Abstract
Combining phylogenetic and network methodologies has the potential to better inform targeted interventions to prevent and treat infectious diseases. This study reconstructed a molecular transmission network for people with recent hepatitis C virus (HCV) infection and modelled the impact of targeting directly acting antiviral (DAA) treatment for HCV in the network. Participants were selected from three Australian studies of recent HCV from 2004-2014. HCV sequence data (Core-E2) from participants at the time of recent HCV detection were analysed to infer a network by connecting pairs of sequences whose divergence was ≤0.03 substitutions/site. Logistic regression was used to identify factors associated with connectivity. Impact of targeting HCV DAAs at both HIV co-infected and random nodes was simulated (1 million replicates). Among 236 participants, 21% (n=49) were connected in the network. HCV/HIV co-infected participants (47%) were more likely to be connected compared to HCV mono-infected participants (16%) (OR 4.56; 95%CI; 2.13–9.74). Simulations targeting DAA HCV treatment to HCV/HIV co-infected individuals prevented 2.5 times more onward infections than providing DAAs to randomly selected individuals. Results demonstrate that genetic distance-based network analyses can be used to identify characteristics associated with HCV transmission, informing targeted prevention and treatment strategies.
Keywords: Hepatitis C virus, Human immunodeficiency virus, Modelling, Molecular epidemiology, Treatment as Prevention
Introduction
Communicable diseases such as HIV and hepatitis C virus (HCV) spread through contacts within injecting (1) and sexual networks (2). The structure of these networks (e.g. number of injecting or sexual partners) governs the spread of infection (3). Important features of social or injecting networks are often characterized through interviews and partner tracing. However, these techniques are laborious, can lead to bias (4), identified links may not represent direct transmission events, and may be less accurate when the infectious disease has a long period between transmission and disease state and a low transmission rate per contact (e.g. HIV (5-8) or HCV (9)).
HIV sequence data have been used to reconstruct molecular transmission networks (10), approximating the transmission network, and reflecting the transmission pathway of the virus between people. Molecular transmission networks represent powerful scientific and clinical tools to study networks through which communicable diseases are transmitted (11). Recently, HIV molecular transmission networks have been proposed to target antiretroviral therapy (12), track the spread of the epidemic (13, 14), model the possible effects on reduction of transmission for different HIV prevention trials (15) and inform real-time public health interventions (16).
A recent study demonstrated that phylogenetic clustering of recent HCV infection in Australia was associated with HIV/HCV co-infection and genotype 1a infection (17), suggesting that understanding the network through which the virus is transmitted is important for the successful implementation of treatment and prevention strategies. It has also been demonstrated that HCV phylogenetic clustering is associated with the injecting network in people who inject drugs (PWID) (18) and that epidemic contact structure can be inferred from phylogenetic trees (19). Recent work in HIV demonstrated the correspondence between communities observed in sexual contact networks and phylogenetic transmission clusters (20). These findings suggest that in the absence of injecting or sexual network data, the construction of a molecular transmission network using HCV sequence data could shed light on the network through which HCV is transmitted.
Modelling of the impact of Directly Acting Antiviral (DAA) therapy to treat HCV has been performed using the injecting network (21) to inform the implementation of treatment and prevention strategies, such as Treatment as Prevention (22). Modelling the impact of DAA therapy in a molecular transmission network using HCV sequences may further validate these strategies and provide insights into the effectiveness of Treatment as Prevention to reduce transmission of HCV among HIV-infected men who have sex with men (MSM).
Here, a partial molecular transmission network was reconstructed for recent HCV using the methodology previously developed for HIV sequence data. The inferred network was used to model the possible impact of treatment of HCV infection using DAAs on reduction of transmission of HCV in the network.
Materials & Methods
Study population and design
Data and specimens from participants in three Australian studies of recent HCV were used. The Australian Trial in Acute Hepatitis C (ATAHC) was a study of recent HCV recruiting participants between 2004 and 2007 (23). The Hepatitis C Incidence and Transmission Study - prison (HITS-p) was a study of PWID in prison that recruited participants from correctional centres between 2005 and 2014 (24). The Hepatitis C Incidence and Transmission Study — community (HITS-c) was a study of community-based PWID, that recruited participants between 2008 and 2013 (25).
For inclusion in this analysis, participants from these cohorts had to have acute or recent HCV defined by an initial positive anti-HCV antibody test and either (1) a negative anti-HCV antibody test within 2 years prior to the initial positive anti-HCV test or (2) acute clinical hepatitis (either jaundice or alanine aminotransferase [ALT] >400 IU/mL) within 12 months of the initial positive anti-HCV result. Participants also had to have a HCV RNA detectable plasma sample (first available sample following the detection of HCV selected). All participants provided written informed consent and protocols were approved by appropriate Human Research Ethics Committees.
