SPHMM/TMHMM): the tool used to predict signal peptides and transmembrane region. Set1 and Set2: proteins predicted by PSORT and Sec-HMMER, respectively; Set3: proteins matched by Sec-HMMER only. Rank0: known secreted proteins in Swiss-Prot; Rank1: predicted by both PSORT and Sec-HMMER; Rank2: predicted by either PSORT or Sec-HMMER; and Rank3: predicted by Sec-HMMER only and with a signal peptide >70 amino acids.
All the 18 152 sequences were annotated via the Protein Centric Annotation System (PCAS), an integrated protein annotation system that we developed previously (16). Functional classification was also performed on all datasets. We extracted all vertebrate secreted proteins from the Swiss-Prot database. Based on the annotation information, they were assigned into 11 classes: antibiotic protein, apolipoprotein, casein, cytokine, hormone, immune system protein, neuropeptide and defense peptide, protease, protease inhibitor, toxin and Wnt protein. Entries without explicit functional annotation were defined as ‘other secreted proteins’. Based on the cross-link information in Swiss-Prot, we obtained representative motifs and domains from Pfam (17), PROSITE (18), SMART (19) and PRINTS (20), representing one of the above eleven functional classes. Taking the Wnt entry in Pfam as an example, if a protein sequence has a Wnt domain, it was classified as a Wnt protein. BLAST searches (21) against the above 11 classes were performed taking all the predicted secreted proteins as queries. If the query sequence has >50% identity with a known protein in class A and the matched sequence length is >80%, we classified this novel secreted protein into class A. For those proteins failing to meet with this cutoff, they were also classified into class A, if they comprise a motif or domain belonging to proteins in class A. An approach with similar ideas to predict sub-cellular location of proteins has been described previously (22). A total of ∼3000 novel secreted proteins were classified into these 11 classes. Details of the domains and protein function assignment can be found at the SPD website. However, the majority of the predicted proteins could not be assigned to these classes, since the number of known representative domains is still very small.
Collection and clustering of the reference dataset
The SPD reference dataset consists of the following data sources: SPDI (5), Riken Mouse Secretome (6), the human and pufferfish secretome (7), Swiss-Prot secreted proteins of vertebrates except for human, mouse and rat (11), secreted proteins extracted from GO assignment (14), DBSubLoc (23), NPD (24), TMPDB (25) and NESbase (26). Most of the data were downloaded from their websites or retrieved from the supplementary materials of related literatures. Vertebrate secreted proteins were extracted from Swiss-Prot according to the annotation. As for GOA, we took all entries as secreted proteins if they were annotated as ‘extracellular matrix’ (ID 0005578) or ‘extracellular space’ (ID 0005615).
All these nine datasets comprise a comprehensive reference dataset. SPDI, Riken, the human and pufferfish secretome, the Swiss-Prot vertebrates and the GO datasets focus on collecting secreted proteins. They were taken as positive controls. On the other hand, NPD, TMPDB and NESbase collect nuclear proteins, transmembrane proteins and proteins with nuclear export signal, respectively. They may serve as negative controls. In total, the SPD core dataset includes 65–75% positive controls and 5–8% negative controls. DBSubLoc is a database of sub-cellular location of proteins, hence, useful in both aspects.
In addition, pairwise BLASTP was performed between each entry of the SPD core dataset and the reference datasets. The output results were processed via TribeMCL, and relevant entries were clustered together. Here BLAST E-value cutoff is set to 1E−10 and inflation value of TribeMCL is set to 5 (27).
WEB INTERFACE
Currently, the SPD web interface includes five modules. (i) Browse: browse SPD proteins according to chromosome and functional classification or GO assignment. (ii) Search: text query from the core dataset with protein IDs, keywords, descriptions and sequence similarity search against both core and reference datasets. (iii) Download: download SPD protein sequences, corresponding cDNAs, etc. (iv) Data statistics: statistics table of the secreted proteins in each division. (v) Help: frequently asked questions about SPD, including descriptions of using the web interface and details of the SPD construction pipeline. The chromosomal browser was designed to show the chromosomal content of proteins with a common feature, for example, proteins from the same data source or with the same confidence rank, etc. (Figure 2A). The GO browser organizes the proteins based on the GO assignments. The text search function supports the Boolean mode, and could be used to look for proteins with special description, length, etc. Sequence similarity search can be used to find the entries similar to the query sequence via BLASTP or BLASTX.
Figure 2.
