Abstract
We describe a comprehensive map of putative transcription factor binding sites (TFBSs) across multiple genomes created using a search method that relies on hidden Markov models built from experimentally determined TFBSs. Using the information in the TRANSFAC and JASPAR databases, we built 1134 models for TFBSs and used them to scan regions 10 kb upstream of the start of the transcript for all known genes in the human, mouse and Drosophila melanogaster genomes. The results, together with homology information on clusters of ortholog genes across the three genomes, were used to create a multi-organism catalog of annotated TFBSs. The catalog can be queried through a web interface accessible at http://bio.chip.org/mapper that allows the identification, visualization and selection of TFBSs occurring in the promoter of a gene of interest and also the common factors predicted to bind across the cluster of orthologs that includes that gene. Alternatively, the interface allows the user to retrieve binding sites for a single transcription factor of interest in a single gene or in all genes of the human, mouse or fruit fly genomes.
INTRODUCTION
The precisely coordinated temporal and spatial control of gene expression that is key to development and differentiation is accomplished by the interplay of multiple regulatory mechanisms. Transcription factors (TFs)—regulatory proteins that bind short DNA motifs called transcription factor binding sites (TFBSs), play a central role in this context by recruiting the transcriptional machinery at the promoter of the genes and leading to the initiation of their transcription (1). TFBSs often occur in close proximity to each other forming cis-regulatory modules that suggest the existence of a combinatorial code for transcriptional regulation (2). The importance of understanding this code and the availability of the complete genome sequence for many organisms has motivated the development of algorithms and search engines for the identification of TFBSs (3–7) and the creation of databases containing information about the TFs and their binding sites in target genes (8–10). Moreover, sequence conservation of regions containing regulatory elements in ortholog genes across species was used to select regulatory elements that are more likely to be functional, a method called ‘phylogenetic footprinting’ that has been widely used in computational approaches for TFBS identification (11–14).
One of the most popular ways of abstracting the characteristics of a TFBS is to use a multiple sequence alignment of experimentally determined binding sites for a given TF to generate a nucleotide weight matrix (NWM) that describes the probability distribution of the four nucleotides at each position in the site (15). The NWM model assumes that nucleotides that form a binding site are independent of each other and their contribution to the specificity of the site is additive (16). To test this assumption experimentally, key positions in the binding sites for two TFs [nucleotides at positions 16 and 17 in the Mnt repressor protein binding site (17), and the central nucleotide triplet in the mouse EGR1 protein binding site (18)] were systematically mutated to all possible combinations and the binding affinity of the respective TFs (or its mutants) for these sites was determined. These data pointed out that nucleotides within a TFBS are not independent and that even though NWMs do not capture those dependences they represent a good enough approximation for modeling the site (16,17). However, it is generally recognized that using NWMs to search for putative TFBSs often leads to the retrieving of a very high number of false positives (15).
An alternative way of modeling a TFBS is by using profile hidden Markov models (HMMs) that, in addition to capturing the probability distribution of the nucleotides at each position, can model insertions or deletions and allow fragment matches to the model in the search procedure (19). HMMs have been used in several bioinformatics applications owing to their increased statistical power in capturing the characteristics of a motif. These applications include generating the extensive Pfam database of conserved protein motifs (20) and, for a very limited number of cases, modeling TFBSs (21–23).
To develop the MAPPER database, we used the curated information on experimentally determined binding sites contained in the TRANSFAC and JASPAR databases (8,9) in conjunction with the statistical power of HMMs to generate a library of 1134 models corresponding to 863 TFs with distinct names. We then used these models to scan the upstream sequences of all known genes in the human, mouse and Drosophila genomes, and we collected the resulting predicted TFBSs in a relational database. The database can be queried via a web-based interface publicly available at http://bio.chip.org/mapper.
MATERIALS AND METHODS
Building the profile HMM database
The flat files of the TRANSFAC Professional database (version 8.1) were parsed to extract the sequences of the binding sites used to generate the TRANSFAC nucleotide weight matrices. We called the alignments generated based on these sites matrix-derived alignments; the HMM models based on them have identifiers starting with ‘M’ and are referred to as matrix-derived models. The TRANSFAC flat files were also parsed to extract the binding sites referenced in the description of the TRANSFAC factors. We called the alignments generated based on these sites factor-derived alignments; the HMM models based on them have identifiers starting with ‘T’ and are referred to as factor-derived models.