The estimated date of infection for subjects who presented with acute clinical hepatitis was calculated as six weeks prior to onset of symptoms. For subjects identified by recent positive HCV antibody test with a negative test in the prior two years, the estimated date of infection was calculated as the midpoint between the first positive test and the last negative test. Among individuals HCV antibody-negative and HCV RNA-positive at the time of acute HCV detection, the estimated date of HCV infection was 4 weeks prior to diagnosis date
Transmission network reconstruction
HCV nucleotide sequences were obtained from the first available sample at the time of HCV detection. Sequences obtained spanned the HCV genome from Core to the beginning of Envelope-2 (Core-E2) nucleotides 347–1750 in H77 reference sequence (GenBank ascension no. NC_004102) and were amplified using a method previously described (26) (Supplementary Methods 1). Sequence curation and alignment was performed using ClustalX (27) and the final sequence fragment analysed was 1221 bp long following the removal of the hypervariable region one (HVR1) due to the extreme genetic variation seen between individuals in this region (26). Molecular transmission network inference was performed using freely available software HIV-TRACE (www.hivtrace.org; (28)) and adapted to the Core-E2 region of HCV by changing the distance threshold.
The transmission network was inferred based on the nucleotide genetic distance between HCV sequences from each participant. Two participants (nodes) were linked by an edge in the network whenever their Core-E2 sequences divergence was ≤0.03 substitutions/site (Tamura-Nei 93 genetic distance; (29)). As per previous studies, the analysis was repeated at 0.02 and 0.025 substitutions per site to (Supplementary Table 1) to characterise the most epidemiologically relevant cut off. The presence of a link between two participants does not imply direct transmission, since there may be unsampled members of the network, resulting in missing nodes. The presence of an edge indicates only an epidemiological relationship between the connected nodes. Clusters were assembled by connecting all nodes with shared edges, so that every individual in a cluster is connected to at least one other individual in the cluster.
Edges (links) were directed in the network based on estimated date of infection of participants. An outbound edge (arrow pointing away) would be assigned from one participant (A) to another (B), when the estimated date of infection of A was earlier than the estimated date of infection of B. This directionality does not indicate that person A infected person B, but rather that person B could not have infected person A (because person B was infected after person A). To account for potential uncertainty in the method used to calculate estimated date of infection, edges were not assigned any directionality if the estimated date of infection of two nodes was within 90 days of each other.
Network simulations
The impact of treatment of HCV with DAAs was simulated over the molecular transmission network, inferred using a 0.03 substitutions/site genetic distance threshold. We explored the effect of targeting DAA therapy at HIV co-infected nodes and DAA therapy provided to random nodes. For each replicate, a node was randomly selected from either the subset of HIV co-infected nodes or from the set of all nodes. If this node belonged to a cluster, a minimum spanning tree for the cluster was constructed using Wilson’s algorithm (30) for efficiently sampling from a uniform distribution of possible spanning trees. These trees were always rooted on the sequence with the oldest estimated date of infection. Because this cohort comprised predominantly acutely infected individuals, the spanning tree constructed for each replicate was taken to be the path of HCV transmission.
As infection progressed over the spanning tree, the HCV infection would be eliminated in the target node at a given efficacy (80%, 90%, 100% of the time). Nodes with eliminated infection could not infect their descendants in the spanning tree, propagating the effect of DAA therapy in a Treatment as Prevention framework. The directionality of edges in the network was crucial for these simulations, as this directionality dictated the potential number of infections that could be prevented for treating each node. For a treated node with only inbound edges, no potential infections would have been prevented. However for a treated node with outbound edges, any of their potential outbound connections were considered to have been potentially protected from infection. We counted the number of prevented onward transmission events. For each targeting scheme (HIV co-infected and random), we performed one million replicate simulations.
Importantly, the directionality of some edges made it sometimes impossible for infection to progress to all parts of a given cluster. This feature of the simulation strategy highlights the importance of unsampled nodes in these clusters and how they present an obstacle to propagation of treatment effects across a cluster. The effect of unsampled nodes will be equally present in both simulations of targeted DAA therapy to those with HIV/HCV co-infection and simulations of untargeted DAA therapy to random nodes. Further, if a targeted node was not clustered, prevention of onward infections is not possible under this framework.
Study outcomes
The primary study outcome was network connectivity, as defined by being connected to at least one other participant in the molecular transmission network and participants were defined as either unconnected or connected. Participants who were connected in the network were further categorised by the number of connections they had and how many of those connections were inbound or outbound. To investigate highly connected components of the network, participants were classified as highly connected if they had three or more outbound edges. The secondary outcome was the median number of potential onward transmission events that were prevented by each treatment strategy that was modelled in the network.
Statistical analyses
Unadjusted logistic regression analysis was used to identify factors associated with being connected in the network. All variables with P <0.20 in the unadjusted analysis were considered in the adjusted logistic regression model, using a backwards stepwise approach with factors sequentially eliminated according to the result of a likelihood ratio test. To account for potential unmeasured confounding introduced by the different cohort characteristics, adjusted logistic regression analysis was performed using mixed modelling, with a random intercept for cohort. For all analyses, statistically significant differences were assessed at P <0.05; P-values are two-sided. All analyses were performed using STATA software (version 12.1; StataCorp L.P., College Station, Texas, USA).
