Abstract
InterPro, an integrated documentation resource of protein families, domains and functional sites, was created to integrate the major protein signature databases. Currently, it includes PROSITE, Pfam, PRINTS, ProDom, SMART, TIGRFAMs, PIRSF and SUPERFAMILY. Signatures are manually integrated into InterPro entries that are curated to provide biological and functional information. Annotation is provided in an abstract, Gene Ontology mapping and links to specialized databases. New features of InterPro include extended protein match views, taxonomic range information and protein 3D structure data. One of the new match views is the InterPro Domain Architecture view, which shows the domain composition of protein matches. Two new entry types were introduced to better describe InterPro entries: these are active site and binding site. PIRSF and the structure-based SUPERFAMILY are the latest member databases to join InterPro, and CATH and PANTHER are soon to be integrated. InterPro release 8.0 contains 11 007 entries, representing 2573 domains, 8166 families, 201 repeats, 26 active sites, 21 binding sites and 20 post-translational modification sites. InterPro covers over 78% of all proteins in the Swiss-Prot and TrEMBL components of UniProt. The database is available for text- and sequence-based searches via a webserver (http://www.ebi.ac.uk/interpro), and for download by anonymous FTP (ftp://ftp.ebi.ac.uk/pub/databases/interpro).
INTRODUCTION
The genome sequencing centres are generating raw sequence data at an alarming rate, and the result is a need for automated sequence analysis methods. The automatic analysis of protein sequences is possible through the use of ‘protein signatures’, which are methods for diagnosing a domain or characteristic region of a protein family in a protein sequence. A number of protein signature databases have been developed, each using a variation on the handful of signature methods available, which include patterns, profiles and hidden Markov models (HMMs). These databases are most effective when used together, rather than in isolation. InterPro (1) integrates into one resource the major protein signatures databases: PROSITE (2), which uses regular expressions and profiles, PRINTS (3), which uses position-specific scoring matrix-based (PSSM-based) fingerprints, ProDom (4), which uses automatic sequence clustering, and Pfam (5), SMART (6), TIGRFAMs (7), PIRSF (also known as PIR SuperFamily) (8) and SUPERFAMILY (9), all of which use HMMs.
Signatures from the member databases are integrated manually as they are developed. A team of biologists have this responsibility, as well as that of annotating the new or existing entries. Each InterPro entry is described by one or more signatures, and corresponds to a biologically meaningful family, domain, repeat or site, e.g. post-translational modification (PTM). Not every entry will contain a signature from each member database, only those that correspond to each other are united. Entries are assigned a type to describe what they represent, which may be family, domain, repeat, PTM, active site or binding site. The last two are new entry types, which were introduced to better describe the signatures in some of the entries. Entries may be related to each other through two different relationships: the parent/child and contains/found in relationship. Parent/child relationships are used to describe a common ancestry between entries, whereas the contains/found in relationship generally refers to the presence of genetically mobile domains. InterPro entries are annotated with a name, an abstract, mapping to Gene Ontology (GO) terms and links to specialized databases. InterPro groups all protein sequences matching related signatures into entries. All hits of the protein signatures in InterPro against a composite of the Swiss-Prot and TrEMBL components of UniProt (10) are precomputed. The matches are available for viewing in each InterPro entry in different formats.
The number of entries and coverage of protein space by InterPro is continuing to grow. The beta release of InterPro in 1999 contained 2423 entries, while the latest release of the database contains 11 007 entries, representing nearly a 5-fold increase in 5 years. In its infancy, InterPro covered ∼66% of all proteins in Swiss-Prot and TrEMBL, and this has increased to over 90% for Swiss-Prot, 76% for TrEMBL and 78% for UniProt (Swiss-Prot and TrEMBL). A number of new features have been added to the InterPro database since its publication in Nucleic Acids Research in 2003. These include additional protein match views, the InterPro Domain Architectures Viewer, taxonomic range information, additional database links and protein 3D structural information. New members databases that have been integrated are the full-length sequence-based PIRSF database and the structure-based SUPERFAMILY. These are described in more detail below.
NEW FEATURES OF INTERPRO
Protein match views
For each protein signature, a list of proteins in UniProt that it matches is precomputed. This list gets updated when new proteins enter UniProt or if the signatures themselves change. The match lists may be viewed in a number of different formats including a table view, a detailed view and an overview. There are new options for the ordering of proteins within these views. For example, the views can all be displayed either ordered by Swiss-Prot ID or for only those proteins of known structure. The overview and detailed view can also be ordered by UniProt accession number, and the former can be ordered by taxonomy too. In the overview, clicking on the protein accession number takes the user to the detailed view for that protein. Searching for a protein accession number in the InterPro text search with the ‘find protein matches’ option returns the overview of matches for the protein. Similarly, the detailed view is retrieved through the accession number link (see Figure 1). For the graphical views, a mouse-over displays the actual positions of the matches on the sequence.