The matrix-derived alignments were used directly to build HMMs, while the factor-derived ones were first aligned with ClustalW. As a consequence, the factor-derived models usually have a lower quality than the matrix-derived ones; however, they increase considerably the search power compared to TRANSFAC as the sites they are based on are not used in TRANSFAC to build NWMs but are only listed in the description of the factors. In addition, the binding sites used to generate several of the high-quality JASPAR matrices were downloaded from http://jaspar.cgb.ki.se and were aligned with ClustalW. The hmmbuild and hmmcalibrate functions of the HMMER package (24) were used to build HMMs based on all retrieved alignments.
Large-scale multi-genome scans
Genome sequences and annotations for human (version hg17), mouse (version mm5) and Drosophila (version dm1) were downloaded from the UCSC Genome Bioinformatics site at http://hgdownload.cse.ucsc.edu/downloads.html (25). The scanned upstream sequences span a gene region of variable length starting from 10 000 bp upstream of the transcript start up to 50 bp downstream of the ATG, thus possibly including the initial introns and non-coding exons. In order to compare the TFBS profile in upstream sequences of ortholog genes, homology information describing clusters of orthologs across the three genomes was obtained from the HomoloGene database (build 30) available at http://www.ncbi.nlm.nih.gov/HomoloGene/ (26). The GoldenPath annotation and chromosome files were used to extract upstream sequences for all annotated genes in the human, mouse and Drosophila genomes. The HMMER hmmpfam function, called with a cut-off value of 16 for the E-value, was used to search for matches for all models in all upstream sequences collected. The hmmpfam output is normally presented in a readable format that is not well suited for automated processing. Since the source code for the program is freely available, we modified it to output its results in an alternative format based on structured lists, which made subsequent parsing and processing much easier.
All HMMER hits returned by the large-scale runs were filtered to flag as redundant the ones that occur in the same sequence, have the same positions (more than 50% overlap) and were retrieved by both matrix-derived and factor-derived models that were linked to each other in the TRANSFAC entries, thus eliminating them from the displayed results.
Database construction
Based on the modifications to the hmmpfam output, the database construction process was completely automated: for each organism the modified hmmpfam program was used to scan a file containing the upstream sequences of all its genes against all models; the resulting output file was read by a Common Lisp script and fed directly into a relational database based on the MySQL database server. This pipeline was designed so that it will allow us to easily update or recreate the database when additional TF models or genome annotations become available. To facilitate access to the database, we implemented a publicly available web-based interface using the Allegro Common Lisp programming language.
RESULTS
Profile HMMs
The parsing procedure described retrieved 359 matrix-derived and 718 factor-derived alignments from TRANSFAC, while 57 alignments corresponding to JASPAR matrices were downloaded. All alignments were used to generate HMMs, resulting in a total of 1134 models.
The distribution of the number of sequences used to build the models showed that the average number of sites used was 22 for matrix-derived models, 16 for factor-derived models and 20 for JASPAR-derived models. The minimum number of sites used for the three types of models was 4, 4 and 6 respectively. Although the number of sequences in the training set influences strongly the quality of the model, we did not want to impose an arbitrary cut-off value on this parameter. Instead, we are providing this information for each model (together with other details such as the model length, HMM consensus and HMM logo), allowing the user to decide on the quality and suitability for the desired analysis of those models for which the training set is small.
The 1134 models correspond to 863 TF entries with distinct names in the TRANSFAC and JASPAR databases. However, this number is only an estimate of the true number of factors covered by MAPPER since in the two databases used as data source, entries with different names pertain sometimes to isoforms of the same TF (e.g. HNF-1, HNF-1alpha) or even to the same TF (e.g. p65, RelA). These discrepancies originated at the curation step in the two databases and pose a serious challenge for automation. However, in the absence of a consistent and standard TF nomenclature, we decided to retain the TRANSFAC and JASPAR names and accession numbers as the most effective choice both for the purpose of this work as well as for the user already familiar with the other two databases. Nevertheless, our system is able to accommodate future changes and evolutions in the TF nomenclature.
The full list of transcription factors and associated models in our database is available at http://bio.chip.org/mapperdb/factors.html. The table contains links to model pages that provide details such as the model length, the number of sequences used to train it, the HMM consensus sequence(s) (the most probable nucleotide at that position according to the model; models that allow insertions or deletions may have more than one consensus sequences displayed) and the HMM logo generated using the Logomat-M software (27). In addition, hit statistics for each model, such as the number of hits in each organism and the minimum and maximum score and E-value are provided.
