Abstract
The Mouse Genome Database (MGD) forms the core of the Mouse Genome Informatics (MGI) system (http://www.informatics.jax.org), a model organism database resource for the laboratory mouse. MGD provides essential integration of experimental knowledge for the mouse system with information annotated from both literature and online sources. MGD curates and presents consensus and experimental data representations of genotype (sequence) through phenotype information, including highly detailed reports about genes and gene products. Primary foci of integration are through representations of relationships among genes, sequences and phenotypes. MGD collaborates with other bioinformatics groups to curate a definitive set of information about the laboratory mouse and to build and implement the data and semantic standards that are essential for comparative genome analysis. Recent improvements in MGD discussed here include the enhancement of phenotype resources, the re-development of the International Mouse Strain Resource, IMSR, the update of mammalian orthology datasets and the electronic publication of classic books in mouse genetics.
INTRODUCTION
The Mouse Genome Database (MGD) provides a comprehensive and integrated view of genetic, genomic and biological information for the laboratory mouse (1,2). MGD contains information on mouse genes, genetic markers and genomic features as well as the associations of these features with sequence sets, reagents, alleles and mutant phenotypes. MGD integrates sequence with biology through the curated association of genome, transcript and protein sequence sets with mouse genes—work done in collaboration with other large genome informatics resources.
MGD is updated daily and there are weekly data exchanges with other major genomics resources such as NCBI and Swiss-Prot. A recent snapshot of MGD content is listed in Table 1. Since the first release of this database in 1994, MGD has continued to evolve, expanding its data coverage, improving data access and providing new data query, analysis and display tools.
Table 1. Snapshot of data content in MGD: October 7, 2004.
| MGD data statistics | October 7, 2004 |
|---|---|
| Number of genes with sequence data | 28 287 |
| Number of genes (including unmapped mutants) | 33 207 |
| Number of markers (including genes) | 57 521 |
| Number of markers mapped | 53 082 |
| Number of genes with links to Swiss-Prot | 7769 |
| Number of genes with GO annotations | 15 309 |
| Number of mouse/human curated orthologies | 14 893 |
| Number of mouse/rat curated orthologies | 12 679 |
| Number of genes with one or more phenotypic alleles | 4996 |
| Number of cataloged phenotypic alleles | 10 949 |
| Number of references | 87 527 |
| Number of mouse nucleotide sequences integrated into the MGI system (includes ESTs) | >7 600 000 |
MGD is the core component of the Mouse Genome Informatics (MGI) database resource (http://www.informatics.jax.org) hosted at The Jackson Laboratory (http://www.jax.org). Other projects and resources that are part of the MGI system include the Gene Expression Database (GXD) (3) and the Mouse Tumor Biology Database (MTB) (4) (http://tumor.informatics.jax.org). All MGI component groups participate actively in the development and application of the Gene Ontology (GO) (5) (http://www.geneontology.org).
IMPROVEMENTS DURING 2004
Hosting of the International Mouse Strain Resource
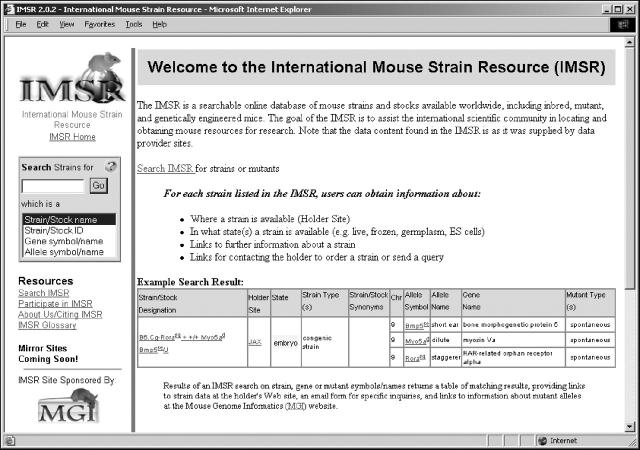
The International Mouse Strain Resource (IMSR) (http://www.informatics.jax.org/imsr/) has as its goal to provide and maintain a worldwide catalog of resources for mouse strains and stocks. The IMSR has developed a searchable database with a web front-end to assist researchers in locating and obtaining the mouse resources they need (Figure 1).