Results
Study population
In total, 293 participants were initially included in this study, however, a Core-E2 sequence was obtainable from 81% (n=236) of participants (ATAHC, n=121; HITS-p, n=92; and HITS-c=, n=23). HCV genotype (G) prevalence was: G1a: 38 % (n=90), G1b: 5% (n=12), G2a: 1% (n=4), G2b: 5% (n=11), G3a: 48% (n=114), G6a: 1% (n=2) and G6l 1% (n=3). The characteristics of those with obtainable HCV sequencing (n=236), and characteristics stratified by being connected in the network, are shown in Table 1. Overall, 41% (n=92) of participants were in prison at sample collection, all of these participants being from the HITS-p cohort. Of the 16% of participants with HIV infection (n=36), all were males from the ATAHC cohort and reported homosexual exposure as a risk factor for HIV acquisition. Of this group, 17% also reported recent (in the last 6 months) injecting drug use (n=6).
Table 1.
Characteristics of participants overall and according to being in the network and logistic regression of factors associated with being in the network at ≤0.03 substitutions/site cut off for participants with recently acquired HCV infection in Australia between 2004 and 2014.
|
Characteristic Total n (%) |
Overall (n=236) |
Not in network (n=187) |
In network (n=49) |
Connected in network Unadjusted |
||
|---|---|---|---|---|---|---|
| Odds ratio | 95% CI | P | ||||
| Age | ||||||
| ≥25 (vs. <25) | 67 (28%) | 52 (30%) | 15 (30%) | 1.07 | 0.54, 2.12 | 0.838 |
| ≤30 (vs. >30) | 101 (45%) | 79 (45%) | 22 (44%) | 1.00 | 0.53, 1.90 | 0.999 |
| ≤35 (vs. >35) | 77 (32%) | 57 (32%) | 20 (40%) | 1.46 | 0.77, 2.78 | 0.248 |
| Female sex (vs. male sex) | 70 (31%) | 57 (32%) | 13 (26%) | 0.74 | 0.36, 1.51 | 0.408 |
| HIV infection (vs. none) | 36 (16%) | 19 (11%) | 17 (34%) | 4.40 | 2.06, 9.36 | <0.001 |
| Injection drug use | ||||||
| Ever (vs. never) | 204 (90%) | 160 (91%) | 44 (88%) | 0.825 | 0.28, 2.39 | 0.723 |
| Recent ab (vs. none) | 130 (59%) | 106 (60%) | 24 (48%) | 0.60 | 0.32, 1.15 | 0.125 |
| High school or higher (vs. less than high school) |
138 (61%) | 103 (59%) | 35 (70%) | 1.86 | 0.92, 3.75 | 0.085 |
| Unstable housing c (vs. stable) Incarceration |
101 (45%) | 82 (47%) | 19 (38%) | 0.67 | 0.35, 1.29 | 0.232 |
| Ever (vs. never) | 114 (50%) | 94 (53%) | 20 (40%) | 0.57 | 0.30, 1.09 | 0.089 |
| Currently b (vs. not) | 92 (41%) | 77 (44%) | 15 (30%) | 0.57 | 0.29, 1.12 | 0.100 |
| Study | ||||||
| ATAHC (vs. other) | 121 (51%) | 92 (49%) | 29 (59%) | 1.38 | 0.73, 2.60 | 0.323 |
| HITS-p (vs. other) | 92 (39%) | 78 (42%) | 14 (29%) | 0.57 | 0.28, 1.15 | 0.118 |
| HITS-c (vs. other) | 23 (10%) | 17 (9%) | 6 (12%) | 1.12 | 0.40, 3.11 | 0.828 |
| HCV Gt 1a (vs. other) | 90 (38%) | 65 (35%) | 25 (51%) | 1.96 | 1.04, 3.69 | 0.039 |
Percentages indicate column percentages
Within last 3-6 months prior to sample date.