Database content
All 1134 models were used to scan a total of 57906 sequences from the human, mouse and Drosophila genomes. The relational database was used to store information on the genomic sequences (gene name and symbol, Locuslink, RefSeq, GenBank and Swiss-Prot identifiers, etc.) and their orthologs (HomoloGene cluster numbers and homology percent), information on the factors and matrices retrieved from TRANSFAC and JASPAR, and the results of the HMMER searches. Each hit in the database is described by its position, a score (a probabilistic measure of the match between the hit and the model—the higher the score the better the match), an E-value (a measure of the likelihood of the hit being retrieved by chance—the lower the E-value the more likely it is that the hit is ‘real’), and the alignment between the model and the sequence at the putative site. The cumulative results characterizing our database are presented in Table 1. As explained in the HMMER documentation (24), and as we have observed by running hmmpfam on a number of control sequences, experimentally validated sites can sometimes be retrieved with a negative score. Therefore, we used ‘relaxed’ scores and E-values for the search in order to obtain a comprehensive representation of the TFBS map in the sequences scanned. Since the total number of hits depends on the stringency of the score and E-value parameters, we present both the figures obtained using relaxed threshold values and those obtained with more realistic thresholds, namely score > 0 or score > 0 and E-value < 1.
Table 1. Cumulative results characterizing the MAPPER database.
| H.sapiens | M.musculus | D.melanogaster | Total | |
|---|---|---|---|---|
| Sequences scanned | 21 735 | 17 218 | 18 953 | 57 906 |
| Base pairs scanned (Mb) | ∼371 | ∼263 | ∼218 | ∼852 |
| Putative TFBS found | ||||
| All | 17 357 280 | 11 093 651 | 10 032 839 | 38 483 770 |
| With score > 0 | 13 503 672 | 8 335 765 | 7 112 552 | 28 951 989 |
| With score > 0 and E-value < 1 | 846 509 | 311 802 | 82 716 | 1 241 027 |
Interface capabilities
We designed and implemented a web-based system called MAPPER (Multi-genome Analysis of Positions and Patterns of Elements of Regulation), publicly available at http://bio.chip.org/mapper, to facilitate access to the catalog of putative TFBSs.
The entry page allows the user to query a sequence based on a gene identifier (e.g. NCBI Gene ID, RNA accession number or CG symbol for Drosophila) and to retrieve all putative binding sites that satisfy the input parameters specified: thresholds on the score (default value is 0) and E-value (default value is 10) and position relative to the ATG or the start of the transcript. For each putative site the interface displays its position according to three different coordinate systems (absolute position on chromosome, distance from the ATG, distance from the transcript start), its score and E-value, and the gene region it belongs to (upstream region, intron or exon). The list of hits can be sorted by position, score, E-value, factor name or factor accession number. For each hit a pop-up window displays the alignment between the putative site on the query sequence and the model. The results page highlights hits retrieved in close proximity (i.e. within a window of 50 bp) for factors known to physically interact with each other (based on the TRANSFAC annotation), the TRANSFAC classes to which the factors for which hits were found belong and the common hits retrieved for the orthologs of the selected gene across the three genomes (if present in the Homologene database). The set of putative hits can be saved as a text file with or without alignments, displayed graphically with respect to the translation start, or exported as a custom track to the GoldenPath browser, thus allowing the user to visualize the putative TFBSs in the context of the genomic region in which they are found and to take advantage of the wealth of information provided by the GoldenPath Genome browser (25).
Figure 1 presents the results displayed by MAPPER for the promoter of the B99/gtse1 (G2 and S phase-expressed-1) gene containing an experimentally characterized p53-responsive element located between positions −126 and −96 from the ATG. This element consists of three half-site decamers and is responsible for the p53-mediated upregulation of the gene following DNA damage (28). As shown in Figure 1A, three models for p53 (T00671, T04997 and M00761) retrieve these sites. The predicted TFBSs in this region are displayed as a list (Figure 1A) and can be exported as text (with or without alignments), in graphical form along the promoter of the gene (Figure 2A) or as a custom track in the GoldenPath Browser (Figure 2B). Figure 1B shows details on the p53 model M00761 whose match to the plus strand of the sequence is shown in the pop-up window in Figure 1A. It was experimentally demonstrated that the p53 binding site contains two copies of the sequence 5′-RRRC(A/T)(T/A)GYYY-3′ (where R = A/G and Y = C/T) separated by a spacer region (29). The HMM consensus sequences for model M00761 displayed in Figure 1B are in agreement with the p53 consensus and show that the model can accommodate up to two insertions or deletions (represented by a dot character) between the two half-sites but not elsewhere. Based on our observations, if the number of insertions in the spacer region was higher the model would identify instead a half-site by matching only one fragment to the model.
