Abstract
The SilkSatDb (silkmoth microsatellite database) (http://www.cdfd.org.in/silksatdb) is a relational database of microsatellites extracted from the available expressed sequence tags and whole genome shotgun sequences of the silkmoth, Bombyx mori. The database has been rendered with a simple and robust web-based search facility, developed using PHP. The SilkSatDb also stores information on primers developed and validated in the laboratory. Users can retrieve information on the microsatellite and the protocols used, along with informative figures and polymorphism status of those microsatellites. In addition, the interface is coupled with Autoprimer, a primer-designing program, using which users can design primers for the loci of interest.
INTRODUCTION
Microsatellites are short tandem repeats of 1–6 bp, which are ubiquitous in both prokaryotes and eukaryotes both in protein-coding and non-coding regions (1). Microsatellites are used widely in a variety of applications including genetic distance measures and phylogeny reconstruction, population genetics, genetic mapping, elucidating evolutionary history, artificial selection and forensics (2,3). As the whole genome sequence data of many organisms are now available, an unambiguous picture of occurrence and genomic distribution of microsatellites is emerging, and the study of microsatellites has attained importance to address various biological questions.
The silkworm, Bombyx mori is an important economic insect and lepidopteran molecular model (4). Study of microsatellites in this insect will help in genetic fingerprinting of diverse silkmoths, construction of molecular linkage map, marker assisted selection, in addition to the basic understanding of microsatellites (5,6). Evolutionarily conserved microsatellite loci of B.mori can further be extended to study other lepidopteran insects, which include some of the most destructive agricultural pests. With expressed sequence tag (EST) sequences published (7) and sequenced whole genome shotgun (WGS) sequences prepared for silkworm (8), an effort has been put forth to create a database of microsatellites called SilkSatDb. The data generated will be useful for the research community working on insect genetics, particularly lepidopteran geneticists across the globe.
STRUCTURE OF THE DATABASE
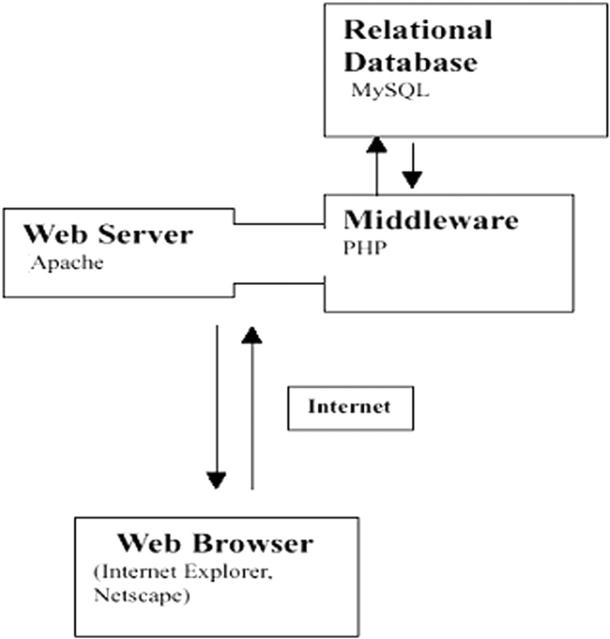
The SilkSatDb is an online relational database that catalogues information about the microsatellite repeats of the silkworm. The design of the database schema basically follows the ‘Three-level schema architecture’ as shown in Figure 1. The database stores three kinds of data: the microsatellite repeats found in B.mori EST and WGS sequences, sequence details and the primers developed for these microsatellites. Currently, the database encompasses information pertaining to 12 000 WGS contigs downloaded from DDBJ (http://www.ddbj.nig.ac.jp/anoftp-e.html), 9300 EST and 50 genomic sequences of B.mori. Each entry in the sequence information table includes an identifier for the sequence type and the accession number to which it belongs. The accession number is a number, possibly with a few characters in front, that uniquely identifies the sequence stored in the flat-file format. Since ESTs are a large collection, information is stored with respect to their cDNA library clone names (based on the annotation from http://www.ab.a.u-tokyo.ac.jp/silkbase/).
Figure 1.
An illustration of three level architecture used in the development of SilkSatDb.