Among total population
Defined as living in prison, a shelter or hostel, or having no fixed address in the last 6 months
Molecular transmission network composition
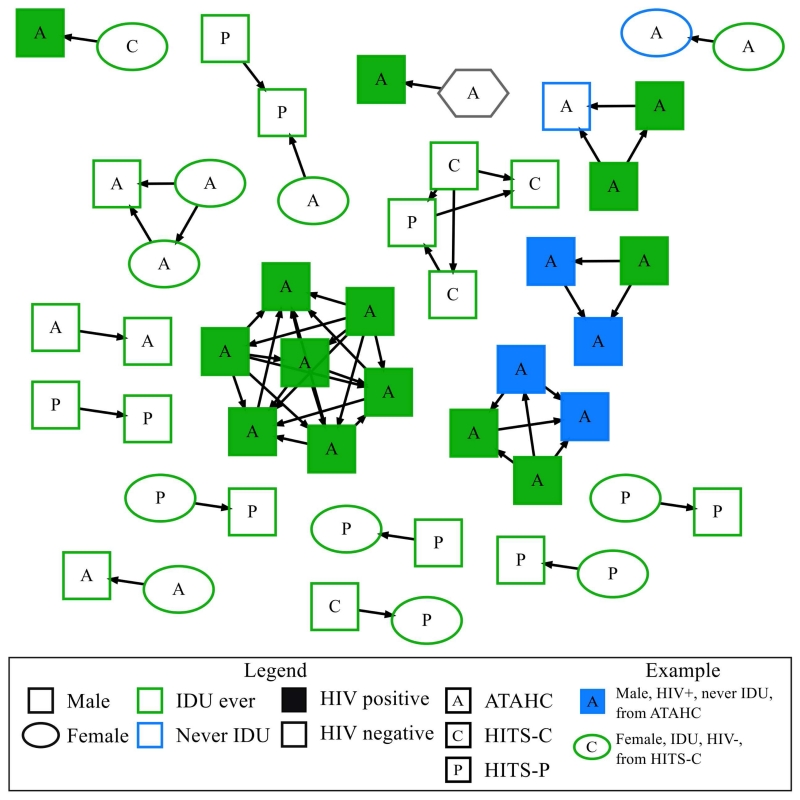
A molecular transmission network was constructed (Figure 1). Overall, 21% of participants (n=49) were connected to at least one other individual in the network. Of those connected in the network, 45% (n=22) were connected to only one other individual, 25% (n=12) were connected to two other individuals and 12% (n=13) were connected to three or more individuals. The largest number of connections observed for one individual in the network was six connections. The number of outbound edges observed for individuals in the network ranged from zero to six. In the network, 70% (n=34) of participants had no outbound edges, 22% (n=11) had one outbound edge and 10% (n=5) were classified as highly connected, having three or more outbound edges. Of the highly connected participants with 3 or more outbound edges, 80% (n=4) were HIV co-infected. There were 18 individual clusters observed in the network ranging in size from two to seven nodes. Of the clusters observed, 61% (n=11) were made of up two connected nodes, 22% (n=4) were made up of three connected nodes, 11% (n=2) were made up of four connected nodes and one cluster of seven connected nodes was observed. The participant characteristics that made up these clusters display both discrete features and mixed characteristics. For example, three discrete clusters were observed that contained only HCV/HIV co-infected males and three clusters with mixed characteristics were observed that linked males and females from prison with individuals outside prison (Figure 1). Six individuals with more than three outbound edges were observed and 80% were HIV co-infected (n=4).
Figure 1.
Molecular transmission network of recently acquired HCV infection in Australia (excluding unconnected individuals) constructed with HCV nucleotide sequences from a 1104 bp region encompassing the Core to E2 (minus HVR1) region. An edge was created between any pair of sequences whose divergence was ≤0.03 substitutions/site. Only clustered individuals (nodes) within the network are shown (28%). Oval shaped nodes denote female sex, square shaped nodes denote male sex, and hexagonal shaped nodes denote unknown gender. Nodes are coloured according to participant’s history of injecting drug use (IDU) (blue, never IDU; green, ever IDU). Filled nodes denote HCV/HIV-coinfection; unfilled nodes denote HCV mono infection. The study of origin of participants is denoted by the initials P (HITS-P, prison), C (HITS-C, community) & A (ATAHC) in the node. The direction of edges is determined by estimated date of infection, with an arrow pointing to the most recent estimated date of infection. A threshold was placed on direction of edges where any pair of nodes that had estimated date of infection within 90 days of each other, the edge remained undirected.
Factors associated with network connectivity
Several factors were considered for their association with various aspects of connectivity in the network (Table 1). In adjusted logistic regression analysis, after adjusting for participant cohort, the only factor that remained independently associated with being connected in the network was HIV/HCV co-infection (Table 1). In total, 47% (17/36) of HCV/HIV co-infected participants were connected in the network compared with 16% (32/200) with HCV alone [Adjusted Odds Ratio (AOR) 4.56; 95% Confidence Interval (CI) 2.13–9.74]. In analyses stratified by HIV infection status, there were no factors significantly associated with connection in the network (Supplementary Table 2). Unadjusted logistic regression analysis was carried out to assess factors associated with having 3 or more outbound edges (Supplementary Table 3). People with HIV/HCV co-infection were more likely to have 3 or more outbound edges (OR 5.64; 95% CI 1.09, 29.13).
Modelling of impact of targeting treatment in the network
Simulations targeting DAA therapy (with 90% efficacy) to HCV/HIV co-infected individuals prevented 2.5 times more onward infections than simulations providing DAA therapy to the same number of random individuals (Table 2). By curing one HIV co-infected node, an average of 0.35 follow-on infections were prevented, compared with 0.14 follow-on infections prevented by curing a random node. Varying the efficacy of DAA therapy (from 80% to 100%) did not have a substantial impact on the observed findings (Table 2).
Table 2.