Figure 1.
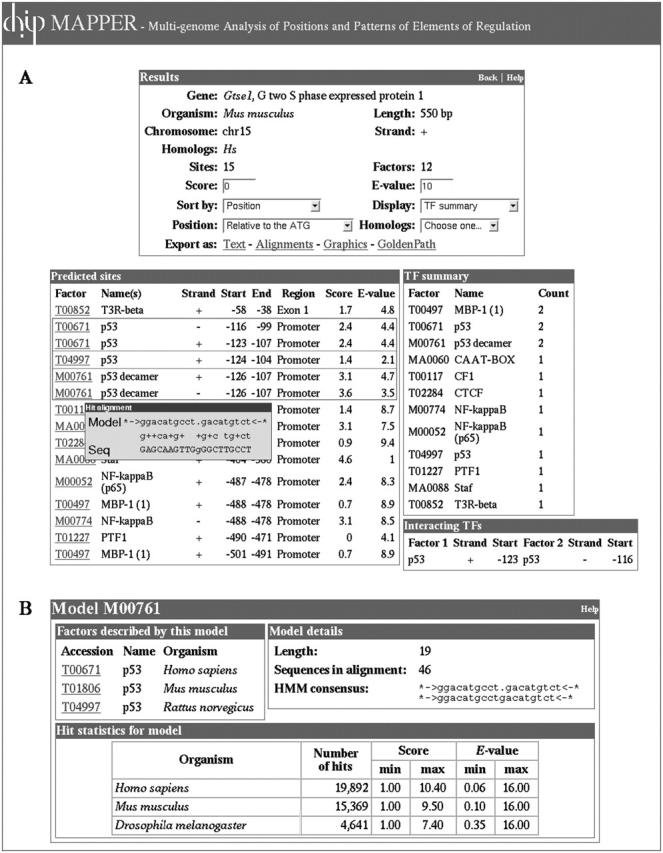
MAPPER output for the promoter of the mouse gtse1 gene. (A) The MAPPER query for putative TFBSs found within 500 bp upstream of the ATG in the mouse gtse1 gene identifies an experimentally characterized p53-responsive element composed of three half sites situated between positions −126 and −96 (28). The models retrieving these sites are boxed. For each hit, a pop-up window displays the alignment between the sequence and the model at the putative site (the match between the model M00761 and the plus strand of the sequence is shown). (B) The consensus sequence displayed in the page for model M00761 shows that the model can accommodate insertion or deletions (represented by a dot character) between the half-sites but not elsewhere. Additional information on the model and its hit statistics over the entire database are also provided.
Figure 2.
Graphical export options for the hit set found in the promoter of the mouse gtse1 gene. The hit set presented as a list in Figure 1A is displayed graphically along the promoter of the gene (A) or exported in the GoldenPath browser (B).
An alternative way of using MAPPER consists in specifying the accession number of a single TFBS model, and retrieving the hits for that model only. This analysis can be performed on a single gene or on all genes. In the latter case the results returned are limited to a fixed number of best-scoring hits for efficiency, and the graphical display options are not available since the genes containing these hits will usually be scattered on different chromosomes.
The web interface design is based on ease of use and flexibility. The page layout was intentionally kept simple, minimizing graphics and striving to present complete and clear information in each page. Context-sensitive menus allow the user to quickly switch between alternative views of the data (e.g. different ordering of the hits, different reference point for coordinates), and to filter the data by changing the score and E-value thresholds. For each page, an on-line help provides step-by-step instructions regarding the format of the input required by each field as well as explanations regarding the meaning of the output fields and the options available for sorting and exporting them.
DISCUSSION
MAPPER represents both a catalog of models for TFBSs and a catalog of putative sites retrieved for them in all human, mouse and Drosophila genes that offers several advantages over other available similar resources.