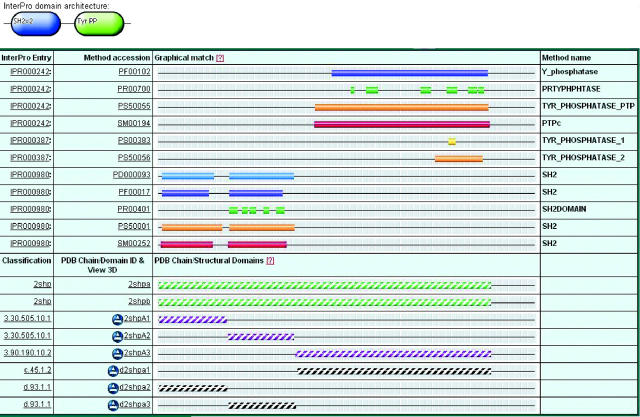
Figure 1.
Illustration of the detailed view for protein Q06124, the human protein-tyrosine phosphatase, non-receptor type 11. From an InterPro entry page, clicking on a protein accession number in the ‘Examples’ field takes you to this view for that protein. The oval shapes at the top of the figure display the InterPro Domain Architecture (IDA) view for this protein, which represents its domain composition. Each oval shape contains the domain name and the number of its iterations of the domain if greater than one. The InterPro detailed view represents the protein sequence as a series of different lines for each protein signature hit. The bars are colour coded according to the member database. A separate view below the signature matches displays the structural domains from the SCOP and CATH as white-striped bars. This view provides a complete picture of the protein domain composition and where sequence-based domains correspond to known structures.
Where structures are available for proteins, there is a link from the graphical views to the corresponding Protein Data Bank (PDB) structures and a separate line in the display, below the InterPro matches, showing the hyperlinked SCOP (11), CATH (12) and PDB (13) matches on the sequence as white striped bars (see Figure 1). This shows where the protein signatures correspond with structural chains. An Astex icon is available for structures, and clicking on which, loads the AstexViewer™ Java applet page displaying the PDB structure, with the residues included in the CATH or SCOP domain definition highlighted on the PDB chain.
InterPro Domain Architecture viewer
The InterPro Domain Architecture (IDA) viewer is a graphical representation of protein domain architecture, where the domain architecture of a protein sequence is displayed as a series of non-overlapping domains (see Figure 1). These domains are calculated by a method that identifies a subset of InterPro entries/methods, representing non-overlapping domains within proteins. If two domains overlap slightly, their centres are used to order the domains, and domain boundaries are discarded to enhance a comparison of various architectures. If a parent/child hierarchy exists between InterPro domains, matches with the children are represented as those from the parent entry. For each InterPro entry, a graphical representation of unique IDA(s) is provided and each kind of IDA is displayed with an example protein and total number of proteins, sharing this architecture, next to it. Clicking on the count of proteins retrieves all proteins sharing a common architecture. Although domains should not overlap, inserted domains (e.g. nested domains) are still shown in the IDA viewer, as this provides more accurate comparison between IDAs.
Taxonomy viewer
A new feature in InterPro is the ‘Taxonomy’ field, which aims to provide an ‘at a glance’ view of the taxonomic range of the sequences associated with each InterPro entry. This is represented as a circular display with the taxonomy-tree root as its centre. The lineages populating the nodes were selected to provide a view of the major groups of organisms with the model organisms on the outer most circle. Nodes of the taxonomy-tree are placed on the inner circles and radial lines lead to the description for each node. No significance is attached to the position of the node on a particular inner-circle, although some attempt has been made to group nodes. The nodes themselves are either true taxonomy nodes or artificial nodes, of which there are three: ‘Unclassified’, ‘Other Eukaryota (Non-Metazoa)’ and the ‘Plastid Group’. The number of sequences associated with each lineage is displayed, and clicking the number retrieves the graphical overview for proteins within that taxonomic group.
Database cross-references
In addition to cross-referencing the member database signatures and GO (14) terms, there is a separate field in InterPro entries, ‘Database Links’, to provide cross-references to other databases. These included cross-references to corresponding Blocks accession numbers, PROSITE documentation, the CArbohydrate-Active EnZymes (CAZy) website and the Enzyme Commission (EC) Database. New links have been produced to the IUPHAR Receptor Database, the MEROPS Peptidase Database (15) and COMe. The bioinorganic motif database, COMe, is an attempt to classify metalloproteins and some other complex proteins using the concept of bioinorganic motifs.