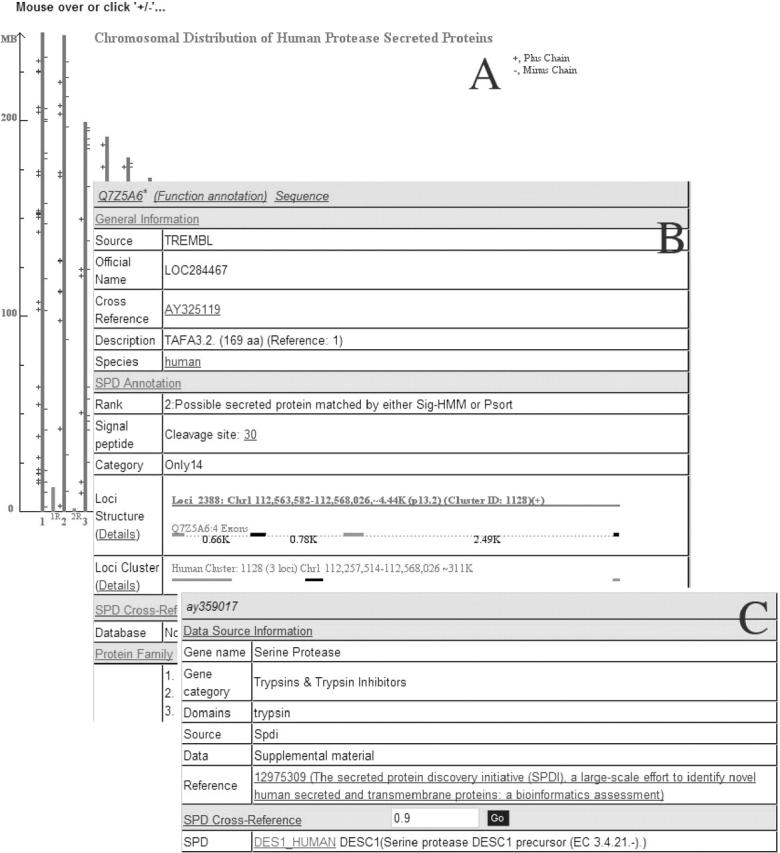
Screen snapshots of the SPD database, only partially drawn (A and B) for clarity. (A) An example page of the chromosomal location of human protease secreted proteins. The clickable ‘+’ and ‘−’ symbols denotes the plus and minus strands as indicated in the browser at the SPD website. (B) An example of an SPD core dataset entry (TAFA 3.2) showing four divisions of the entry format and various fields with links to detail information and original data source. (C) An example of an SPD reference dataset entry (AY359017) showing two divisions with general information.
Entry format of the core dataset
The data fields of the entries of the core dataset was designed as four major divisions and displayed as separate parts within a table on the web page: the general information, the SPD annotation, the SPD cross-reference and the protein family (Figure 2B). Each entry starts with a header line at the top with links to the PCAS annotation and original websites of this protein (16).
The ‘General information’ section is designed to show names, descriptions, reference papers, cDNAs and the GO assignment, etc., retrieved from original data sources. Names of Swiss-Prot/TrEMBL/RefSeq entries were taken from the ucsc_kgXref table (28). Ensembl names were retrieved by the EnsMart batch query (29). For the ‘GO’ field, if multiple GO entries were assigned to a protein, one schematic figure can be shown to display the relationship between these GO entries. The ‘SPD annotation’ includes confidence rank, signal peptide cleavage site, functional category, domain structure, chromosomal loci, loci cluster, homolog clusters, etc. The ‘Domain structure’ field displays possible domain architectures derived from the PCAS system. The ‘Loci structure’ and ‘Loci cluster’ fields show the chromosomal content of this entry obtained from BLAT search against the UCSC HG16, mm4 and rn3, respectively (30,31). Users can browse the detailed information such as the intron/exon structure, synteny pair information, genetic band, etc. The ‘Homolog cluster’ field displays similar sequences with overall identity cutoff >90% and overall length coverage >90%. The ‘SPD Cross-Reference’ field can be used to show similar sequences from different reference datasets. By default, only sequences with overall identity >90% are shown. The ‘Protein Family’ field gives the corresponding cluster yielded via TribeMCL.
Entry format of the reference dataset
Comparing with the format of the core dataset, the layout of the entries of the reference dataset is relatively simple and can be divided into two large sections. The first section shows the original information, such as the data source, and the second part lists the similar sequences from the core dataset with a certain overall identity cutoff (Figure 2C).