Figure 1.
The new IMSR is a searchable online database of mouse strains and stocks available worldwide, including inbred, mutant and genetically engineered mice. The goal of the IMSR is to assist the international scientific community in locating and obtaining mouse resources for research. The data content found in the IMSR is as it was supplied by data provider sites.
An initial version of the IMSR was developed in 1999 (6) as a collaborative effort with the Medical Research Council (MRC) Mammalian Genetics Unit (Harwell, UK) and contained a searchable resource for mouse stocks and strains held at The Jackson Laboratory (JAX) and at the MRC Harwell sites. While this proved to be a useful resource, it was severely limited in containing only information for these two major mouse laboratories.
With the establishment of multiple mutagenesis centers, gene trap centers, and the increasing use of genetic engineering technologies, the number of mouse stocks and strains and the specialization of genotype and their characterization has exploded. A number of new repositories and distribution centers have been established worldwide to cope with the exponential increase in specialized mouse stocks. The new pressing need for a central cataloging of stocks and strains prompted us to re-develop the IMSR in a more robust fashion, such that it could easily accommodate data from multiple sites, provide a better search interface for users, and enable links to phenotype searching and to specific stock data from each site that distributed mouse resources.
Users can search IMSR by strain, gene or allele designations, strain state(s) and strain classes. For each strain satisfying the search criteria, IMSR provides users with data on where a strain is available from, in what state(s) the strain exists (e.g. live, cryopreserved embryos or gametes, ES cell lines), the class of strain and mutant alleles carried by the strain. Hypertext links are provided (i) from each strain designation to its strain information page at the holding site, (ii) to an auto-generated email form to the holder's designated representative for obtaining additional information or ordering the mouse resource and (iii) from each mutant phenotypic allele carried by a strain to the detailed characterization of that allele in the MGI.
Additional links from the IMSR homepage provide instructions for participating in IMSR by listing one's mouse resources, for searching MGI for additional mouse genetic, genomic and biological information, and for checking the official mouse nomenclature guidelines from the International Committee on Genetic Nomenclature for Mice.
Current centers with mouse resources included in IMSR are as follows: The Jackson Laboratory (JAX), the Mouse Mutant Regional Resources Centers (MMRRC), the Center for Animal Resources and Development (CARD), the Oak Ridge National Laboratory (ORNL), the European Mouse Mutant Archive (EMMA) and the BayGenomics Gene Trap Resource. In progress is the incorporation of stocks from the MRC Genetics Unit, Harwell (Har), the Beta Cell Biology Consortium (BCBC), Neuromice (NMICE) and the Mouse Models of Human Cancer Consortium (MMHCC). Interest has been expressed by several other sites, including additional mouse mutagenesis centers, additional gene trap resources, and other distribution centers. IMSR also accepts stock listings from individuals. All strains and stocks listed in IMSR should be available to the research community and regular updating from sites is required to keep the IMSR current.
Enhanced orthology resources
MGD provides a curated set of mammalian orthologs for the research community. Although MGD supports orthology annotations to over 20 mammalian genomes, the priority effort focuses on the creation of orthology sets among mouse, human and rat. This set is constructed through an iterative process using both computational and manual approaches. This year, we worked with the HomoloGene resource at the NCBI (7) to reciprocally incorporate some of the HomoloGene computational three-way reciprocal best-hit sets into the MGI system. HomoloGene incorporates MGD-curated mammalian orthology sets in their resources. In addition, we continue to work with the research community to carefully curate gene family sets, usually at the instigation of the research community (8,9).