Effect of DAA therapy targeted at HIV-coinfected individuals versus random individuals.
| Treatment efficacy |
HIV coinfection- targeted1 |
Randomly targeted1 |
Prevention yield improvement2 |
|---|---|---|---|
| 80% | 0.308 | 0.122 | 2.525 |
| 90% | 0.346 | 0.137 | 2.530 |
| 100% | 0.384 | 0.153 | 2.514 |
Median number of prevented infections per targeted individual
Ratio of median number of prevented infections between HIV-coinfection targeted and randomly delivered DAA therapy
Discussion
This study characterised a molecular transmission network for participants from three cohorts of people with recent HCV in Australia between 2004 and 2014. Overall, 21% of participants were connected in the network, and HIV infection was independently associated with network connectivity. It was found that 2.5 times more onward infections would be prevented in this network by directing DAA therapy to people with HIV/HCV co-infection, compared to people treated at random. This study demonstrates that computationally undemanding network methods incorporating phylogenetic data can be used to identify characteristics associated with higher connectivity and potential transmission risk, with potential utility towards better understanding transmission in other settings and/or emerging HCV epidemics.
This study is the first to construct a molecular transmission network for HCV to identify putative transmission. However, comparisons may be made to other phylogenetic studies of HCV, where the proportion with phylogenetic clustering have been found to vary from 22% (17) to >30% (18, 31, 32). These studies differ in the methods and cut offs used to classify clustering or putative transmission, which may account for the variation in the prevalence of clustering observed. The study populations also differed (e.g. the current study only included those with recent infection) which may have accounted for the differences in observed results.
HIV/HCV co-infection was independently associated with having a putative transmission link and being connected in the transmission network. In this study population, HIV infection was present exclusively in homosexual males. However, 17% of individuals with HIV/HCV co-infection also reported recent injecting drug use, indicating the co-existence of both injecting and sexual risk behaviours in components of the network. Previous studies have shown the association between phylogenetic clustering of HCV infection among HIV positive MSM (17, 31, 33, 34). This study demonstrates that acute to acute transmission of HCV occurs among HIV positive MSM through a distinct network, suggesting the need for tailored interventions to reduce transmission of HCV among this group.
This study further demonstrated the complex patterns of transmission of recently acquired HCV infection. Several putative transmission links were observed linking participants in prison and in the community who had histories of recent injecting drug use. Evidence of transmission of HCV in prison has previously been demonstrated (24, 35), however our results indicate that transmission of HCV in prison should not be considered as a discrete network, given the observed connections between PWID participants in prison and in the community. Due to the inclusion of only newly acquired infections in this study, it is likely that many connections among PWID both in prison and in the community with chronic HCV infection may have been excluded. Therefore the degree of connectivity among PWID may be under represented in this study.
Recent meta analyses have demonstrated that HCV prevalence among HIV co-infected MSM is continuing to rise (36). In one study modelling HCV transmission among HIV/HCV co-infected MSM in the UK, 94% of infections were attributable to high-risk individuals comprising only 7% of the population (37). To investigate the presence of high-risk individuals in this study, highly connected components of the network were identified. Participants were classified as highly connected if they had three or more outbound edges. While 70% of participants in the network had no outbound edges at all, several individuals did exhibit a high degree of connectivity. Interestingly, of the participants who were classified as highly connected (having three or more outbound edges), 80% were HIV co-infected (n=4). HIV/HCV co-infection was also associated with having three or more outbound edges. This suggests that the HIV/HCV network is more highly connected than those with HCV mono-infection, highlighting the importance of identifying and targeting treatment and prevention interventions toward this high transmission risk group.
Given the finding that HIV/HCV co-infection may be associated with a greater potential for HCV transmission among people with recent HCV, a strategy to reduce potential transmission among this group was investigated. This study modelled targeting DAA therapy to people with HIV/HCV co-infection compared to randomly treating people in the molecular transmission network. Given limitations on the network, such as missing potential transmission links, this modelling strategy took a comparative approach. It was found that directing DAA therapy to people with HIV/HCV co-infection prevented 2.5 times more onward infections compared to randomly treating people in the network. By curing one HIV co-infected node, an average of 0.35 follow-on HCV infections were prevented, compared with 0.14 follow-on infections prevented by curing a random node. To prevent transmission of one infection, targeted treatment to only three people with HIV/HCV co-infection would be required, as compared to seven people treated at random. Recent studies modelling the impact of DAA therapy among key populations have all demonstrated that the prevalence of chronic HCV could be reduced with only modest levels of treatment per annum (38, 39). Another recent study modelling the impact of DAA therapy on reduction of transmission of HCV among HIV co-infected MSM in the UK suggested that treatment could reduce rates of HCV transmission among this group (37) but that rates of treatment would need to be high. This study further supports the finding that DAA therapy could reduce transmission of HCV among people with HIV/HCV co-infection.
This novel study demonstrates that sequence data from people with recent HCV infection can be used to characterise highly connected transmission networks. These transmission networks are highly disease relevant, as they reflect actual transmission, not just social connections or injecting behaviour. They can therefore be used to model the impact of different treatment interventions on transmission.
