Abstract
Taiwan Polymorphic Marker Database (TPMD) (http://tpmd.nhri.org.tw/) is a marker database designed to provide experimental details and useful marker information allelotyped in Taiwanese populations accompanied by resources and technical supports. The current version deposited more than 372 000 allelotyping data from 1425 frequently used and fluorescent-labeled microsatellite markers with variation types of dinucleotide, trinucleotide and tetranucleotide. TPMD contains text and map displays with searchable and retrievable options for marker names, chromosomal location in various human genome maps and marker heterozygosity in populations of Taiwanese, Japanese and Caucasian. The integration of marker information in map display is useful for the selection of high heterozygosity and commonly used microsatellite markers to refine mapping of diseases locus followed by identification of disease gene by positional candidate cloning. In addition, our results indicated that the number of markers with heterozygosity over 0.7 in Asian populations is lower than that in Caucasian. To increase accuracy and facilitate genetic studies using microsatellite markers, we also list markers with genotyping difficulty due to ambiguity of allele calling and recommend an optimal set of microsatellite markers for genotyping in Taiwanese, and possible extension of genotyping in other Mongoloid populations.
INTRODUCTION
Microsatellites, the short (1–13 bp) tandem nucleotide repeats, are found to comprise ∼3% but ubiquitously throughout the human genome (1). The average density of microsatellites in human genome is approximately one microsatellite per 2 kb. However, only a small fraction of microsatellites with properties such as high heterozygosity, known position along the genetic map, strong and specific fragment after PCR and easy scoring of allele sizes are suitable for developing as genotyping markers. Genetic markers based on PCR-amplified microsatellites have been extremely important in human genetic studies owing to their high degree of length polymorphism among individuals in human populations (2–4). The use of fluorescent-labeled microsatellite markers and fragment analysis by automatic DNA sequencer has led to widespread applications in studies of population genetics (5) and in biomedical researches including human disease mapping studies for positional cloning (6,7), detection of allelic imbalance or loss of heterozygosity on cancer genome (8,9), diagnosis of expanded triplet repeat loci in several neurodegenerative diseases (10) and human identification in forensic analysis or parentage testing (11,12). In large-scale genotyping studies, although consistent and accurate allele sizing and data handling required careful selection of markers with high heterozygosity in selected population, genotyping by using microsatellite markers remained the most convenient and low cost technology. Since length polymorphism of microsatellites is population-dependent and since estimation of allele frequency is essential for successful linkage analysis (13,14), a shared database and resources for genotyping with microsatellite markers will not only reduce cost and errors of genotyping but also facilitate disease gene mapping in human populations. To achieve these aims, the Taiwan Polymorphie Marker Database (TPMD) was formally launched in public domain in June 2001. In addition, the Applied Taiwan Genotyping Consortium (ATGC) was formed from four qualified laboratories with identical training, experimental protocols and conventional quality control DNAs (CEPH 1331-01 and 1331-02) for data expansion of TPMD and from several laboratories for promoting genetic studies of prevalent inherited disease in Taiwan. The newly launched TPMD website contains two major sections including the database for sharing useful microsatellite marker information and the resources for supporting consistent genotyping experiments.
DATABASE OF MICROSATELLITE MARKERS
Currently, the database contains more than 364 440 allelotyping data genotyped in Han Chinese of Taiwan from 1425 commonly used microsatellite markers with average marker density ∼2.3 cM and variation types of dinucleotide (856, 60%), trinucleotide (97, 6.8%) and tetranucleotide (470, 33.2%). Majority of microsatellite markers which are commercially available are obtained from ABI PRISM Linkage Mapping Set-version 1 and 2 (dinucleotide markers), and Research Genetics CHLC Human Screening Set/Weber Version 8 and Single Chromosome Human Screening Set (tri- and tetranucleotide markers). There are 148 (10.4%) markers synthesized in our laboratories based on UniSTS to meet the needs of specific experiments or selection process of high heterozygosity markers for constructing optimal marker set. Two web interfaces with text and map displays were presented in the TPMD. The text display provides searches of microsatellite marker either by marker name or by chromosomal location with hyperlink to UniSTS and to TPMD for detail allelotyping information such as heterozygosity, range of allele sizes (base pair), genetic position (cM) and fluorescent labels. To facilitate marker selection in refine mapping and positional candidate cloning of disease genes in mapped loci, interfaces for a graphic map display and retrieving markers from an interesting chromosomal locus are developed. It features the marker selection by position in cytogenetic, physical and genetic maps with comparison of marker heterozygosity in Caucasian, Japanese and Taiwanese populations (2,15,16). With the integration of other microsatellite markers from The Center for Medical Genetics at Marshfield Clinic and DeCODE Genetics into human physical map display (2,17), a total of 7932 commonly used microsatellite markers with average marker density of 0.4 cM are integrated and should be useful for marker selection in refine mapping of disease loci and positional candidate cloning of disease genes.
