Abstract
2-epi-5-epi-Valiolone synthase (EEVS), a C7-sugar phosphate cyclase (SPC) homologous to 3-dehydroquinate synthase (DHQS), was discovered during studies of the biosynthesis of the C7N-aminocyclitol family of natural products. EEVS was originally thought to be present only in certain actinomycetes, but analyses of genome sequences showed that it is broadly distributed in both prokaryotes and eukaryotes, including vertebrates. Another SPC, desmethyl-4-deoxygadusol synthase (DDGS), was later discovered as involved in the biosynthesis of mycosporine-like amino acid sunscreen compounds. Current database annotations are quite unreliable, with many EEVSs reported as DHQS, and most DDGSs reported as EEVS, DHQS, or simply hypothetical proteins. Here, we identify sequence features useful for distinguishing these enzymes, report a crystal structure of a representative DDGS showing the high similarity of the EEVS and DDGS enzymes, identify notable active site differences, and demonstrate the importance of two of these active site residues for catalysis by point mutations. Further, we functionally characterized two representatives of a distinct clade equidistant from known EEVS and known DDGS groups, and show them to be authentic EEVSs. Moreover, we document and discuss the distribution of genes that encode EEVS and DDGS in various prokaryotes and eukaryotes, including pathogenic bacteria, plant symbionts, nitrogen-fixing bacteria, myxobacteria, cyanobacteria, fungi, stramenopiles, and animals, suggesting their broad potential biological roles in nature.
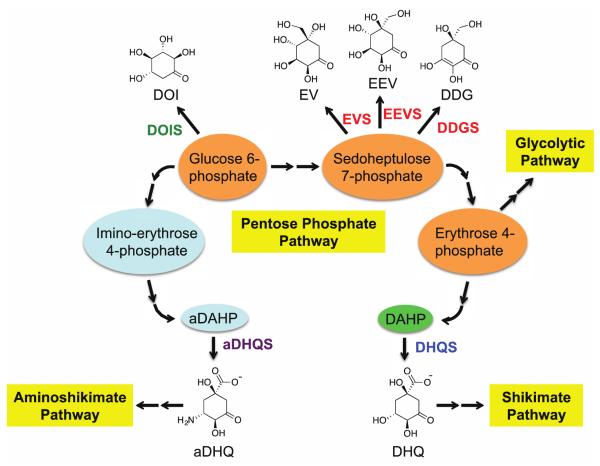
2-epi-5-epi-Valiolone synthase (EEVS) is a member of the sugar phosphate cyclases (SPCs), a group of homologous enzymes that catalyze the cyclization of sugar phosphates to cyclic compounds in primary and secondary metabolism.1, 2 Members of the SPC family (Figure 1) share significant sequence and structural similarity with 3-dehydroquinate synthase (DHQS).3, 4 DHQS converts 3-deoxy-D-arabinoheptulosonate 7-phosphate (DAHP) to 3-dehydroquinate (DHQ), the first committed step in the shikimate pathway, which leads to aromatic amino acids, folates, ubiquinones, and many secondary metabolites. Other members of the SPC family include aminodehydroquinate synthase (aDHQS, a variant of DHQS),5 2-deoxy-scyllo-inosose synthase (DOIS),6 desmethyl-4-deoxygadusol synthase (DDGS)1, 7 and 2-epi-valiolone synthase (EVS).8, 9 aDHQS converts amino-DAHP to amino-DHQ, the precursor of 3-amino-5-hydroxybenzoic acid, which is involved in the biosynthesis of important polyketide antibiotics, such as rifamycin, geldanamycin, mitomycin, and ansamitocin.10-13 DOIS catalyzes the conversion of glucose 6-phosphate to 2-deoxy-scyllo-inosose, which is the precursor of deoxystreptamine-containing aminoglycoside antibiotics, e.g., butirosin, neomycin, kanamycin, and tobramycin.14 Finally, EEVS, DDGS, and EVS use sedoheptulose 7-phosphate (SH7P; a pentose phosphate pathway intermediate) as substrate to give 2-epi-5-epi-valiolone, 2-desmethyl-4-deoxygadusol, and 2-epi-valiolone, respectively. 2-epi-5-epi-Valiolone and 2-epi-valiolone are precursors of aminocyclitol natural products, such as the antidiabetic drug acarbose and the antifungal agent validamycin A,9, 15, 16 whereas desmethyl-4-deoxygadusol is the precursor of the mycosporine-like amino acid sunscreen compounds.7 Of particular interest is that EEVS is also involved in the formation of gadusol, another sunscreen-like compound found in fish, and possibly in other vertebrates, e.g., amphibians, reptiles, and birds, but not mammals.17
Figure 1.
Six sugar phosphate cyclase family members, their substrates and products, and their connections to the Pentose Phosphate Pathway. DOIS, 2-deoxy-scyllo-inosose synthase; EVS, 2-epi-valiolone synthase; EEVS, 2-epi-5-epi-valiolone synthase; DDGS, desmethyl-4-deoxygadusol synthase; aDHQS, aminodehydroquinate synthase; DHQS, 3-dehydroquinate synthase.
For catalysis, all SPC superfamily members require NAD+ and a metal ion, either Zn2+ or Co2+, as prosthetic groups. Among them, DHQS has been particularly well-studied because of its involvement in primary metabolism; thus, it is a potential target for antibacterial drug development. The primary amino acid sequences of EEVSs and DDGSs are highly similar to each other, and to some extent to those of DHQSs,1, 8 and we have noticed that these enzymes are often misannotated in genome databases, particularly involving the misassignment of DDGS as EEVS or DHQS. This inconsistent/inaccurate functional assignment of these enzymes has hampered correct prediction of their roles in nature.