DISCUSSION
SPD is comprehensive
SPD tends to be inclusive, not exclusive. In other words, SPD collects as many secreted proteins as possible. First, all distinct sequences are kept in the database, including entries with >90% identity, for it is difficult to decide on the correct variant sequence. Second, nine reference datasets were also introduced to help increase the coverage. For example, most members of a recently identified secreted protein family, TAFA, have been covered by SPD (32). In fact, reference datasets may also help users to gain some information, such as similarity relationships between entries, to discern possible redundancy. In addition, the rank system can be implemented to show the prediction confidence as well.
Distinguish true positives from false positives
Four modules are provided to help users to judge the entries that are true positives. (i) The rank system: proteins of Rank0 or Rank1 tend to be more convincing than those of Rank2 and Rank3. (ii) The category assignment: proteins classified into relevant functional categories are usually more reliable. (iii) Clustering information: users may make some judgment according to the clustering information. For example, an SPD secreted protein might be more reliable if it is grouped into a cluster comprising many entries from GO secreted proteins, Riken mouse secretome, etc. (iv) GO assignment: proteins with GO assignment like ‘extracellular space’ or ‘extracellular matrix’ are likely to be true positives. In contrast, proteins like ‘integral to membrane’ tend to be false positive.
SPD is tuned for biologists
SPD has been tuned for biologists looking for novel secreted proteins. First, mRNA or cDNA sequences can be found in the ‘cross-reference’ field. Second, reference number is shown in the ‘description’ field, which reflects whether this protein is novel or not.
Conflicts with GO assignments
Based on the GO browser, users could find that many proteins with some GO assignments have conflicts, such as membrane, intracellular, etc. This could be explained in three aspects: (i) some proteins can be sorted to multiple locations; (ii) some proteins have low prediction confidence, such as those in rank3; and (iii) the GO assignment might be not much convincing, for example, cellular component information labeled with IEA (inferred from electronic annotation) or NR (not recorded) tends to be not very reliable.
FUTURE DEVELOPMENT
The current SPD database has data source from three model organisms. We plan to add secretomes from other organisms, when their completed genome sequences are available. Moreover, evolutionary analysis to construct the ortholog groups is underway to provide useful information for wet lab biological experiments.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by grants from China National High-Tech 863 Program and Beijing Municipal Committee of Science and Technology. The authors appreciate the anonymous referees for their helpful comments and suggestions.
REFERENCES
- 1.Damas J.K., Gullestad,L. and Aukrust,P. (2001) Cytokines as new treatment targets in chronic heart failure. Curr. Control Trials Cardiovasc. Med., 2, 271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blobel G. (2000) Protein targeting (Nobel lecture). Chembiochem, 1, 86–102. [DOI] [PubMed] [Google Scholar]
- 3.Walter P., Gilmore,R. and Blobel,G. (1984) Protein translocation across the endoplasmic reticulum. Cell, 38, 5–8. [DOI] [PubMed] [Google Scholar]
- 4.Sakaguchi M. (1997) Eukaryotic protein secretion. Curr. Opin. Biotechnol., 8, 595–601. [DOI] [PubMed] [Google Scholar]
- 5.Clark H.F., Gurney,A.L., Abaya,E., Baker,K., Baldwin,D., Brush,J., Chen,J., Chow,B., Chui,C., Crowley,C. et al. (2003) The secreted protein discovery initiative (SPDI), a large-scale effort to identify novel human secreted and transmembrane proteins: a bioinformatics assessment. Genome Res., 13, 2265–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grimmond S.M., Miranda,K.C., Yuan,Z., Davis,M.J., Hume,D.A., Yagi,K., Tominaga,N., Bono,H., Hayashizaki,Y., Okazaki,Y. et al. (2003) The mouse secretome: functional classification of the proteins secreted into the extracellular environment. Genome Res., 13, 1350–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klee E.W., Carlson,D.F., Fahrenkrug,S.C., Ekker,S.C. and Ellis,L.B. (2004) Identifying secretomes in people, pufferfish and pigs. Nucleic Acids Res., 32, 1414–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y., Yu,P., Luo,J. and Jiang,Y. (2003) Secreted protein prediction system combining CJ-SPHMM, TMHMM, and PSORT. Mamm. Genome, 14, 859–865. [DOI] [PubMed] [Google Scholar]