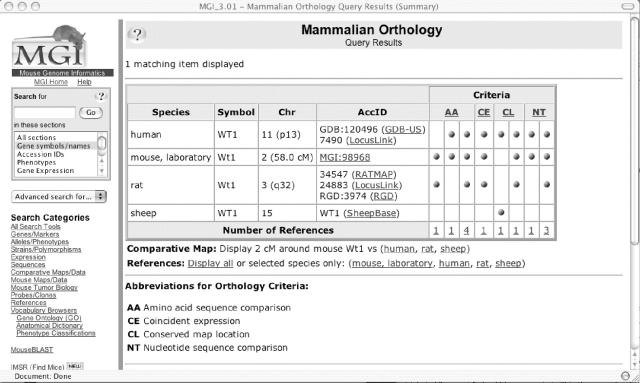
The Orthology Detail Page in MGD (Figure 2) for the gene Wt1 illustrates the paradigm for orthology data. All assertions of orthology are supported by a statement of evidence and a citation. There are links to comparative mapping visualizations and links to genomics resources for the other species represented.
Figure 2.
Mammalian Orthology Detail Page. The Mammalian Orthology Query Results page presents a table of results from MGI orthology curation. The table includes species, symbol, chromosome, external and internal accession IDs and criteria for the assertions. The criteria include both a statement of evidence and a citation. Hypertext links are incorporated as appropriate. Comparative chromosome map visualizations between any two of the species can be accessed from this page. These data are updated nightly.
Electronic publication of classic books in mouse genetics
MGD has responded to user requests in making electronic copies of popular out-of-print books available online. Two such books have been developed as online versions, Mouse Genetics by Lee Silver (Oxford University Press, 1995) at http://www.informatics.jax.org/silver/ and The Coat Colors of Mice by Willys K. Silvers (Springer Verlag, 1979) at http://www.informatics.jax.org/wksilvers/ (Figure 3). To develop these online books, publisher and author copyrights were obtained, text was re-developed and hypertext links placed within the text for cross-referencing and to provide direct access to MGI for enhanced gene and reference data. Photographs and graphics were scanned into electronic form or in some cases, redrawn. Both books have been welcomed by MGD users and permission to include additional out-of-print books is being sought.
Figure 3.
Electronic publication of classic books in mouse genetics. MGI offers electronic versions of key out-of-print books in mouse genetics, Mouse Genetics by Lee M. Silver (http://www.informatics.jax.org/silver/) and The Coat Colors of Mice by Willys K. Silvers (http://www.informatics.jax.org/wksilvers/). Gene symbols in both books link to MGI gene detail pages where readers can access all the information MGI has assembled for that gene, including phenotypic alleles, nucleotide and protein sequence, mapping and expression data and GO annotations. References cited in the books are linked to PubMed or MGI reference detail pages, which, in turn are linked to additional curated information in MGI.
OTHER INFORMATION
User input
MGD encourages user input into its gene and allele annotation efforts. On each gene detail and allele detail page, a clickable button (‘Your Input Welcome’) brings the user to a web-based form for submitting updates to the information being viewed.
Mouse gene nomenclature
The MGD gene annotation group assigns unique symbols and names to mouse genes under the guidelines set by the International Committee on Standardized Genetic Nomenclature for mouse (http://www.informatics.jax.org/mgihome/nomen/index.shtml). Through curation of shared links between MGI and other bioinformatics resources, the official nomenclature for mouse genes is becoming widely disseminated. The MGI nomenclature group works closely with human (http://www.gene.ucl.ac.uk/cgi-bin/nomenclature/searchgenes.pl) and rat (http://rgd.mcw.edu) nomenclature specialists to provide consistent nomenclature for mammalian species. Scientists can reserve symbols prior to publication using the electronic nomenclature submission form (http://www.informatics.jax.org/mgihome/nomen/nomen_submit_form.shtml) or by contacting the MGD nomenclature coordinator by email (nomen@informatics.jax.org). The MGD nomenclature coordinator can also assist with other nomenclature issues, such as revision of gene family designations.
Electronic data submission
Any type of data that MGD maintains can be submitted as an electronic contribution. Over the last year, the most frequent submissions have been of mutant and phenotypic allele information originating from the large mouse mutagenesis centers. Other common types of submission include gene and strain nomenclature, mutant and QTL mapping data, polymorphisms and mammalian homologies. Each electronic submission receives a permanent database accession ID. All datasets are associated with either an electronic submission reference or a published paper. These reference pages provide links to associated datasets. Online information about data submission procedure is found at http://www.informatics.jax.org/mgihome/submissions/submissions_menu.shtml.