This has prompted us to evaluate the genes and their encoded protein sequences with the goal of establishing parameters for more accurately annotating EEVS, DDGS, and other SPCs. Here, we describe a bioinformatics study and an approach to more accurately assign putative functions to EEVS and DDGS encoding genes in databases. We also report a crystal structure of a representative DDGS, Ava_3858 from Anabaena variabilis (AvDDGS),8 compare it with those of EEVS and DHQS, identify notable active site differences, and demonstrate the importance of two of these active site residues for catalysis.4, 18 In addition, we confirm through biochemical experiments the function of two putative EEVS proteins that form a separate clade from the previously characterized EEVSs, and demonstrate the broad distribution of EEVS and DDGS in prokaryotes and eukaryotes.
RESULTS AND DISCUSSION
Bioinformatics analysis and reassignment of EEVS and DDGS
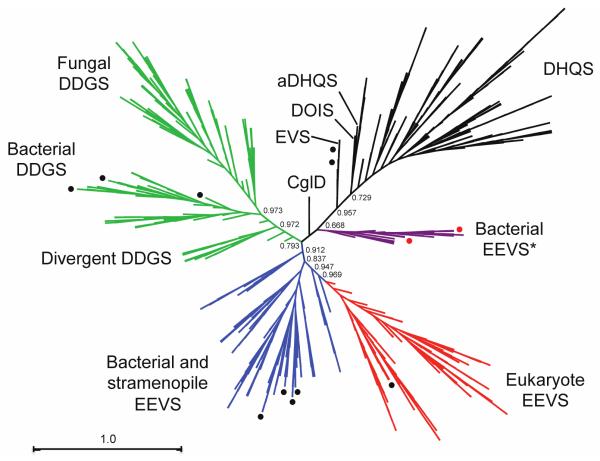
During the past decade, there has been a significant upsurge in deposition of genes annotated as EEVS or DHQS-like proteins in public databases, and as noted in the introduction, many of them have been incorrectly annotated. To address this in a comprehensive manner, we have gathered for evaluation all genes that had e-values < 7e–68 in a BLAST search of the NCBI database using the known EEVS from the validamycin A pathway (ValA or ShEEVS) and the DDGS from the shinorine pathway (Npun_R5600) as queries. The 630 amino acid sequences were aligned using MUSCLE and the phylogenetic tree (Figure 2) was constructed using FastTree, employing the JTT model of protein evolution and the CAT approximation. From the 630 proteins studied, we preliminarily concluded based on the phylogenetic tree (Figure 2) that 335 of them are EEVS and 295 are DDGS. As expected, ValA groups together with other known EEVS proteins: AcbC, CetA, PyrA, BE-Orf9, and SalB.1, 19-22 About 30% of the EEVS proteins had been annotated as DHQS or hypothetical proteins (Table S1). All (i.e. 100%) of the DDGS proteins had been misannotated, most commonly as DHQS, EEVS, or hypothetical proteins (Table S2; the new annotations are also available on our website at http://people.oregonstate.edu/~mahmudt/?page_id=396).
Figure 2.
Radial cladogram of the sugar phosphate cyclase superfamily. A group of putative bacterial EEVS (drawn in purple and designated “EEVS*”) occurs as a separate clade from the known EEVS clades (drawn in blue and red). Numbers show local support values. Black circles show sedoheptulose 7-phosphate cyclases (EEVS, EVS, and DDGS) that have been biochemically characterized. Red circles identify the two bacterial EEVS* (GacC and Staur_1386) that are characterized in this study.
Conserved motifs for EEVS and DDGS
Through detailed comparative bioinformatics analysis of these preliminary groupings, we identified two stretches of residues that appeared most useful for distinguishing the enzymes from each other. In these segments, the conserved amino acid sequences for EEVS were MLEELxPNLxE and xxRxxDxGH, which were recognizably different from the DHQS sequences, and also adequately distinguishable from those of DDGS [MLELExPNLHE and LDRVIAxGH] (Figure 3). Specifically, in the first conserved regions, the EEVS proteins contain an MLEEL motif, whereas the DDGS proteins contain a MLELE motif. In the second conserved regions, the EEVS proteins contain an Asp, in place of an Ala in the DDGS proteins. Some proteins have slight variations from these conserved motifs, for example KL instead of EL for EEVS. However, the overall conserved motif can still sufficiently distinguish EEVS and DDGS enzymes despite the sequence variations.
Figure 3.
Comparisons of partial amino acid sequences of the sugar phosphate cyclases. Conserved amino acid residues in two fingerprint segments are shown from bacterial DHQS, aminoDHQS, DOIS, and EVS and various EEVS and DDGS groups as labeled. Black circles above the sequences indicate residues found in the catalytic pocket of the enzymes. Stars above the EEVS and DDGS sequence groups indicates residues that differentiate DDGS from EEVS.
Independent analysis and comparison of crystal structures of ShEEVS and AvDDGS also identified these characteristic residue differences as a means to differentiate EEVS and DDGS. Leu267 in ShEEVS versus Glu254 in AvDDGS (part of the first conserved region MLEEL/MLELE) and Asp281 in ShEEVS versus Ala268 in AvDDGS (part of the second conserved region xxRxxDxGH/xxRxxAxGH) contribute to the active site pocket where they may also be responsible for the different activities of these enzymes (see below).
Additionally, a new putative DDGS gene whose product shows low sequence similarity to the known DDGS was recently reported in the halotolerant cyanobacterium Aphanothece halophytica.23 Inactivation of this gene in this organism resulted in mutants that no longer produce MAAs. In our phylogenetic analysis, forty-one proteins formed a new clade with the A. halophytica DDGS (Figure 2) with their predicted protein sequences having some conserved motifs (e.g. YxxxEY(G)xNxxET and QC(D)RPHA(G)YGHTWSP) distinct from the mainstream DDGS sequences (Figure 3). This “divergent DDGS” clade includes proteins from cyanobacteria, algae, and marine invertebrates (Table S3). Some of the divergent DDGS genes (e.g. that of A. halophytica23) are not clustered with the other MAA biosynthetic genes, but others are (e.g. that of Anabaena sp. 90).