However, there are several limitations to this study. This study only included people with recent HCV in Australia. Therefore, these findings may not be generalizable to chronic HCV or epidemics in other countries. These findings may also under represent the effect of targeting treatment to prevent transmission, as there is likely to be potential transmission links in the network missing due to limited sampling. Further, the majority of transmissions observed in this network occurred in those with recent HCV/HIV co-infection, who only represent a small proportion of the total HCV burden in Australia. With an estimated 230,000 individuals with chronic HCV infection in Australia (40), and only an estimated 2,500 of those living with HIV/HCV co-infection (40), the population level impact of targeting DAA therapy to people with HIV/HCV co-infection is likely to be small. However, the impact of targeted treatment may vary according to the network structure within a population. The impact of targeted treatment within HCV/HIV co-infected individuals may be further limited due to the possible impact of re-infection. As this network is evidently highly connected, this creates the possibility of elevated risk of reinfection. With sufficiently high coverage of targeted treatment, this risk could be reduced. Further, it is possible that there is some bias towards a higher proportion being connected in the network among people with HIV, than would be expected in the general population with acute HCV infection, given that recruitment was somewhat limited to geographical areas with higher cases of HIV/HCV co-infection (e.g. Sydney and Melbourne) and people with HIV/HCV co-infection make up a much smaller proportion of those with HCV infection, compared to those with HCV mono-infection.
Molecular transmission network based studies can provide useful knowledge and insights into HCV transmission where injecting or sexual network data is not available or not feasible to obtain, therefore this methodology could be applied in many other settings to understand and prevent transmission by identifying characteristics of groups with higher risk of transmission. This study supported previous findings that among people with acute or recent HCV infection in Australia, those with HIV/HCV co-infection were associated with a higher transmission risk (17, 33). The data obtained from molecular transmission network studies can be used to model the impact of different treatment and prevention strategies. This study demonstrated that targeted use of DAA therapy may reduce network transmission more efficiently compared to random use of therapy. Targeted interventions of DAA HCV therapy to networks of people with HCV infection may be useful to prevent new infections in Treatment as Prevention strategies, especially among people with HIV/HCV co-infection.
Supplementary Material
Acknowledgements and Disclosures
The cooperation of study staff and the participants in the HITS cohorts and the ATAHC study is gratefully acknowledged.
Statement of Interests
Dr. Grebely is a consultant/advisor and has received research grants from AbbVie, Bristol Myers Squibb, Gilead Sciences, Merck. Dr. Dore is a consultant/advisor and has received research grants from Abbvie, Bristol Myers Squibb, Gilead, Merck, Janssen and Roche. Dr. Hellard has received research grants from Gilead Sciences, AbbVie and BMS. Dr. Wertheim is a consultant for the United States Centers for Disease Control and Prevention.
This work was supported by the United States National Institutes of Health [R01 DA 15999-01] and the Australian Government Department of Health. The views expressed in this publication do not necessarily represent the position of the United States National Institutes of Health or the Australian Government. JOW is supported by a Career Development Award from the United States National Institutes of Health (K01AI110181). GD and AL are supported by an National Health and Medical Research Council (NHMRC) Program Grant (APP1053206). JG is supported by a NHMRC Career Development Fellowship (APP1035383), GD is supported by a NHMRC Practitioner Research Fellowship (APP455355), MH is supported by a NHMRC Principal Research Fellowship (APP1062877), AL is supported by a NHMRC Practitioner Research Fellowship (APP1043067), GM is supported by a NHMRC Career Development Fellowship (APP1051859) and LM is supported by a NHMRC Senior Research Fellowship (APP1060443).
