Abstract
Introduction
Many COL4A5 splice region variants have been described in patients with X-linked Alport syndrome, but few have been confirmed by functional analysis to actually cause defective splicing. We sought to demonstrate that a novel COL4A5 splice region variant in a family with Alport syndrome is pathogenic using functional studies. We also describe an alternative method of diagnosis.
Methods
We analyzed targeted next-generation sequencing results of an individual with Alport syndrome and confirmed results by Sanger sequencing in family members. A splicing reporter minigene assay was used to examine the variant’s effect on splicing in transfected cells. Plucked hair follicles from patients and controls were examined for collagen IV proteins using immunofluorescence microscopy.
Results
A novel splice region mutation in COL4A5, c.1780-6T>G, was identified and segregated with disease in this family. This variant caused frequent skipping of exon 25, resulting in a frameshift and truncation of collagen α5(IV) protein. We also developed and validated a new approach to characterize the expression of collagen α5(IV) protein in the basement membranes of plucked hair follicles. We demonstrated reduced collagen α5(IV) protein in affected male and female individuals in this family, supporting frequent failure of normal splicing.
Conclusions
Differing normal to abnormal transcript ratios in affected individuals carrying splice region variants may contribute to variable disease severity observed in Alport families. Examination of plucked hair follicles in suspected X-linked Alport syndrome patients may offer a less invasive alternative method of diagnosis and serve as a pathogenicity test for COL4A5 variants of uncertain significance.
Keywords: Alport Syndrome, COL4A5, Splice site mutation, mmunofluorescence, hair follicle
Introduction
Alport syndrome (AS) is a hereditary disease with a classical clinical presentation of renal failure, hearing loss, and ocular manifestations [1, 2]. The original descriptions of AS were in males who inherited disease in an X-linked Mendelian pattern, but AS can also be inherited in an autosomal fashion. The phenotype can vary in severity and age of presentation, both between and within families [3–6]. Early large-scale genetic studies of AS suggested a correlation between mutation type and phenotype [7, 8]. Hundreds of variants in the AS genes (COL4A3, COL4A4 and COL4A5) have now been described, and approximately 13 – 16 % are splice site mutations [9]. However, the functional consequences of many splice region variants in AS patients have not been investigated. Furthermore, the correlation between genotype and phenotype at the molecular level has not been investigated for the vast majority of variants reported as pathogenic.
This report describes a novel splice region variant upstream of COL4A5 exon 25 in a family with both males and females affected. We examined the consequences of this variant on exon 25 splicing using in silico and in vitro approaches. We also tested a novel non-invasive method to characterize the variable deposition of collagen α5(IV) protein in the basement membranes of plucked hair follicles from this family by simple immunofluorescence staining. This method could be generally applicable to help diagnose X-linked Alport syndrome.
Materials and methods
Patients and specimens
Patient recruitment and specimen collection and processing were performed by the Washington University Kidney Translational Research Core. Human studies were conducted according to protocols approved by the Washington University Human Research Protection Office. Informed consent was obtained from all individual participants included in the study.
Clinical data
A diagnosis of Alport Syndrome was made by kidney and/or skin biopsy in at least one individual in each generation of this kindred. All affected family members had documented persistent hematuria. Clinical histories and data were obtained from referring physicians after all documents were deidentified. Biopsies were clinically indicated and were performed by the treating physicians.
Custom targeted next-generation and direct Sanger sequencing
Targeted next-generation sequencing of a customized kidney disease gene panel was performed under IRB approval (Schulman IRB, PI: C. Larsen, Nephropath/Arkana Laboratories, Little Rock, AR) as a research test for one affected family member. This assay used next-generation sequencing to analyze all coding exons and at least 10 bp of flanking intronic sequence of a panel of 301 genes to a minimum depth of X15 [10, 11]. Target enrichment was performed using oligonucleotide-based targeted capture (Agilent Rapid Capture Enrichment) of whole-genome shotgun sequencing libraries. Sequencing was performed on the Illumina MiSeq using a paired-end, 150 base-pair configuration. Analysis and interpretation was performed using DNAstar software. All analyses were based on the human reference sequence UCSC build hg19 (NCBI build 37.2).