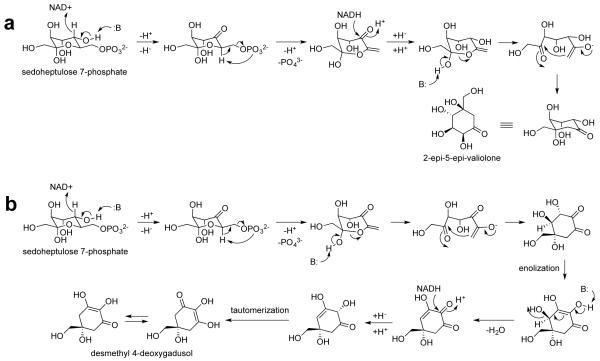
As can be seen in Figure 3, there is more variation in the conserved regions of bacterial EEVSs than DDGSs. This could be due to the differences in the reaction performed by these enzymes. EEVSs perform a cyclization reaction through a mechanism similar to those of DHQS and DOIS, involving five step reactions (Figure 4).18, 19, 24 On the other hand, DDGSs perform a cyclization and a dehydration reaction, involving a more complex mechanism.7 Due to this, more residues in the conserved region of DDGS may play key roles, and/or the spatial requirements for DDGS catalysis may be more stringent. Thus mutations in DDGS enzymes could negatively impact its catalysis more so than would changes in the EEVS enzymes. On the other hand, the relatively low sequence divergence observed among the vertebrate EEVSs is consistent with a recent report describing decelerated amino acid substitution in modern vertebrates.25 Also, the transfer of EEVS gene from microorganisms to vertebrates is predicted to occur later during evolution.17
Figure 4.
Proposed catalytic mechanisms for EEVS (a) and DDGS (b).
Crystal structure of AvDDGS
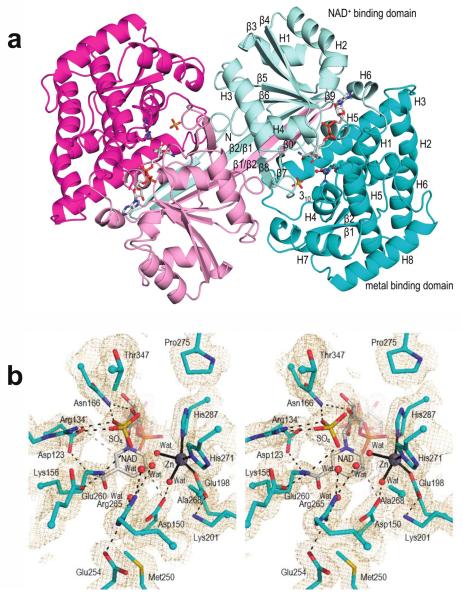
To investigate the catalytic pocket and unique features of a representative DDGS, we solved the X-ray crystal structure of AvDDGS. Crystals of recombinant His-tagged AvDDGS yielded diffraction data to 1.7 Å resolution and the structure was easily solved by molecular replacement using the ShEEVS structure. The final refined model contains two chains, making up one dimer, in the asymmetric unit (Figure 5a); each chain included 400 of the 444 expected residues, one Zn2+, one NAD+, and a sulfate in the active site with a final R/Rfree of 15.6/18.3% (Table S5). The N-terminal tag and residues 1–2 and 403–410 are not modeled in either chain, but otherwise the main chain is well-ordered with clear density in both chains. Zn2+ and NAD+ were not added during sample preparation or crystallization, but fortuitously are both present and have clear, unambiguous electron density in the electron density maps (Figure 5b). In both chains, the estimated occupancies of Zn2+ and NAD+ are 0.5 and 0.75, respectively, and a sulfate (at occupancy 0.25) binds at the NAD+ pyrophosphate position when NAD+ is not there.
Figure 5.
Overall structure and active site of AvDDGS. a. Ribbon diagram of the two chains of the AvDDGS dimer are show in cyan and magenta tones, respectively, with the N-terminal NAD+-binding domains in light hues and the C-terminal metal-binding domains in dark hues. The NAD+, Zn2+ with its coordinating residues, and sulfate bound in the active site are shown. Secondary structural elements and domains of one monomer are labeled. The extended β-strands involved in the domain-swapped interaction (labeled β1/β2 and β2/β1) represent β1 and β2, respectively, in the labeled monomer. b. Stereoview of the AvDDGS active site residues (cyan carbons), waters (red spheres), sulfate bound in the active site (yellow), NAD+ (grey carbons), and Zn2+ (silver sphere). Coordination bonds (black lines), select hydrogen bonds (black dashes), or approaches of interest (grey dashes) and 2Fo-Fc electron density (orange, contoured at 1ρrms) are also shown.
The dimer seen in the asymmetric unit (Figure 5a) is consistent with those observed in other SPC family enzymes and thus is thought to be biologically relevant. The overall structure is highly similar to ShEEVS (rmsd = 0.9 A for 353 Cα atoms). All core secondary structural elements are conserved (Figure S1), and this includes a domain-swapped interaction observed in ShEEVS (PDB 4P53). In what appears to be a common feature of this subset of sedoheptulose 7-phosphate cyclases (SH7PCs), instead of having a β-hairpin near position 31 in AvDDGS, the N-terminal residues continue in a linear direction, making an extended β-strand (here called β1/β2 as it combines what are two β-strands in other SPC enzymes like DHQS) that reaches across the back of the dimer, and effectively contributes a β-strand to each monomer of the dimer (Figure 5A). In AvDDGS, a β-hairpin near residue 12 means that one short β-strand (β0) adds an 8th strand to the typically 7-stranded β-sheet observed in other SPC family members.
Each chain of AvDDGS consists of the expected N-terminal NAD+-binding domain and C-terminal metal-binding domain seen in the SPC family (Figure S1). The N-terminal NAD+-binding domain has a core 8-stranded β-sheet surrounded by 5 α-helices, 2 short 310 helices, and 1 β-hairpin (strands 7 and 8). The C-terminal metal-binding domain is primarily α-helical and consists of 8 α-helixes, 1 β-hairpin and 1 310 helix. The Zn-coordinating residues as well as most of the residues making up the active site cavity come from the metal-binding domain although the active site itself is located in the cleft between the two domains.