References
- 1.Hellard M, Rolls D, Davis RS, Robins G, Pattison P, Higgs P, et al. The Impact of Injecting Networks on Hepatitis C Transmission and Treatment in People Who Inject Drugs. Journal of hepatology. 2013;58:S23–S4. doi: 10.1002/hep.27403. [DOI] [PubMed] [Google Scholar]
- 2.Anderson RM, Gupta S, Ng W. The significance of sexual partner contact networks for the transmission dynamics of HIV. Journal of acquired immune deficiency syndromes. 1990;3(4):417–29. [PubMed] [Google Scholar]
- 3.Robinson K, Cohen T, Colijn C. The dynamics of sexual contact networks: effects on disease spread and control. Theoretical population biology. 2012;81(2):89–96. doi: 10.1016/j.tpb.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biernacki P, Waldorf D. Snowball Sampling - Problems and Techniques of Chain Referral Sampling. Sociol Method Res. 1981;10(2):141–63. [Google Scholar]
- 5.Kaplan EH, Heimer R. A Model-Based Estimate of HIV Infectivity via Needle Sharing. JAIDS Journal of Acquired Immune Deficiency Syndromes. 1992;5(11):1116–8. [PubMed] [Google Scholar]
- 6.Hudgens MG, Longini IM, Vanichseni S, Hu DJ, Kitayaporn D, Mock PA, et al. Subtype-specific Transmission Probabilities for Human Immunodeficiency Virus Type 1 among Injecting Drug Users in Bangkok, Thailand. American Journal of Epidemiology. 2002;155(2):159–68. doi: 10.1093/aje/155.2.159. [DOI] [PubMed] [Google Scholar]
- 7.Henderson DK, Fahey BJ, Willy M, Schmitt JM, Carey K, Koziol DE, et al. Risk for Occupational Transmission of Human Immunodeficiency Virus Type 1 (HIV-1) Associated with Clinical ExposuresA Prospective Evaluation. Annals of Internal Medicine. 1990;113(10):740–6. doi: 10.7326/0003-4819-113-10-740. [DOI] [PubMed] [Google Scholar]
- 8.Patel P, Borkowf CB, Brooks JT, Lasry A, Lansky A, Mermin J. Estimating per-act HIV transmission risk: a systematic review. Aids. 2014;28(10):1509–19. doi: 10.1097/QAD.0000000000000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moloughney BW. Transmission and postexposure management of bloodborne virus infections in the health care setting: Where are we now? Can Med Assoc J. 2001;165(4):445–51. [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis F, Hughes GJ, Rambaut A, Pozniak A, Leigh Brown AJ. Episodic sexual transmission of HIV revealed by molecular phylodynamics. PLoS medicine. 2008;5(3):e50. doi: 10.1371/journal.pmed.0050050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mehta SR, Wertheim JO, Brouwer KC, Wagner KD, Chaillon A, Strathdee S, et al. HIV Transmission Networks in the San Diego-Tijuana Border Region. EBioMedicine. 2015;2(10):1456–63. doi: 10.1016/j.ebiom.2015.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Little SJ, Kosakovsky Pond SL, Anderson CM, Young JA, Wertheim JO, Mehta SR, et al. Using HIV networks to inform real time prevention interventions. PloS one. 2014;9(6):e98443. doi: 10.1371/journal.pone.0098443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oster AM, Wertheim JO, Hernandez AL, Ocfemia MC, Saduvala N, Hall HI. Using Molecular HIV Surveillance Data to Understand Transmission Between Subpopulations in the United States. Journal of acquired immune deficiency syndromes. 2015;70(4):444–51. doi: 10.1097/QAI.0000000000000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wertheim JO, Oster AM, Hernandez AL, Saduvala N, Banez Ocfemia MC, Hall HI. The International Dimension of the U.S. HIV Transmission Network and Onward Transmission of HIV Recently Imported into the United States. AIDS Res Hum Retroviruses. 2016 doi: 10.1089/aid.2015.0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wertheim JO, Kosakovsky Pond SL, Little SJ, De Gruttola V. Using HIV transmission networks to investigate community effects in HIV prevention trials. PloS one. 2011;6(11):e27775. doi: 10.1371/journal.pone.0027775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poon AF, Gustafson R, Daly P, Zerr L, Demlow SE, Wong J, et al. Near real-time monitoring of HIV transmission hotspots from routine HIV genotyping: an implementation case study. Lancet HIV. 2016;3(5):e231–8. doi: 10.1016/S2352-3018(16)00046-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartlett SR, Jacka B, Bull RA, Luciani F, Matthews GV, Lamoury FM, et al. HIV infection and hepatitis C virus genotype 1a are associated with phylogenetic clustering among people with recently acquired hepatitis C virus infection. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2016;37:252–8. doi: 10.1016/j.meegid.2015.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sacks-Davis R, Daraganova G, Aitken C, Higgs P, Tracy L, Bowden S, et al. Hepatitis C virus phylogenetic clustering is associated with the social-injecting network in a cohort of people who inject drugs. PloS one. 2012;7(10):e47335. doi: 10.1371/journal.pone.0047335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leventhal GE, Kouyos R, Stadler T, Wyl V, Yerly S, Boni J, et al. Inferring epidemic contact structure from phylogenetic trees. PLoS computational biology. 2012;8(3):e1002413. doi: 10.1371/journal.pcbi.1002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villandre L, Stephens DA, Labbe A, Gunthard HF, Kouyos R, Stadler T, et al. Assessment of Overlap of Phylogenetic Transmission Clusters and Communities in Simple Sexual Contact Networks: Applications to HIV-1. PloS one. 2016;11(2) doi: 10.1371/journal.pone.0148459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rolls DA, Wang P, Jenkinson R, Pattison PE, Robins GL, Sacks-Davis R, et al. Modelling a disease-relevant contact network of people who inject drugs. Soc Networks. 2013;35(4):699–710. [Google Scholar]