First, the set of TFBS models in MAPPER is very comprehensive. The models are based on curated information on experimentally determined binding sites contained in the TRANSFAC and JASPAR databases, but use HMMs instead of NWMs to model the characteristics of the sites. While some of these models have an equivalent NWM in the TRANSFAC and JASPAR databases, many others (the factor-derived models) are specific to MAPPER. In addition to capturing the probability distribution of nucleotides at each position in the binding site, HMMs allow modeling insertions, deletions and fragment matches to the model thus attaining a higher level of generality than the corresponding NWM built on the same alignment. Many TFs bind the DNA as dimers or tetramers and thus the characterized binding sites are composed of functional half-sites separated by spacers of variable lengths. While in the functional moieties of the TFBSs insertions and deletions are uncommon, they can occur in the more divergent spacer regions (30). Moreover, in the search procedure HMMER allows fragment matches to the model that in this context will lead to the retrieval of half-sites. In our database, 481 models (42% of the total) allowed at least one insertion or deletion during the large-scale searches.
We recognize that profile HMMs also have limitations. In order to convert the observed counts into probabilities HMMER combines the actual counts with priors, in this case single-component Dirichlet priors (24). These priors introduce pseudocounts that weight heavier if the training set is low thus biasing the model. A small training set would represent a problem for any statistical approach (including NWMs) and, due to the fact that in some cases the number of known or available sequences for a binding site is small, the quality of the model has to be evaluated by different means. For these reasons, the MAPPER model pages provide details such as the length of the model, the number of sequences in the training set, the HMM consensus, the HMM logos and hit statistics over the entire database (minimum and maximum scores and E-values) so that the user can make an informed evaluation of the quality of any given model. Moreover, even if some models are trained on a small number of sequences this does not usually represent a problem in MAPPER since most TFs are described by more than one model.
Second, in contrast to TRANSFAC and JASPAR that require the user to supply the nucleotide sequence of the gene of interest or the profiles to be included in the search, MAPPER needs as little information as a gene identifier and outputs all putative binding sites found for all its models that satisfy the cut-off parameters. The sequences scanned to build the database are likely to contain the regulatory regions of a large number of genes, according to the most up-to-date annotations. Having a pre-computed list of putative TFBSs in three genomes instead of simply offering the ability to scan a user-supplied sequence, represents in our opinion a good trade-off between generality and efficiency: the results can be retrieved very quickly (both for a single organism or when analyzing multiple genomes), and, as our analysis covered a wide range of scores and E-values, it allows the user to experiment efficiently with very different settings of these parameters when querying the database. Moreover, the static database allows the retrieval of the best scoring hits for a given transcription factor of interest by supplying only a model identifier as input.
Finally, the MAPPER interface was designed to provide comprehensive and detailed information to the users, and offers powerful options for the visualization and the export of the data.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Sven Rahmann and Benjamin Schuster-Böckler for making available the source code for generating the HMM logos locally, and the anonymous reviewers for their useful comments.
REFERENCES
- 1.Davidson E.H., Rast,J.P., Oliveri,P., Ransick,A., Calestani,C., Yuh,C.H., Minokawa,T., Amore,G., Hinman,V., Arenas-Mena,C. et al. (2002) A genomic regulatory network for development. Science, 295, 1669–1678. [DOI] [PubMed] [Google Scholar]
- 2.Levine M. and Tjian,R. (2003) Transcription regulation and animal diversity. Nature, 424, 147–151. [DOI] [PubMed] [Google Scholar]
- 3.Bailey T.L. and Noble,W.S. (2003) Searching for statistically significant regulatory modules. Bioinformatics, 19 (Suppl. 2), II16–II25. [DOI] [PubMed] [Google Scholar]
- 4.Sinha S., Van Nimwegen,E. and Siggia,E.D. (2003) A probabilistic method to detect regulatory modules. Bioinformatics, 19 (Suppl. 1), I292–I301. [DOI] [PubMed] [Google Scholar]