Active Site of AvDDGS
The active site is well defined (Figure 5b) with the Zn2+ and NAD+ bound as in DHQS, DOIS, and ShEEVS, with some key residues briefly noted here (and highlighted in Figure S1). The Zn2+ ion is coordinated by Glu198, His271, His287, and two waters. For NAD+ binding, the adenosine ribose O2’ hydroxyl hydrogen bonds with Asp56 and Asn58, located at the end of β3, and the adenine forms hydrogen bonds with the Thr186 and Thr143 side chains and backbone carbonyls of Thr143 and Leu183. The pyrophosphate oxygens (and the sulfate oxygens found in their place) hydrogen bond with the backbone amides of Gly119 and Leu120 in the glycine-rich turn connecting β5 and H3, as well as with the Thr144 side chain. The nicotinamide ribose hydroxyls hydrogen bond with the side chains of Glu87, Lys90, Lys165, and Asn166, and the nicotinamide amide group hydrogen bonds with the Asp123 carboxylate and the backbone carbonyl of Lys156. Also, as observed in ShEEVS,4 the carboxylates of Asp150 and Asp123 (Figure 5b) are roughly in the plane of the nicotinamide ring where they can make interactions with atoms C2 and C4 of the positively charged NAD+. In addition, the nicotinamide amide oxygen and pyrophosphate oxygens hydrogen bond with ordered waters.
The SH7P Binding Site in AvDDGS
Although this structure of AvDDGS is unliganded, we are able to make inferences about substrate binding via a comparison to DHQS with its bound substrate analog, carbaphosphonate (CBP) (Figure 6a). Supporting the value of this comparison, the bound sulfate in AvDDGS overlays well with the CBP phosphate and AvDDGS ordered water sites overlay well with the CBP C2, C4, and C5 hydroxyls. As these substituents are also present in AvDDGS’s substrate, SH7P, we hypothesize that it will bind in a similar way.
Figure 6.
Comparisons of the AvDDGS active site region with the DHQS•CBP complex and with ShEEVS. a. Stereoview of select active site residues in AvDDGS (cyan carbons) with NAD+ (grey carbons) overlaid on DHQS in complex with CBP (salmon carbons; PDB 1DQS). Hydrogen bonding interactions (dashed lines) and coordination bonds with Zn2+ (solid lines) in the AvDDGS (black) and DHQS (grey) active sites are shown. The sulfate (yellow) and waters (red spheres) in the active site of AvDDGS in agreement with components of CBP or water (salmon sphere) in the active site of DHQS as well as the zinc ions (silver spheres) are shown. Labels for Arg265 (AvDDGS) and Arg264 (DHQS) are separated due to their disparate positions in the active site and in the sequence alignment. A prime on a residue number means it is from the other subunit of the dimer. View of active site is rotated roughly 90° counter-clockwise with respect to the view in Figure 5b and 6b. b. Stereoview of active site residues in AvDDGS (cyan carbons) with NAD+ (grey carbons), sulfate bound in the active site (yellow), and select waters (red spheres) overlaid on ShEEVS with NAD+ (both purple carbons) and select waters (purple spheres). Hydrogen bonds (dashed) and coordination bonds (solid) are shown in the AvDDGS active site (black lines) and ShEEVS active site (light pink lines). A prime on a residue number means it is from the other subunit of the dimer.
One difference between SH7P and CBP occurs at the second substituent of C2, where in place of the CBP carboxylate, SH7P has a hydroxymethyl group. In the AvDDGS structure, one ordered water overlays roughly with one of the CBP carboxylate oxygens, perhaps indicating the position of the hydroxymethyl in SH7P. Notably, this is also the site of major differences between the DHQS and EEVS/DDGS active sites. In DHQS, Arg264 and Lys250 hydrogen bond with the C2-carboxylate (Figure 6a) and in DDGS (and EEVS) neither residue is conserved. The Lys is replaced by Met250 and the residue equivalent to Arg is not positionally conserved (even when it remains an Arg), greatly decreasing the local positive charge. Also the Lys→Met change allows Asp150 to be closer to the pocket and it binds to the water that may mimic the C2-hydroxymethyl substituent (Figure 6a). As we noted in describing the EEVS structure,4 a different Arg that is one position later in the sequence (Arg265 in AvDDGS) is conserved in ShEEVS and AvDDGS. This alternate Arg hydrogen bonds with the water noted above that mimics the C2-hydroxyl in CBP (Figure 6a).
Comparison of AvDDGS with ShEEVS
The active sites of ShEEVS and AvDDGS are quite similar but have key “fingerprint” differences1, 4, 8 that are putatively responsible for their differences in activity. Two binding pocket differences that had been previously identified are Asp281 in ShEEVS vs. Ala268 in AvDDGS and His360 in ShEEVS vs. Thr347 in AvDDGS. As noted above, here we have added a third active site fingerprint residue: Leu267 in ShEEVS vs. Glu254 in AvDDGS. An active site overlay of these two enzymes (Figure 6b) reveals just a few notable differences that essentially involve these three residues. In AvDDGS, the Glu254 side chain vs. Leu267 in ShEEVS displaces an ordered water and hydrogen bonds directly to Arg265. Interestingly, the presence of Ala 268 vs. Asp281 in ShEEVS opens room for a unique ordered water in AvDDGS that also hydrogen bonds to Arg265. The presence of Thr347 vs. His360 in ShEEVS opens room near the putative binding site. For each of these changes it is not immediately obvious how it may contribute to the different catalytic activity of AvDDGS.
Looking beyond the active site, two of these positions, Glu254 and Ala268, are part of the sequence motifs we have used for distinguishing DDGS from EEVS (Figure 3). As seen in Figure S2, despite their variation in amino acid sequence, in the folded protein the two segment have the same secondary structure in DDGS and EEVS and orients the side chains in similar directions.