GENOTYPING RESOURCES
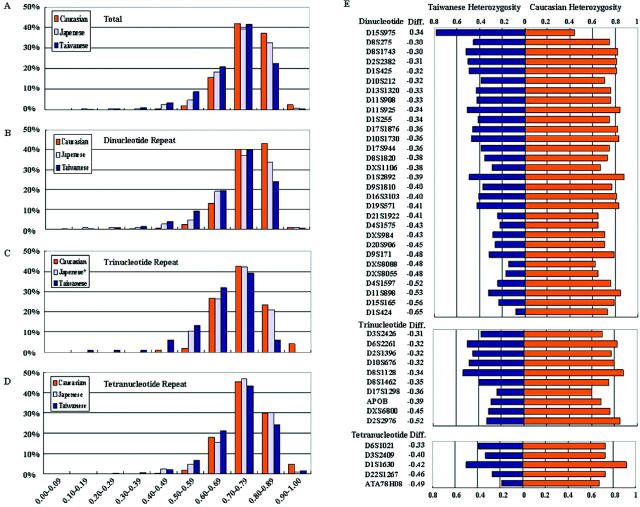
To increase the consistency of allele calling and to avoid redundant microsatellite marker genotyping, we summarized and shared useful information for facilitating genotyping experiments. In the resources pages, we provided our protocols for microsatellite marker genotyping, while a list of markers with atypical variation types resulted in difficulties of consistent allele calling in our laboratories, and hence, we recommend TPMD marker set with 10 cM resolution for optimal genotyping among Taiwanese. Since marker heterozygosity, especially for heterozygosity >0.7, is one of the important factors for successful genotyping and linkage analysis, we compared the distribution of marker heterozygosities genotyped among Taiwanese, Japanese and Caucasian (Figure 1). Our results indicated that the percentages of markers of all variation types with heterozygosity >0.7 was 82, 73 and 65% in Caucasian, Japanese and Taiwanese populations, respectively (Figure 1A). Similar results were observed in panels of dinucleotide, trinucleotide and tetranucleotide microsatellite markers genotyped in the mentioned earlier three populations (Figure 1B–D). Indeed, except dinucleotide marker D15S975 with higher heterozygosity in Taiwanese (0.78) than that in Caucasian (0.44), the remaining 44 microsatellite markers show higher heterozygosity in Caucasian than that in Taiwanese with a difference of heterozygosity over 0.3 (Figure 1E). Our results further suggested that the development of a set of microsatellite markers with high heterozygosity would facilitate genotyping and genetic studies of human diseases in Taiwanese. For establishing the optimal set of microsatellite markers in Taiwanese in use of high-throughput and low cost protocol, we selected tri- and tetranucleotide markers with high heterozygosity and well-separated alleles in electrophoresis as the majority of markers in the optimal set. After careful experimental evaluation of more than 760 microsatellite markers, a table with optimal microsatellite marker set contained 363 microsatellite markers (56 dinucleotide, 34 trinucleotide and 273 tetranucleotide markers) with average heterozygosity >0.77 and average marker density of 9.71 cM is included for genotyping in Taiwanese population. Based on migration of human populations in Asia, the development of an optimal microsatellite marker set should be potentially suitable for genotyping in Mongoloid populations (18,19).
Figure 1.
Comparison of heterozygosity of microsatellite markers in human populations. The distribution of marker heterozygosity in percentage of total markers (A), dinucleotide repeats (B), trinucleotide repeats (C) and tetranucleotide repeats (D) were shown. The asterisk represents only 19 trinucleotide markers genotyped in Japanese that were available in the literature (16). (E) The 45 microsatellite markers with a difference of heterozygosity over 0.3 in comparison between Caucasian and Taiwanese population.
FUTURE DEVELOPMENT
We aim to expand microsatellite marker genotyping data in TPMD and increase marker density to 5 cM resolution for optimal set of microsatellite markers in Taiwanese. The qualified genotyping data from other populations including aborigines of Taiwan will be included in the future. With maturation of single nucleotide polymorphism (SNP) genotyping technologies, we will incorporate SNP markers genotyped in Taiwanese started from high minor allele frequency SNP markers.
Acknowledgments
ACKNOWLEDGEMENTS
We are indebted to members in Department of Research Resources of NHRI for TPMD maintenance. This work was supported by the National Research Program for Genomic Medicine of National Science Council and by the intramural funding of National Health Research Institutes (NHRI). Many authors in Applied Taiwan Genotyping Consortium (ATGC) received contract grants from Division of Molecular Genomic Medicine with help from Department of Intramural Research Affairs of NHRI.