3D Structural information
A separate field, called ‘Structural links’, provides information on curated structure links. Structural domains from SCOP (11) and CATH (12) are made up of one or more protein chains in a PDB entry. These may include the full chain or region(s) of chain(s). The links to the curated structural domains in this field of InterPro entries are based on the correspondence between the proteins matching the InterPro entry and those proteins of known structure belonging to SCOP or CATH superfamilies. In addition, they include only those links where the structural domains overlap considerably with one or more of the InterPro signatures on the protein sequence. The structural domains are also displayed at the protein level in the graphical views, as described above. Here, all the representative domains at the SCOP/CATH family level are displayed, showing the location of the structural domain(s) in the protein. This enables the user to directly access the SCOP/CATH classification for that particular domain from the protein's detailed graphical view. Mapping between UniProt and PDB entries can be many-to-many, so the ‘Structure’ link displays all the PDB entries associated with that particular protein. The user is able to view the residue-by-residue mapping between UniProt and a PDB chain of interest. This is a useful tool, quite unique in its nature, to show such relationships in a compact way.
New member databases
Two of the newest member databases to join the InterPro Consortium are PIRSF (8) and the structure-based SUPERFAMILY (9). PIRSF is a network classification system that accommodates a flexible number of levels from superfamily to subfamily to reflect varying degrees of sequence conservation. Members of a PIRSF homeomorphic family share full-length sequence similarity with a common domain architecture (homeomorphic) and have common evolutionary origin (monophyletic). PIRSF HMMs are designed to cover the full length of a protein sequence, and thus to include all domains within the sequence. In this way, PIRSF homeomorphic families tend to encompass one or more of the existing InterPro domain entries and show the domain composition of UniProt sequences. Classification based on full-length proteins allows annotation of both generic biochemical and specific biological functions, identification of domain and family relationships, and classification of multidomain proteins. SUPERFAMILY is the first member database that is based solely on structural protein families rather than sequence-based protein families. SUPERFAMILY is a collection of HMMs built from members of SCOP structural superfamilies. This facilitates comparison of protein families based on structure and sequence and adds a new dimension to InterPro entries. Many of the SUPERFAMILY HMMs actually correspond to Pfam HMMs, but also, unsurprisingly, to the structural links in InterPro generated from the SCOP and CATH links to proteins in the PDB.
DISCUSSION
The amalgamation of PROSITE, PRINTS, Pfam, ProDom, SMART, TIGRFAMs, PIRSF and SUPERFAMILY into InterPro has provided a useful tool for protein sequence analysis and characterization. InterPro has a number of applications and databases dependent on its continued success. It is the tool of choice for the annotation of new genomes and is used extensively for the automatic annotation of TrEMBL entries. The mappings of InterPro to GO terms (14) provide a means of large-scale mapping of proteins onto GO terms. This accounts for the bulk of the UniProt proteins that are mapped to GO terms. In addition, InterPro is used for the Proteome Analysis Database (16), to provide statistical analyses of whole proteomes for the completely sequenced genomes. For each proteome, the database provides tables of all the InterPro matches ordered by the number of proteins matching the entries, the top 30 InterPro hits, the top 200 hits, the 15 most common families, etc. A tool is also available to perform proteome comparisons between two or more organisms of choice through InterPro analyses.
The InterPro database is available via a webserver (http://www.ebi.ac.uk/interpro) and anonymous FTP (ftp://ftp.ebi.ac.uk/pub/databases/interpro). The webserver facilitates text and sequence searches, and the FTP site provides regular releases of an XML file for downloading. Future plans for InterPro involve the integration of the next two member databases, CATH HMMs and the PANTHER database. In addition, SWISS-MODEL 3D structure homology models (17) will be displayed in protein graphical views to provide predicted structural information where proteins do not have their structures solved. InterPro is growing along with its member databases, and has increased coverage of the UniProt protein sequence database. The resource continues to expand and provide up-to-date data and new features and thus increases its use to the scientific community as a powerful protein classification tool.
Acknowledgments
ACKNOWLEDGEMENTS
The InterPro project is supported by the ProFuSe grant (number QLG2-CT-2000-00517) and the Integr8 grant (number QLRI-CT-2001000015) of the European Commission.