Point mutations in ShEEVS and AvDDGS
To test the importance of the three active site residues noted in the previous section as being characteristic to ShEEVS or AvDDGS, we generated a total of 14 mutants of ShEEVS (ValA) and AvDDGS (Ava_3858), consisting of six single point mutants (L267E, D281A, and H360T for ShEEVS; A268D, E254L, and T347H for AvDDGS), six double point mutants (D281A/H360T, L267E/D281A, and L267E/H360T for ShEEVS; E254L/A268D, E254L/T347H, and A268D/T347H for AvDDGS), and two triple point mutants (L267E/D281A/H360T for ShEEVS; E254L/A268D/T347H for AvDDGS). All proteins were recombinantly produced in Escherichia coli and characterized for their activity under the conditions previously described.8 For consistency between the proteins, they were tested fresh upon cell disruption and centrifugation without additional purification steps (Figure S3). Also, we developed a thin layer chromatography (TLC) protocol that separates the EEV and DDG products, and a staining reagent, p-anisaldehyde, that differentiates EEV and DDG as yellow and purple spots, respectively (Figure S4). The results revealed that ShEEVS Leu267 and Asp281 are critical for its activity, and the equivalent AvDDGS residues Ala268 and Glu254 are as well (Figure S4). This is consistent with their high conservation in the two sequence motifs we used for distinguishing DDGS from EEVS. Interestingly, we found that ShEEVS His360 and the equivalent AvDDGS Thr347 do not directly contribute to their respective catalytic activity. Yet, our bioinformatics studies revealed that these residues are highly conserved among proteins from their respective classes (94% of EEVS have His360 and 96% of DDGS have Thr347).
The lack of activity of A268D and E254L mutants of AvDDGS may suggest that one or both of these residues play a role in the early ring opening and aldol cyclization steps or in the unique dehydratase reaction by AvDDGS (Figure 4b). However, as there are no detectable intermediates produced by either of these mutants, their actual role(s) in AvDDGS catalytic activity are still unclear. Further elucidation of the DDGS catalytic mechanism, including the residues responsible for its proposed dehydratase activity, will be a subject of future investigations.
Phylogenetically distinct putative EEVS genes in some Gram-(+) and Gram-(−) bacteria
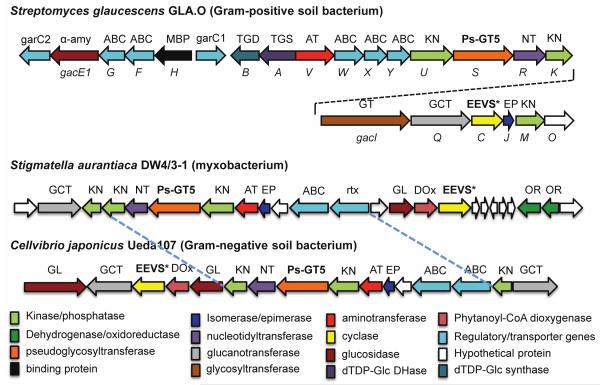
Phylogenetic studies also revealed a group of putative EEVSs (indicated as EEVS* here) that are arranged in a separate clade from the known EEVS clade and more similar to DHQSs (Figure 2). Those include GacC, a putative EEVS in the acarbose pathway from Streptomyces glaucescens GLA.O and Staur_1386, from the myxobacterium Stigmatella aurantiaca DW 4/3–1. A previous report has noted that although the gac cluster in S. glaucescens GLA.O has similarity to the acarbose cluster in Actinoplanes sp. SE50/110, but with enough differences to be uncertain of their equivalence.26 Staur_1386 is part of a cryptic biosynthetic gene cluster in S. aurantiaca DW 4/3–1 with notable similarity to a cluster in a phylogenetically distant bacterium, Cellvibrio japonicus Ueda107 (Figure 7). Our discovery of putative pseudoglycosyltransferase genes within these clusters27-29 has allowed us to predict their involvement in pseudo-oligosaccharide biosynthesis, but the exact end products remain unknown.
Figure 7.
Biosynthetic gene clusters in Streptomyces glaucescens GLA.O, Stigmatella aurantiaca DW4/3–1, and Cellvibrio japonicus Ueda107 (previously known as Pseudomonas fluorescens subsp. cellulosa).
To confirm the catalytic function of GacC and Staur_1386, we cloned the corresponding genes from S. glaucescens GLA.O and S. aurantiaca DW 4/3–1 and heterologously expressed them in E. coli. There are two possible start codons in the gacC gene that would result in proteins with 388 and 410 amino acids. As the smaller protein is similar in size to AcbC from the acarbose pathway19 and the larger protein is similar to ValA from the validamycin pathway,30 we produced both versions of GacC. The recombinant GacC-388, GacC-410, and Staur_1386 proteins (unpurified, Figure S5) were characterized using SH7P as substrate in the presence of NAD+ and Zn2+ or Co2+. Analysis of the products by TLC and GC-MS revealed the production of 2-epi-5-epi-valiolone by all of these proteins (Figure S6), and this EEVS activity was further confirmed by assays of the purified recombinant GacC-388 and Staur_1386.
Unexpectedly, our phylogenetic analysis revealed that S. hygroscopicus subsp. hygroscopicus strain NRRL B-1477 had an EEVS* (WP_030827434.1) (Table S4), and only shared 41% identity with ValA from S. hygroscopicus subsp. jinggangensis. A recent analysis of Streptomyces lineages showed that Streptomyces from this clade descended from multiple lineages.31 So we propose that the EEVS* gene or the gene cassette has spread through horizontal gene transfer (HGT)32 and, since homologous recombination is not uncommon in Streptomyces, possibly formed through homologous recombination events. Interestingly, 14 of the total 51 known EEVS* proteins have a common insertion (Figure S7), and all were from Gram-(−) bacteria (i.e. 14 of the 19 Gram-(−) bacterial EEVS*).
In our phylogenetic analysis and in previous analyses,17 this EEVS* clade branches off before the main EEVS clade and DDGS branches split. This implies a gene duplication event followed by divergence of paralogs, which may explain why DDGS appears to be more closely related to one EEVS clade than the other. However, alternative routes of evolution and/or convergent evolution cannot be entirely ruled out. For example, another plausible scenario is that the ancestor of EEVS and DDGS underwent a gene duplication event, followed by those paralogs diverging into EEVS and DDGS enzymes. Whereas the evolutionary history of EEVS and DDGS remain uncertain, based on their unequal distributions across kingdoms of life (see Figures 2 and 8), it appears that both enzymes may have spread through multiple speciation and HGT events.