The extraction of microsatellite repeats is done using Simple Sequence Repeat Finder (SSRF) program (9,10), which is written in C. This program scans a given DNA sequence and identifies non-redundant, perfect microsatellite tracts. Repeats information extracted is categorized (except EST) as mono, di, tri, tetra, penta and hexa repeats and placed in their respective sequence groupings. The EST sequence repeats are classified according to their library type.
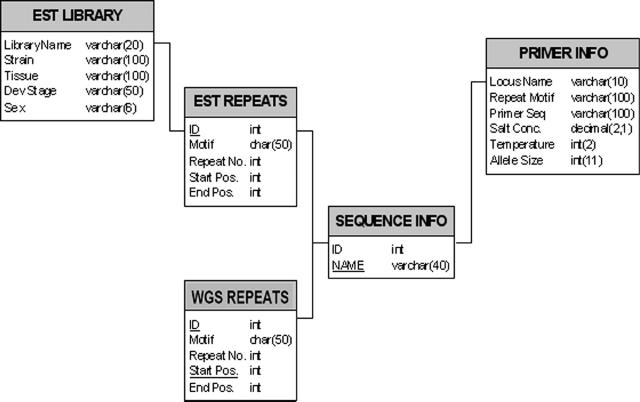
In addition to the sequence information as mentioned above the database also includes a list of primers, designed and tested in our laboratory, for about 200 loci, with their respective PCR amplification conditions. Figure 2 illustrates the entire relational schema of SilkSatDb.
Figure 2.
The schema of the SilkSatDb relational database.
Besides storing data on microsatellites, SilkSatDb hosts other useful details of microsatellites such as their frequencies, different types of mutations prevalent in them, their allelic frequency, mapping populations and their evolutionary conservation in heterologous silkmoths (11). The protocols for microsatellite analysis and the recently improvised methodology for the inter simple sequence repeats (ISSRs)-based genotyping are also listed with experimental details. Thus, the whole website forms an integrated site wherein the in silico analysis is extended to bench, providing the user everything about silkmoth microsatellites and microsatellite-based markers. To the best of our knowledge, this is the first site on microsatellites and microsatellite-based genetic markers in insects outside Drosophila.
DATA EXTRACTION
The user-friendly interface for the database has been developed using PHP, a server side scripting language. This acts as a comprehensive and integrated resource for retrieval of the information from SilkSatDb. The user can query for microsatellites, extracted from WGS and EST sequences, using the repeat type/motif and the number of occurrences of the repeat.
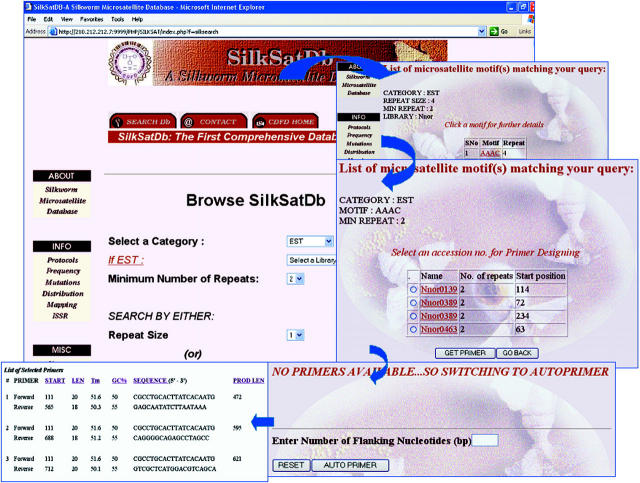
The query results are displayed in a tabular format showing the accession number, the motif, its frequency and location in the sequence. Hyper links are provided for the motif and the accession number in the table. The motif link takes the user to the detailed description of the motif occurrence in each sequence. The accession number link connects to its respective sequence information. Selection buttons are provided for each of these accession numbers in the table, by which the user can get the available primers corresponding to the microsatellite tracts. If the primers are not available, the interface links to ‘Autoprimer’ (9,10), an automated primer design software that takes the sequence data along with flanking regions and other primer parameters such as primer length, GC content, Tm etc., as an input for the designing of the primers (Figure 3).
Figure 3.