REFERENCES
- 1.Apweiler R., Attwood,T.K., Bairoch,A., Bateman,A., Birney,E., Biswas,M., Bucher,P., Cerutti,L., Corpet,F., Croning,M.D. et al. (2001) The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res., 29, 37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hulo N., Sigrist,C.J., Le Saux,V., Langendijk-Genevaux,P.S., Bordoli,L., Gattiker,A., De Castro,E., Bucher,P. and Bairoch,A. (2004) The PROSITE database, its status in 2002. Nucleic Acids Res., 30, 235–238.11752303 [Google Scholar]
- 3.Attwood T.K., Bradley,P., Flower,D.R., Gaulton,A., Maudling,N., Mitchell,A.L., Moulton,G., Nordle,A., Paine,K., Taylor,P., Uddin,A. and Zygouri,C. (2003) PRINTS and its automatic supplement pre-PRINTS. Nucleic Acids Res., 31, 400–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Servant F., Bru,C., Carrere,S., Courcelle,E., Gouzy,J., Peyruc,D. and Kahn,D. (2002) ProDom: automated clustering of homologous domains. Brief Bioinformatics, 3, 246–251. [DOI] [PubMed] [Google Scholar]
- 5.Bateman A., Coin,L., Durbin,R., Finn,R.D., Hollich,V., Griffiths-Jones,S., Khanna,A., Marshall,M., Moxon,S., Sonnhammer,E.L., Studholme,D.J., Yeats,C. and Eddy,S.R. (2004) The Pfam protein families database. Nucleic Acids Res., 32, 138–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Letunic I., Copley,R.R., Schmidt,S., Ciccarelli,F.D., Doerks,T., Schultz,J., Ponting,C.P. and Bork,P. (2004) SMART 4.0: towards genomic data integration. Nucleic Acids Res., 32, 142–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haft D.H., Selengut,J.D. and White,O. (2003) The TIGRFAMs database of protein families. Nucleic Acids Res., 31, 371–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu C.H., Nikolskaya,A., Huang,H., Yeh,L.S., Natale,D.A., Vinayaka,C.R., Hu,Z.Z., Mazumder,R., Kumar,S., Kourtesis,P., Ledley,R.S., Suzek,B.E., Arminski,L., Chen,Y., Zhang,J., Cardenas,J.L., Chung,S., Castro-Alvear,J., Dinkov,G. and Barker,W.C. (2004) PIRSF: family classification system at the Protein Information Resource. Nucleic Acids Res., 32, 112–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madera M., Vogel,C., Kummerfeld,S.K., Chothia,C. and Gough,J. (2004) The SUPERFAMILY database in 2004: additions and improvements. Nucleic Acids Res., 32, 235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Apweiler R., Bairoch,A., Wu,C.H., Barker,W.C., Boeckmann,B., Ferro,S., Gasteiger,E., Huang,H., Lopez,R., Magrane,M., Martin,M.J., Natale,D.A., O'Donovan,C., Redaschi,N. and Yeh,L.S. (2004) UniProt: the Universal Protein knowledgebase. Nucleic Acids Res., 32, 115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andreeva A., Howorth,D., Brenner,S.E., Hubbard,T.J., Chothia,C. and Murzin,A.G. (2004) SCOP database in 2004: refinements integrate structure and sequence family. Nucleic Acids Res., 32, 226–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orengo C.A., Pearl,F.M. and Thornton,J.M. (2003) The CATH domain structure database. Methods Biochem. Anal., 44, 249–271. [DOI] [PubMed] [Google Scholar]
- 13.Berman H., Henrick,K. and Nakamura,H. (2003) Announcing the worldwide Protein Data Bank. Nature Struct. Biol., 10, 980. [DOI] [PubMed] [Google Scholar]
- 14.Harris M.A., Clark,J., Ireland,A., Lomax,J., Ashburner,M., Foulger,R., Eilbeck,K., Lewis,S., Marshall,B., Mungall,C., Richter,J., Rubin,G.M., Blake,J.A., Bult,C., Dolan,M., Drabkin,H., Eppig,J.T., Hill,D.P., Ni,L., Ringwald,M., Balakrishnan,R., Cherry,J.M., Christie,K.R., Costanzo,M.C., Dwight,S.S., Engel,S., Fisk,D.G., Hirschman,J.E., Hong,E.L., Nash,R.S., Sethuraman,A., Theesfeld,C.L., Botstein,D., Dolinski,K., Feierbach,B., Berardini,T., Mundodi,S., Rhee,S.Y., Apweiler,R., Barrell,D., Camon,E., Dimmer,E., Lee,V., Chisholm,R., Gaudet,P., Kibbe,W., Kishore,R., Schwarz,E.M., Sternberg,P., Gwinn,M., Hannick,L., Wortman,J., Berriman,M., Wood,V., de la Cruz,N., Tonellato,P., Jaiswal,P., Seigfried,T. and White,R. (2004) The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res., 32, 258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rawlings N.D., Tolle,D.P. and Barrett,A.J. (2004) MEROPS: the peptidase database. Nucleic Acids Res., 32, 160–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pruess M., Fleischmann,W., Kanapin,A., Karavidopoulou,Y., Kersey,P., Kriventseva,E., Mittard,V., Mulder,N., Phan,I., Servant,F. and Apweiler,R. (2003) The Proteome Analysis database: a tool for the in silico analysis of whole proteomes. Nucleic Acids Res., 31, 414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kopp J. and Schwede,T. (2004) The SWISS-MODEL Repository of annotated three-dimensional protein structure homology models. Nucleic Acids Res., 32, 230–234. [DOI] [PMC free article] [PubMed] [Google Scholar]