Community outreach and user support
MGD provides extensive user support through online documentation and easy email or phone access to User Support Staff. User Support WWW access: http://www.informatics.jax.org/mgihome/support/support.shtml
Email access: mgi-help@informatics.jax.org
Telephone access: +1 207 288 6445
FAX access: +1 207 288 6132
Other outreach
MGI-LIST (http://www.informatics.jax.org/mgihome/lists/lists.shtml) is a moderated and active email bulletin board supported by the MGI User Support group. Other outreach includes Online Tutorials and answers to Frequently Asked Questions, available at http://www.informatics.jax.org/userdocs/helpdocs_menu.shtml.
IMPLEMENTATION
MGD is implemented in the Sybase relational database system, version 12.5. A large set of CGI scripts and Java Servlets mediate the user's interaction with the database. For computational users, direct SQL access can be requested through User Support. User-requested database reports and a number of widely used data files (generated daily) are available on the ftp site (ftp://ftp.informatics.jax.org/pub/reports/index.html).
CITING MGD
The following citation format is suggested when referring to datasets specific to the MGD component of MGI: Mouse Genome Database (MGD), Mouse Genome Informatics, The Jackson Laboratory, Bar Harbor, Maine (http://www.informatics.jax.org). [Type in date (month, year) when you retrieve the data cited.] For general citation of the MGI resource please cite this article.
Acknowledgments
ACKNOWLEDGEMENTS
The Mouse Genome Database is supported by NIH/NHGRI grant HG00330.
REFERENCES
- 1.Bult C.J., Blake,J.A., Richardson,J.E., Kadin,J.A., Eppig,J.T. and the Mouse Genome Database Group (2004) The Mouse Genome Database (MGD): integrating biology with the genome. Nucleic Acids Res., 32, D476–D481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blake J.A., Richardson.J.E., Bult,C.J., Kadin,J.A., Eppig,J.T. and the Mouse Genome Database Group (2003) MGD: the Mouse Genome Database. Nucleic Acids Res., 31, 193–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hill D.P., Begley,D.A., Finger,J.H., Hayamizu,T.F., McCright,I.J., Smith,C.M., Beal,J.S., Corbani,L.E., Blake,J.A., Eppig,J.T., Kadin,J.A., Richardson,J.E. and Ringwald,M. (2004) The Mouse Gene Expression Database (GXD): updates and enhancements. Nucleic Acids Res., 32, D568–D571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Näf D., Krupke,D.M., Sundberg,J.P., Eppig,J.T. and Bult,C.J. (2002) The mouse tumor biology database: a public resource for cancer genetics and pathology of the mouse. Cancer Res., 62, 1235–1240. [PubMed] [Google Scholar]
- 5.The Gene Ontology Consortium (2004) The Gene Ontology (GO) Database and Informatics Resource. Nucleic Acids Res., 32, D258–D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eppig J.T. and Strivens,M. (1999) Finding a mouse: the International Mouse Strain Resource (IMSR). Trends Genet., 15, 81–82. [DOI] [PubMed] [Google Scholar]
- 7.Wheeler D.L., Church,D.M., Edgar,R., Federhen,S., Helmberg,W., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Sequeira,E. et al. (2004) Database resources of the National Center for Biotechnology Information: update. Nucleic Acids Res., 32, D35–D40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mashek D.G., Bornfeldt,K.E., Coleman,R.A., Berger,J., Bernlohr,D.A., Black,P., DiRusso,C.C., Farber,S.A., Guo,W., Hashimoto,N. et al. (2004) Revised nomenclature for the mammalian long-chain acyl-CoA synthetase gene family. J. Lipid Res., 45, 1958–1961. [DOI] [PubMed] [Google Scholar]
- 9.Nelson D.R., Zeldin,D.C., Hoffman,S.M.G., Maltais,L.J., Wain,H.M. and Nebert,D.W. (2004) Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Phamacogenetics, 14, 1–18. [DOI] [PubMed] [Google Scholar]