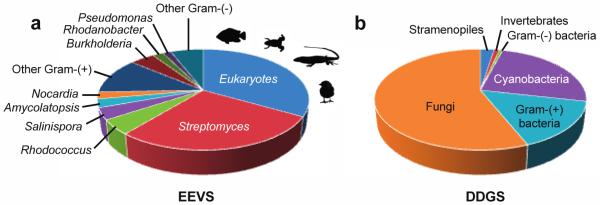
Figure 8.
Distribution of EEVS and DDGS in the sequenced microorganisms. With the exception of a fungus and seven stramenopiles, all eukaryotic EEVS are from vertebrates (fish, amphibians, reptiles, and birds).
Distribution of EEVS and DDGS genes in bacteria and eukaryotes
EEVS catalyzes the first committed step in the biosynthesis of bacterial-derived C7N-aminocyclitol natural products,2, 33-35 and was long thought to be present only in certain secondary metabolite-producing bacteria. However, the present study reveals a broad distribution of EEVS in various Gram-(+) and Gram-(−) bacteria, including gliding bacteria (myxobacteria) and cyanobacteria (Table S1). Some of them are clustered with other major biosynthetic enzymes such as terpene synthases, polyketide synthases, and non-ribosomal peptide synthetases, suggesting involvement in diverse natural products biosynthesis (data not shown). Strikingly, EEVS genes are also present in vertebrates, e.g., fish, amphibians, reptiles, and birds.17 These genes are always paired with a methyltransferase-oxidase gene and together they are responsible for the biosynthesis of the sunscreen compound gadusol.17 Interestingly, despite that many reports of putative EEVS genes in fungal genomes, based on our bioinformatics analysis EEVS is conspicuously absent in fungi. Except a putative EEVS in the yeast Saitoella complicate, we would reclassify all of the annotated fungal EEVS proteins as DDGSs (Table S2).
Microbial DDGS are mostly distributed in cyanobacteria, fungi, and Gram-(+) bacteria. However, we identified Gram-(−) bacteria of the genus Lewinella and Halomonas as containing the divergent DDGS protein (Table S3). Recent reports also suggest the presence of DDGS in some marine invertebrates and stramenopiles (Figure 8).36 In our analysis, the putative marine invertebrate DDGS groups with the divergent DDGS clade. However, the stramenopiles have representatives in both the main DDGS clade and the divergent DDGS clade.
Figure 8 also shows that EEVS and DDGS enzymes are distributed quite differently. For example, while DDGS is widely distributed among fungi, there is only a single fungus, S. complicate, with an EEVS (Table S1 and S2). The EEVS encoded by this fungus, clades with the vertebrate EEVS branch. Given that most fungi lack EEVS genes and the vertebrate EEVS appears to be gained through HGT, it seems reasonable to suggest that S. complicate also gained its EEVS gene through HGT.
Interestingly, the available genomes usually either contain an EEVS or a DDGS instead of having both. The few exceptions include Rhodococcus fascians (Gram-(+) bacteria), Chondrus crispus (alga), and Aureococcus anophagefferens (alga). It is not uncommon for organisms to have multiple SPCs, as many organisms have some combination of DHQS, DOIS, EVS, or aDHQS in addition to an EEVS or DDGS. This strong anti-correlation between the presence of EEVS and DDGS genes within the same organism is an oddity that warrants further investigation.
METHODS
Molecular Phylogenetic Analysis
Publically available amino acid sequences were obtained from the NCBI. Sequences were aligned using MUSCLE.37 Approximate maximum likelihood phylogenetic analysis was performed using FastTree 2.1.3 with a JTT+CAT model.38 MUSCLE and FastTree were performed on the Center for Genome and Biocomputing (Oregon State University) server. Sources of proteins for the analyses are listed on Tables S1–S4. Archaeopteryx was used to view and edit the phylogenetic tree.39 Amino acid sequences were analyzed and viewed using the software Geneious (Biomatters).
Expression, Purification, and Crystallization of AvDDGS
The protein was expressed and purified as previously described.8 For details of AvDDGS crystallization, see the Supporting Information.
X-ray Diffraction Data Collection
For diffraction data collection at –170 °C, crystals were briefly passed through a solution containing 30% (v/v) glycerol and then cryo-cooled by being plunged into liquid nitrogen. Data were collected from three crystals using λ = 0.976 Å and Δφ=1° steps at beamline 5.0.3 at the Advanced Light Source (Berkeley, CA). For details, see the Supporting Information. The atomic coordinates have been deposited in the Protein Data Bank (PDB entry 5PTR).
Structure Determination
The phase problem was initially solved by molecular replacement using MR Rosetta with default settings.40 The search model was ShEEVS (PDB entry 4P53), the closest structurally known homolog with 39% sequence identity, and resulted in a preliminary solution with R and Rfree values of 0.20 and 0.22 with 772 residues built (two chains in the asymmetric unit). All manual model building was done in Coot.41 Refinements were done using Phenix42 with TLS refinement and riding hydrogens. For details, see the Supporting Information.
AvDDGS and ShEEVS Mutagenesis and Characterization
For details, see the Supporting Information.
Cloning and Expression of gacC and staur_1386, Protein Purification, Enzyme Assay, and GC-MS Analysis
For details, see the Supporting Information.
Supplementary Material
Acknowledgements
This work was supported in part by grant GM112068 (to TM) from the National Institute of General Medical Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health (NIH).
Footnotes
The Supporting Information is available free of charge on the ACS Publication Website at DOI:…
Supporting methods, figures and tables (PDF and Microsoft Excel).
Notes
The authors declare no competing financial interest.