Symbol display of query form for repeat motif search, output format for microsatellite loci and the primer designing using ‘Autoprimer’ primer-designing program for the selected locus.
FUTURE PERSPECTIVES
Plans for future database releases include incorporation of additional microsatellite loci to the existing data as and when they are developed and validated. The construction of the microsatellite linkage map of silkworm is underway and the mapping data will be hyperlinked to the SilksatDb when it becomes available. The EST sequencing projects from various lepidopterans are underway in several laboratories. In future, the database will be updated with microsatellite loci extracted from the genome and EST sequences. Also, the microsatellite loci of silkworm are being tested in heterologous species, for their evolutionary conservation. These data too will be uploaded when they become available, so that the community of investigators from insect molecular biology and molecular ecology can utilize this resource to address various issues.
ACCESS
This is a new molecular genetic resource freely accessible for research purposes for non-profit and academic organizations at http://www.cdfd.org.in/silksatdb. Comments, suggestions and questions are welcome and should be directed to jnagaraju@cdfd.org.in.
Acknowledgments
ACKNOWLEDGEMENTS
The authors acknowledge the help of Mrs. Geeta Thanu during the database creation. This work was supported by a grant from Department of Biotechnology, Government of India, New Delhi, to J.N. H.A.N. gratefully acknowledges the core grant support of CDFD. K.P.A. and V.B.S. are recipients of CSIR research fellowship.
REFERENCES
- 1.Toth G., Gaspari,Z. and Jurka,J. (2000) Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res., 10, 967–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schlotterer C. (2000) Evolutionary dynamics of microsatellite DNA. Chromosoma, 109, 365–371. [DOI] [PubMed] [Google Scholar]
- 3.Ellegren H. (2004) Microsatellites: simple sequences with complex evolution. Nature Rev. Genet., 5, 435–445. [DOI] [PubMed] [Google Scholar]
- 4.Nagaraju J., Klimenko,V. and Couble,P. (2000) The silkworm Bombyx mori, a model genetic system. In Reeves, E. (ed.), Encyclopedia of Genetics. Fitzroy Dearborn, London, UK, pp. 219–239.
- 5.Nagaraju J. and Goldsmith,M.R. (2002) Silkworm genomics—progress and prospects. Curr. Sci., 83, 415–425. [Google Scholar]
- 6.Reddy K.D., Abraham,E.G. and Nagaraju,J. (1999) Microsatellites of the silkworm, Bombyx mori: abundance, polymorphism and strain characterization. Genome, 42, 1057–1065. [PubMed] [Google Scholar]
- 7.Mita K., Morimyo,M., Okano,K., Koike,Y., Nohata,J., Kawasaki,H., Kadono-Okuda,K.,Yamamoto,K., Suzuki,M.G., Shimada,T. et al. (2003) The construction of an EST database for Bombyx mori and its application. Proc. Natl Acad. Sci. USA, 100, 14121–14126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mita K., Kasahara,M., Sasaki,S., Nagayasu,Y., Yamada,T., Kanamori,H., Namiki,N., Kitagawa,M., Yamashita,H., Yasukochi,Y. et al. (2004) The genome sequence of silkworm, Bombyx mori. DNA Res., 11, 27–35. [DOI] [PubMed] [Google Scholar]
- 9.Sreenu V.B., Vishwanath,A., Nagaraju,J. and Nagarajaram,H.A. (2003) MICdb: database of prokaryotic microsatellites. Nucleic Acids Res., 31, 106–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sreenu V.B., Ranjitkumar,G., Swaminathan,S., Priya,S., Bose,B., Pavan,M.N., Geetha Thanu, Nagaraju,J. and Nagarajaram,H.A. (2003) MICAS: a fully automated web server for microsatellite extraction and analysis from prokaryote and viral genomic sequences. Appl. Bioinformatics, 2, 165–168. [PubMed] [Google Scholar]
- 11.Prasad M.D., Muthulakshmi,M., Madhu,M., Archak,S., Mita,K. and Nagaraju,J. (2004) Survey and analysis of microsatellites in the silkworm, Bombyx mori: frequency, distribution, mutations, marker potential and their conservation in heterologous species. Genetics, doi:10.1534/genetics.104.031005. [DOI] [PMC free article] [PubMed] [Google Scholar]