- 9.Sonnhammer E.L., von Heijne,G. and Krogh,A. (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol., 6, 175–182. [PubMed] [Google Scholar]
- 10.Nakai K. and Horton,P. (1999) PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci., 24, 34–36. [DOI] [PubMed] [Google Scholar]
- 11.Boeckmann B., Bairoch,A., Apweiler,R., Blatter,M.C., Estreicher,A., Gasteiger,E., Martin,M.J., Michoud,K., O'Donovan,C., Phan,I. et al. (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res., 31, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pruitt K.D. and Maglott,D.R. (2001) RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res., 29, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Birney E., Andrews,D., Bevan,P., Caccamo,M., Cameron,G., Chen,Y., Clarke,L., Coates,G., Cox,T., Cuff,J. et al. (2004) Ensembl 2004. Nucleic Acids Res., 32, D468–D470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camon E., Magrane,M., Barrell,D., Lee,V., Dimmer,E., Maslen,J., Binns,D., Harte,N., Lopez,R. and Apweiler,R. (2004) The Gene Ontology Annotation (GOA) Database: sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res., 32, D262–D266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enright A.J., Van Dongen,S. and Ouzounis,C.A. (2002) An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res., 30, 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y., Yin,Y., Chen,Y., Gao,G., Yu,P., Luo,J. and Jiang,Y. (2003) PCAS—a precomputed proteome annotation database resource. BMC Genomics, 4, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L. (2002) The Pfam protein families database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falquet L., Pagni,M., Bucher,P., Hulo,N., Sigrist,C.J., Hofmann,K. and Bairoch,A. (2002) The PROSITE database, its status in 2002. Nucleic Acids Res., 30, 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Letunic I., Goodstadt,L., Dickens,N.J., Doerks,T., Schultz,J., Mott,R., Ciccarelli,F., Copley,R.R., Ponting,C.P. and Bork,P. (2002) Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res., 30, 242–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Attwood T.K., Blythe,M.J., Flower,D.R., Gaulton,A., Mabey,J.E., Maudling,N., McGregor,L., Mitchell,A.L., Moulton,G., Paine,K. et al. (2002) PRINTS and PRINTS-S shed light on protein ancestry. Nucleic Acids Res., 30, 239–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mott R., Schultz,J., Bork,P. and Ponting,C.P. (2002) Predicting protein cellular localization using a domain projection method. Genome Res., 12, 1168–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo T., Hua,S., Ji,X. and Sun,Z. (2004) DBSubLoc: database of protein subcellular localization. Nucleic Acids Res., 32, D122–D124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dellaire G., Farrall,R. and Bickmore,W.A. (2003) The Nuclear Protein Database (NPD): sub-nuclear localisation and functional annotation of the nuclear proteome. Nucleic Acids Res., 31, 328–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ikeda M., Arai,M., Okuno,T. and Shimizu,T. (2003) TMPDB: a database of experimentally-characterized transmembrane topologies. Nucleic Acids Res., 31, 406–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.la Cour T., Gupta,R., Rapacki,K., Skriver,K., Poulsen,F.M. and Brunak,S. (2003) NESbase version 1.0: a database of nuclear export signals. Nucleic Acids Res., 31, 393–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forrest A.R., Ravasi,T., Taylor,D., Huber,T., Hume,D.A. and Grimmond,S. (2003) Phosphoregulators: protein kinases and protein phosphatases of mouse. Genome Res., 13, 1443–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karolchik D., Hinrichs,A.S., Furey,T.S., Roskin,K.M., Sugnet,C.W., Haussler,D. and Kent,W.J. (2004) The UCSC Table Browser data retrieval tool. Nucleic Acids Res., 32, D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilbert D. (2003) Shopping in the genome market with EnsMart. Brief Bioinformatics, 4, 292–296. [DOI] [PubMed] [Google Scholar]
- 30.Karolchik D., Baertsch,R., Diekhans,M., Furey,T.S., Hinrichs,A., Lu,Y.T., Roskin,K.M., Schwartz,M., Sugnet,C.W., Thomas,D.J. et al. (2003) The UCSC Genome Browser Database. Nucleic Acids Res., 31, 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kent W.J. (2002) BLAT—the BLAST-like alignment tool. Genome Res., 12, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tom Tang Y., Emtage,P., Funk,W.D., Hu,T., Arterburn,M., Park,E.E.J. and Rupp,F. (2004) TAFA: a novel secreted family with conserved cysteine residues and restricted expression in the brain. Genomics, 83, 727–734. [DOI] [PubMed] [Google Scholar]