REFERENCES
- 1.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 2.Broman K.W., Murray,J.C., Sheffield,V.C., White,R.L. and Weber,J.L. (1998) Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am. J. Hum. Genet., 63, 861–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weissenbach J., Gyapay,G., Dib,C., Vignal,A., Morissette,J., Millasseau,P., Vaysseix,G. and Lathrop,M. (1992) A second-generation linkage map of the human genome. Nature, 359, 794–801. [DOI] [PubMed] [Google Scholar]
- 4.Gyapay G., Morissette,J., Vignal,A., Dib,C., Fizames,C., Millasseau,P., Marc,S., Bernardi,G., Lathrop,M. and Weissenbach,J. (1994) The 1993–94 Genethon human genetic linkage map. Nature Genet., 7, 246–339. [DOI] [PubMed] [Google Scholar]
- 5.Schlotterer C. and Pemberton,J. (1994) The use of microsatellites for genetic analysis of natural populations. EXS., 69, 203–214. [DOI] [PubMed] [Google Scholar]
- 6.Dib C., Faure,S., Fizames,C., Samson,D., Drouot,N., Vignal,A., Millasseau,P., Marc,S., Hazan,J., Seboun,E. et al. (1996) A comprehensive genetic map of the human genome based on 5264 microsatellites. Nature, 380, 152–154. [DOI] [PubMed] [Google Scholar]
- 7.Chen W.M., Liu,Y.F., Lin,M.W., Chen,I.C., Lin,P.Y., Lin,G.L., Jou,Y.S., Lin,Y.T., Fann,C.S., Wu,J.Y. et al. (2004) Autosomal dominant avascular necrosis of femoral head in two taiwanese pedigrees and linkage to chromosome 12q13. Am. J. Hum. Genet., 75, 310–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mao L., Schoenberg,M.P., Scicchitano,M., Erozan,Y.S., Merlo,A., Schwab,D. and Sidransky,D. (1996) Molecular detection of primary bladder cancer by microsatellite analysis. Science, 271, 659–662. [DOI] [PubMed] [Google Scholar]
- 9.Jou Y.S., Lee,C.S., Chang,Y.H., Hsiao,C.F., Chen,C.F., Chao,C.C., Wu,L.S., Yeh,S.H., Chen,D.S. and Chen,P.J. (2004) Clustering of minimal deleted regions reveals distinct genetic pathways of human hepatocellular carcinoma. Cancer Res., 64, 3030–3036. [DOI] [PubMed] [Google Scholar]
- 10.Caskey C.T., Pizzuti,A., Fu,Y.H., Fenwick,R.G., Jr and Nelson,D.L. (1992) Triplet repeat mutations in human disease. Science, 256, 784–789. [DOI] [PubMed] [Google Scholar]
- 11.Edwards A., Civitello,A., Hammond,H.A. and Caskey,C.T. (1991) DNA typing and genetic mapping with trimeric and tetrameric tandem repeats. Am. J. Hum. Genet., 49, 746–756. [PMC free article] [PubMed] [Google Scholar]
- 12.Edwards A., Hammond,H.A., Jin,L., Caskey,C.T. and Chakraborty,R. (1992) Genetic variation at five trimeric and tetrameric tandem repeat loci in four human population groups. Genomics, 12, 241–253. [DOI] [PubMed] [Google Scholar]
- 13.Goring H.H. and Terwilliger,J.D. (2000) Linkage analysis in the presence of errors II: marker-locus genotyping errors modeled with hypercomplex recombination fractions. Am. J. Hum. Genet., 66, 1107–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abecasis G.R., Cherny,S.S. and Cardon,L.R. (2001) The impact of genotyping error on family-based analysis of quantitative traits. Eur. J. Hum. Genet., 9, 130–134. [DOI] [PubMed] [Google Scholar]
- 15.Ikari K., Onda,H., Furushima,K., Maeda,S., Harata,S. and Takeda,J. (2001) Establishment of an optimized set of 406 microsatellite markers covering the whole genome for the Japanese population. J. Hum. Genet., 46, 207–210. [DOI] [PubMed] [Google Scholar]
- 16.Mizutani M., Yamamoto,T., Torii,K., Kawase,H., Yoshimoto,T., Uchihi,R., Tanaka,M., Tamaki,K. and Katsumata,Y. (2001) Analysis of 168 short tandem repeat loci in the Japanese population, using a screening set for human genetic mapping. J. Hum. Genet., 46, 448–455. [DOI] [PubMed] [Google Scholar]
- 17.Kong A., Gudbjartsson,D.F., Sainz,J., Jonsdottir,G.M., Gudjonsson,S.A., Richardsson,B., Sigurdardottir,S., Barnard,J., Hallbeck,B., Masson,G. et al. (2002) A high-resolution recombination map of the human genome. Nature Genet., 31, 241–247. [DOI] [PubMed] [Google Scholar]
- 18.Cavalli-Sforza L.L. (1997) Genes, peoples, and languages. Proc. Natl Acad. Sci. USA, 94, 7719–7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cavalli-Sforza L.L. (1998) The DNA revolution in population genetics. Trends Genet., 14, 60–65. [DOI] [PubMed] [Google Scholar]