- 22.Rolls DA, Sacks-Davis R, Jenkinson R, McBryde E, Pattison P, Robins G, et al. Hepatitis C Transmission and Treatment in Contact Networks of People Who Inject Drugs. PloS one. 2013;8(11) doi: 10.1371/journal.pone.0078286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dore GJ, Hellard M, Matthews GV, Grebely J, Haber PS, Petoumenos K, et al. Effective treatment of injecting drug users with recently acquired hepatitis C virus infection. Gastroenterology. 2010;138(1):123–35. e1–2. doi: 10.1053/j.gastro.2009.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teutsch S, Luciani F, Scheuer N, McCredie L, Hosseiny P, Rawlinson W, et al. Incidence of primary hepatitis C infection and risk factors for transmission in an Australian prisoner cohort. Bmc Public Health. 2010;10:633. doi: 10.1186/1471-2458-10-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White B, Dore GJ, Lloyd AR, Rawlinson WD, Maher L. Opioid substitution therapy protects against hepatitis C virus acquisition in people who inject drugs: the HITS-c study. Med J Australia. 2014;201(6):326–9. doi: 10.5694/mja13.00153. [DOI] [PubMed] [Google Scholar]
- 26.Lamoury FM, Jacka B, Bartlett S, Bull RA, Wong A, Amin J, et al. The Influence of Hepatitis C Virus Genetic Region on Phylogenetic Clustering Analysis. PloS one. 2015;10(7):e0131437. doi: 10.1371/journal.pone.0131437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 28.Wertheim JO, Brown AJL, Hepler NL, Mehta SR, Richman DD, Smith DM, et al. The Global Transmission Network of HIV-1. J Infect Dis. 2014;209(2):304–13. doi: 10.1093/infdis/jit524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamura K, Nei M. Estimation of the Number of Nucleotide Substitutions in the Control Region of Mitochondrial-DNA in Humans and Chimpanzees. Molecular biology and evolution. 1993;10(3):512–26. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 30.Wilson DB. Dimension of the loop-erased random walk in three dimensions. Phys Rev E Stat Nonlin Soft Matter Phys. 2010;82(6 Pt 1):062102. doi: 10.1103/PhysRevE.82.062102. [DOI] [PubMed] [Google Scholar]
- 31.Urbanus AT, van de Laar TJ, Stolte IG, Schinkel J, Heijman T, Coutinho RA, et al. Hepatitis C virus infections among HIV-infected men who have sex with men: an expanding epidemic. Aids. 2009;23(12):F1–F7. doi: 10.1097/QAD.0b013e32832e5631. [DOI] [PubMed] [Google Scholar]
- 32.Aitken CK, McCaw RF, Bowden DS, Tracy SL, Kelsall JG, Higgs PG, et al. Molecular epidemiology of hepatitis C virus in a social network of injection drug users. J Infect Dis. 2004;190(9):1586–95. doi: 10.1086/424678. [DOI] [PubMed] [Google Scholar]
- 33.Matthews GV, Pham ST, Hellard M, Grebely J, Zhang L, Oon A, et al. Patterns and characteristics of hepatitis C transmission clusters among HIV-positive and HIV-negative individuals in the Australian trial in acute hepatitis C. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2011;52(6):803–11. doi: 10.1093/cid/ciq200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bradshaw D, Jacka B, Sacks-Davis R, Lamoury F, Applegate T, Dore G, et al. A novel method comparing sexual networks with the HCV phylogeny in HIV-positive MSM with acute HCV infection identifies two potential intervention targets for permucosally transmitted HCV in Australia. Hiv Med. 2014;15:136. [Google Scholar]
- 35.Bretana NA, Boelen L, Bull R, Teutsch S, White PA, Lloyd AR, et al. Transmission of Hepatitis C Virus among Prisoners, Australia, 2005-2012. Emerg Infect Dis. 2015;21(5):765–74. doi: 10.3201/eid2105.141832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jordan AE, Perlman DC, Neurer J, Smith DJ, Des Jarlais DC, Hagan H. Prevalence of hepatitis C virus infection among HIV+ men who have sex with men: a systematic review and meta-analysis. Int J STD AIDS. 2016 doi: 10.1177/0956462416630910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin NK, Thornton A, Hickman M, Sabin C, Nelson M, Cooke GS, et al. Can Hepatitis C Virus (HCV) Direct-Acting Antiviral Treatment as Prevention Reverse the HCV Epidemic Among Men Who Have Sex With Men in the United Kingdom? Epidemiological and Modeling Insights. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2016;62(9):1072–80. doi: 10.1093/cid/ciw075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin NK, Vickerman P, Grebely J, Hellard M, Hutchinson SJ, Lima VD, et al. Hepatitis C virus treatment for prevention among people who inject drugs: Modeling treatment scale-up in the age of direct-acting antivirals. Hepatology. 2013;58(5):1598–609. doi: 10.1002/hep.26431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin NK, Vickerman P, Dore GJ, Hickman M. The hepatitis C virus epidemics in key populations (including people who inject drugs, prisoners and MSM): the use of direct-acting antivirals as treatment for prevention. Current opinion in HIV and AIDS. 2015;10(5):374–80. doi: 10.1097/COH.0000000000000179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.The Kirby Institute . HIV, viral hepatitis and sexually transmissible infections in Australia Annual Surveillance Report. The Kirby Institute; UNSW Australia, Sydney NSW 20522015: 2015. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.