- 5.Rajewsky N., Vergassola,M., Gaul,U. and Siggia,E.D. (2002) Computational detection of genomic cis-regulatory modules applied to body patterning in the early Drosophila embryo. BMC Bioinformatics, 3, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharan R., Ben-Hur,A., Loots,G.G. and Ovcharenko,I. (2004) CREME: Cis-Regulatory Module Explorer for the human genome. Nucleic Acids Res., 32, W253–W256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alkema W.B., Johansson,O., Lagergren,J. and Wasserman,W.W. (2004) MSCAN: identification of functional clusters of transcription factor binding sites. Nucleic Acids Res., 32, W195–W198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wingender E., Chen,X., Fricke,E., Geffers,R., Hehl,R., Liebich,I., Krull,M., Matys,V., Michael,H., Ohnhauser,R. et al. (2001) The TRANSFAC system on gene expression regulation. Nucleic Acids Res., 29, 281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandelin A., Alkema,W., Engstrom,P., Wasserman,W.W. and Lenhard,B. (2004) JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res., 32,D91–D94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kel-Margoulis O.V., Romashchenko,A.G., Kolchanov,N.A., Wingender,E. and Kel,A.E. (2000) COMPEL: a database on composite regulatory elements providing combinatorial transcriptional regulation. Nucleic Acids Res., 28, 311–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandelin A., Wasserman,W.W. and Lenhard,B. (2004) ConSite: web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res., 32, W249–W252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bigelow H.R., Wenick,A.S., Wong,A. and Hobert,O. (2004) CisOrtho: a program pipeline for genome-wide identification of transcription factor target genes using phylogenetic footprinting. BMC Bioinformatics, 5, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loots G.G. and Ovcharenko,I. (2004) rVISTA 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res., 32, W217–W221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dieterich C., Wang,H., Rateitschak,K., Luz,H. and Vingron,M. (2003) CORG: a database for COmparative Regulatory Genomics. Nucleic Acids Res., 31, 55–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stormo G.D. (2000) DNA binding sites: representation and discovery. Bioinformatics, 16, 16–23. [DOI] [PubMed] [Google Scholar]
- 16.Benos P.V., Bulyk,M.L. and Stormo,G.D. (2002) Additivity in protein–DNA interactions: how good an approximation is it? Nucleic Acids Res., 30, 4442–4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Man T.K. and Stormo,G.D. (2001) Non-independence of Mnt repressor-operator interaction determined by a new quantitative multiple fluorescence relative affinity (QuMFRA) assay. Nucleic Acids Res., 29, 2471–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bulyk M.L., Johnson,P.L. and Church,G.M. (2002) Nucleotides of transcription factor binding sites exert interdependent effects on the binding affinities of transcription factors. Nucleic Acids Res., 30, 1255–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eddy S.R. (1998) Profile hidden Markov models. Bioinformatics, 14, 755–763. [DOI] [PubMed] [Google Scholar]
- 20.Bateman A., Coin,L., Durbin,R., Finn,R.D., Hollich,V., Griffiths-Jones,S., Khanna,A., Marshall,M., Moxon,S., Sonnhammer,E.L. et al. (2004) The Pfam protein families database. Nucleic Acids Res., 32, D138–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conkright M.D., Guzman,E., Flechner,L., Su,A.I., Hogenesch,J.B. and Montminy,M. (2003) Genome-wide analysis of CREB target genes reveals a core promoter requirement for cAMP responsiveness. Mol. Cell, 11, 1101–1108. [DOI] [PubMed] [Google Scholar]
- 22.Ellrott K., Yang,C., Sladek,F.M. and Jiang,T. (2002) Identifying transcription factor binding sites through Markov chain optimization. Bioinformatics, 18 (Suppl. 2), S100–S109. [DOI] [PubMed] [Google Scholar]
- 23.Price C.W., Fawcett,P., Ceremonie,H., Su,N., Murphy,C.K. and Youngman,P. (2001) Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol., 41, 757–774. [DOI] [PubMed] [Google Scholar]
- 24.Eddy S.R. (2003) HMMER User's Guide: Biological sequence analysis using profile hidden Markov models.
- 25.Kent W.J., Sugnet,C.W., Furey,T.S., Roskin,K.M., Pringle,T.H., Zahler,A.M. and Haussler,D. (2002) The human genome browser at UCSC. Genome Res., 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wheeler D.L., Church,D.M., Federhen,S., Lash,A.E., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Sequeira,E., Tatusova,T.A. et al. (2003) Database resources of the National Center for Biotechnology. Nucleic Acids Res., 31, 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuster-Bockler B., Schultz,J. and Rahmann,S. (2004) HMM Logos for visualization of protein families. BMC Bioinformatics, 5, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Utrera R., Collavin,L., Lazarevic,D., Delia,D. and Schneider,C. (1998) A novel p53-inducible gene coding for a microtubule-localized protein with G2-phase-specific expression. EMBO J., 17, 5015–5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.el-Deiry W.S., Kern,S.E., Pietenpol,J.A., Kinzler,K.W. and Vogelstein,B. (1992) Definition of a consensus binding site for p53. Nature Genet., 1, 45–49. [DOI] [PubMed] [Google Scholar]
- 30.Sinha S. and Tompa,M. (2002) Discovery of novel transcription factor binding sites by statistical overrepresentation. Nucleic Acids Res., 30, 5549–5560. [DOI] [PMC free article] [PubMed] [Google Scholar]