References
- (1).Wu X, Flatt PM, Schlorke O, Zeeck A, Dairi T, Mahmud T. A comparative analysis of the sugar phosphate cyclase superfamily involved in primary and secondary metabolism. ChemBioChem. 2007;8:239–248. doi: 10.1002/cbic.200600446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Mahmud T. Progress in aminocyclitol biosynthesis. Curr. Opin. Chem. Biol. 2009;13:161–170. doi: 10.1016/j.cbpa.2009.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Nango E, Kumasaka T, Hirayama T, Tanaka N, Eguchi T. Structure of 2-deoxy-scyllo-inosose synthase, a key enzyme in the biosynthesis of 2-deoxystreptamine-containing aminoglycoside antibiotics, in complex with a mechanism-based inhibitor and NAD+ Proteins. 2008;70:517–527. doi: 10.1002/prot.21526. [DOI] [PubMed] [Google Scholar]
- (4).Kean KM, Codding SJ, Asamizu S, Mahmud T, Karplus PA. Structure of a sedoheptulose 7-phosphate cyclase: ValA from Streptomyces hygroscopicus. Biochemistry. 2014;53:4250–4260. doi: 10.1021/bi5003508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Floss HG. Natural products derived from unusual variants of the shikimate pathway. Nat. Prod. Rep. 1997;14:433–452. doi: 10.1039/np9971400433. [DOI] [PubMed] [Google Scholar]
- (6).Yamauchi N, Kakinuma K. Biochemical studies on 2-deoxy-scyllo-inosose, an early intermediate in the biosynthesis of 2-deoxystreptamine. IV. A clue to the similarity of 2-deoxy-scyllo-inosose synthase to dehydroquinate synthase. J. Antibiot. 1993;46:1916–1918. doi: 10.7164/antibiotics.46.1916. [DOI] [PubMed] [Google Scholar]
- (7).Balskus EP, Walsh CT. The genetic and molecular basis for sunscreen biosynthesis in cyanobacteria. Science. 2010;329:1653–1656. doi: 10.1126/science.1193637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Asamizu S, Xie P, Brumsted CJ, Flatt PM, Mahmud T. Evolutionary divergence of sedoheptulose 7-phosphate cyclases leads to several distinct cyclic products. J. Am. Chem. Soc. 2012;134:12219–12229. doi: 10.1021/ja3041866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Asamizu S, Abugreen M, Mahmud T. Comparative Metabolomic Analysis of an Alternative Biosynthetic Pathway to Pseudosugars in Actinosynnema mirum DSM 43827. ChemBioChem. 2013;14:1548–1551. doi: 10.1002/cbic.201300384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Kim CG, Kirschning A, Bergon P, Zhou P, Su E, Sauerbrei B, Ning S, Ahn Y, Breuer M, Leistner E, Floss HG. Biosynthesis of 3-amino-5-hydroxybenzoic acid, the precursor of mC7N units in ansamycin antibiotics. J. Am. Chem. Soc. 1996;118:7486–7491. [Google Scholar]
- (11).Rascher A, Hu Z, Buchanan GO, Reid R, Hutchinson CR. Insights into the biosynthesis of the benzoquinone ansamycins geldanamycin and herbimycin, obtained by gene sequencing and disruption. Appl. Environ. Microbiol. 2005;71:4862–4871. doi: 10.1128/AEM.71.8.4862-4871.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Mao Y, Varoglu M, Sherman DH. Molecular characterization and analysis of the biosynthetic gene cluster for the antitumor antibiotic mitomycin C from Streptomyces lavendulae NRRL 2564. Chem. Biol. 1999;6:251–263. doi: 10.1016/S1074-5521(99)80040-4. [DOI] [PubMed] [Google Scholar]
- (13).Floss HG, Yu TW, Arakawa K. The biosynthesis of 3-amino-5-hydroxybenzoic acid (AHBA), the precursor of mC7N units in ansamycin and mitomycin antibiotics: a review. J. Antibiot. 2011;64:35–44. doi: 10.1038/ja.2010.139. [DOI] [PubMed] [Google Scholar]
- (14).Flatt PM, Mahmud T. Biosynthesis of aminocyclitol-aminoglycoside antibiotics and related compounds. Nat. Prod. Rep. 2007;24:358–392. doi: 10.1039/b603816f. [DOI] [PubMed] [Google Scholar]
- (15).Mahmud T, Tornus I, Egelkrout E, Wolf E, Uy C, Floss HG, Lee S. Biosynthetic studies on the alpha-glucosidase inhibitor acarbose in Actinoplanes sp.: 2-epi-5-epi-valiolone is the direct precursor of the valienamine moiety. J. Am. Chem. Soc. 1999;121:6973–6983. [Google Scholar]
- (16).Dong H, Mahmud T, Tornus I, Lee S, Floss HG. Biosynthesis of the validamycins: identification of intermediates in the biosynthesis of validamycin A by Streptomyces hygroscopicus var. limoneus. J. Am. Chem. Soc. 2001;123:2733–2742. doi: 10.1021/ja003643n. [DOI] [PubMed] [Google Scholar]
- (17).Osborn AR, Almabruk KH, Holzwarth G, Asamizu S, LaDu J, Kean KM, Karplus PA, Tanguay RL, Bakalinsky AT, Mahmud T. De novo synthesis of a sunscreen compound in vertebrates. Elife. 2015;4:e05919. doi: 10.7554/eLife.05919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Carpenter EP, Hawkins AR, Frost JW, Brown KA. Structure of dehydroquinate synthase reveals an active site capable of multistep catalysis. Nature. 1998;394:299–302. doi: 10.1038/28431. [DOI] [PubMed] [Google Scholar]
- (19).Stratmann A, Mahmud T, Lee S, Distler J, Floss HG, Piepersberg W. The AcbC protein from Actinoplanes species is a C7-cyclitol synthase related to 3-dehydroquinate synthases and is involved in the biosynthesis of the alpha-glucosidase inhibitor acarbose. J. Biol. Chem. 1999;274:10889–10896. doi: 10.1074/jbc.274.16.10889. [DOI] [PubMed] [Google Scholar]
- (20).Wu X, Flatt PM, Xu H, Mahmud T. Biosynthetic gene cluster of cetoniacytone A, an unusual aminocyclitol from the endosymbiotic Bacterium Actinomyces sp. Lu 9419. ChemBioChem. 2009;10:304–314. doi: 10.1002/cbic.200800527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Flatt PM, Wu X, Perry S, Mahmud T. Genetic insights into pyralomicin biosynthesis in Nonomuraea spiralis IMC A-0156. J. Nat. Prod. 2013;76:939–946. doi: 10.1021/np400159a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Choi WS, Wu X, Choeng YH, Mahmud T, Jeong BC, Lee SH, Chang YK, Kim CJ, Hong SK. Genetic organization of the putative salbostatin biosynthetic gene cluster including the 2-epi-5-epi-valiolone synthase gene in Streptomyces albus ATCC 21838. Appl. Microbiol. Biotechnol. 2008;80:637–645. doi: 10.1007/s00253-008-1591-2. [DOI] [PubMed] [Google Scholar]
- (23).Waditee-Sirisattha R, Kageyama H, Sopun W, Tanaka Y, Takabe T. Identification and upregulation of biosynthetic genes required for accumulation of Mycosporine-2-glycine under salt stress conditions in the halotolerant cyanobacterium Aphanothece halophytica. Appl. Environ. Microbiol. 2014;80:1763–1769. doi: 10.1128/AEM.03729-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Hirayama T, Kudo F, Huang Z, Eguchi T. Role of glutamate 243 in the active site of 2-deoxy-scyllo-inosose synthase from Bacillus circulans. Bioorg. Med. Chem. 2007;15:418–423. doi: 10.1016/j.bmc.2006.09.042. [DOI] [PubMed] [Google Scholar]
- (25).Huang S, Chen Z, Yan X, Yu T, Huang G, Yan Q, Pontarotti PA, Zhao H, Li J, Yang P, Wang R, Li R, Tao X, Deng T, Wang Y, Li G, Zhang Q, Zhou S, You L, Yuan S, Fu Y, Wu F, Dong M, Chen S, Xu A. Decelerated genome evolution in modern vertebrates revealed by analysis of multiple lancelet genomes. Nat. Commun. 2014;5:5896. doi: 10.1038/ncomms6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Rockser Y, Wehmeier UF. The gac-gene cluster for the production of acarbose from Streptomyces glaucescens GLA.O: identification, isolation and characterization. J. Biotechnol. 2009;140:114–123. doi: 10.1016/j.jbiotec.2008.10.016. [DOI] [PubMed] [Google Scholar]
- (27).Asamizu S, Yang J, Almabruk KH, Mahmud T. Pseudoglycosyltransferase catalyzes nonglycosidic C-N coupling in validamycin a biosynthesis. J. Am. Chem. Soc. 2011;133:12124–12135. doi: 10.1021/ja203574u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Abuelizz HA, Mahmud T. Distinct Substrate Specificity and Catalytic Activity of the Pseudoglycosyltransferase VldE. Chem. Biol. 2015;22:724–733. doi: 10.1016/j.chembiol.2015.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Cavalier MC, Yim YS, Asamizu S, Neau D, Almabruk KH, Mahmud T, Lee YH. Mechanistic insights into validoxylamine A 7′-phosphate synthesis by VldE using the structure of the entire product complex. PLoS One. 2012;7:e44934. doi: 10.1371/journal.pone.0044934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Yu Y, Bai L, Minagawa K, Jian X, Li L, Li J, Chen S, Cao E, Mahmud T, Floss HG, Zhou X, Deng Z. Gene cluster responsible for validamycin biosynthesis in Streptomyces hygroscopicus subsp. jinggangensis 5008. Appl. Environ. Microbiol. 2005;71:5066–5076. doi: 10.1128/AEM.71.9.5066-5076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Cheng K, Rong X, Huang Y. Widespread interspecies homologous recombination reveals reticulate evolution within the genus Streptomyces. Mol. Phylogenet. Evol. 2016;102:246–254. doi: 10.1016/j.ympev.2016.06.004. [DOI] [PubMed] [Google Scholar]
- (32).Pinto-Carbo M, Sieber S, Dessein S, Wicker T, Verstraete B, Gademann K, Eberl L, Carlier A. Evidence of horizontal gene transfer between obligate leaf nodule symbionts. ISME J. 2016;10:2092–2105. doi: 10.1038/ismej.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Mahmud T, Flatt PM, Wu X. Biosynthesis of unusual aminocyclitol-containing natural products. J. Nat. Prod. 2007;70:1384–1391. doi: 10.1021/np070210q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Sieber S, Carlier A, Neuburger M, Grabenweger G, Eberl L, Gademann K. Isolation and Total Synthesis of Kirkamide, an Aminocyclitol from an Obligate Leaf Nodule Symbiont. Angew. Chem. Int. Ed. Engl. 2015;54:7968–7970. doi: 10.1002/anie.201502696. [DOI] [PubMed] [Google Scholar]
- (35).Mahmud T. The C7N aminocyclitol family of natural products. Nat. Prod. Rep. 2003;20:137–166. doi: 10.1039/b205561a. [DOI] [PubMed] [Google Scholar]
- (36).Shinzato C, Shoguchi E, Kawashima T, Hamada M, Hisata K, Tanaka M, Fujie M, Fujiwara M, Koyanagi R, Ikuta T, Fujiyama A, Miller DJ, Satoh N. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature. 2011;476:320–323. doi: 10.1038/nature10249. [DOI] [PubMed] [Google Scholar]
- (37).Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Price MN, Dehal PS, Arkin AP. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Han MV, Zmasek CM. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics. 2009;10:356. doi: 10.1186/1471-2105-10-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Terwilliger TC, Dimaio F, Read RJ, Baker D, Bunkoczi G, Adams PD, Grosse-Kunstleve RW, Afonine PV, Echols N. phenix.mr_rosetta: molecular replacement and model rebuilding with Phenix and Rosetta. J. Struct. Funct. Genomics. 2012;13:81–90. doi: 10.1007/s10969-012-9129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.