Sanger sequencing was performed on all family members from whom DNA was available. Primers flanking the splice region variant were used for PCR amplification and for forward and reverse sequencing. The primers were: COL4A5_SplMutG_Forward primer, 5′-TGGCTATATCCTTTCCCCAGT-3′ and COL4A5_SplMutG_Reverse primer, 5′-GCCACACCTTGTATGCCTTT-3′. Genomic DNA was amplified using the following temperature cycling protocol: 98°C for 3 min, [95°C for 1 min, 60°C for 45 sec, 72°C for 2 min] × 30 cycles, [95°C for 1 min, 55°C for 45 sec, 72°C for 2 min] × 30 cycles, 72°C for 10 mins then 4°C.
Splicing analysis
Firstly, we used publicly available in silico analysis tools to predict the effects of the c.1780-6T>G variant on COL4A5 exon 25 splicing. These tools included Human Splicing Finder [http://www.umd.be/HSF3/HSF.html] [12], MaxEntScan::score3ss [http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq_acc.html] [13], and NetGene2 Server [http://www.cbs.dtu.dk/services/NetGene2/] [14,15].
Secondly, human COL4A5 exon 25 coding sequence plus 200–300 base-pairs of flanking intronic sequences were synthesized and inserted into the pCAS2.1 splicing vector by GenScript (Piscataway, NJ) using BamHI and MluI restriction sites, as described previously [16–18]. One vector contained the reference sequence (WT), and one contained the c.1780-6T>G variant (VAR).
HEK293T cells were grown to 60 % confluence on 12-well plates and transfected in duplicate with either: no vector, empty pCAS2.1 vector, WT vector, VAR vector, or both WT and VAR vectors (2 ug total plasmid DNA per sample) using Lipofectamine 2000 reagent (Invitrogen). Cells were harvested 15–20 hours later, and RNA was extracted from each sample using microcolumn kits™ with a DNase step (Qiagen). 1.5–2.0 ug of RNA were used to make cDNA using Multiscribe RT polymerase per protocol (AB systems). pCAS2.1 specific primers (pCAS-KO1-Forward primer: 5′-TGACGTCGCCGCCCATCAC-3′ and pCAS-2-Reverse primer: 5′-ATTGGTTGTTGAGTTGGTTGTC-3′) were used to amplify cDNA to determine the splicing patterns. The amplicon should include COL4A5 exon 25 sequence if splicing function remained normal. To amplify cDNA we used KOD Xtreme™ Hot Start DNA polymerase (per protocol), using three-step PCR cycling with a Tm of 53°C for 15 seconds per cycle for a total of 35 cycles.
Splicing patterns were determined by electrophoresis of PCR products on 1 % agarose gels. Bands of interest were purified for Sanger sequencing using the WizardR SV gel system (Promega).
Collagen IV α2 and α5 staining of hair follicles
Fluorochrome-conjugated monoclonal anti-human collagen IV antibodies (Cosmo Bio, Carlsbad, CA) were used to stain plucked hairs. A FITC-conjugated antibody to collagen α5(IV) imperfection III, the epitope of which is a five amino acid sequence (IDVEF) encoded by exon 13 [19], a FITC-conjugated antibody to collagen α5(IV) NC1 domain [20], and a Texas Red-conjugated antibody to collagen α2(IV) imperfection XIII [19] were incubated with hairs at room temperature for 30 minutes. Hairs were then whole-mounted for confocal immunofluorescence microscopy imaging of follicles.
Results
Alport family clinical findings
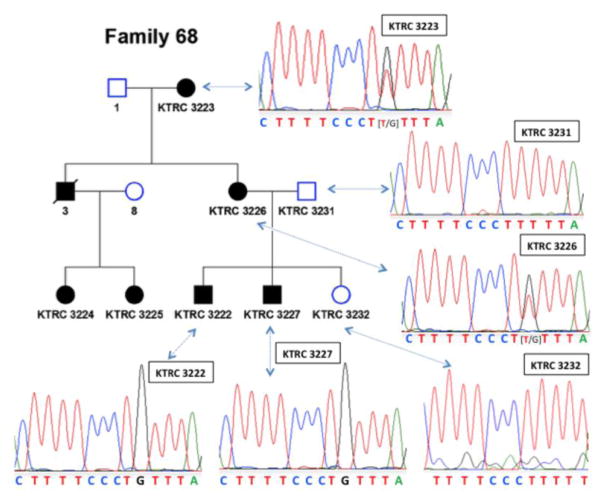
Family 68 is a three-generation pedigree with affected family members in each generation (Fig. 1 and Table 1). There was no male-to-male transmission of disease. No affected members of this family are known to have hearing loss or ocular abnormalities typical of Alport syndrome.
Figure 1. Pedigree of Alport Family 68 with chromatograms showing that the c.1780-6T>G splice region variant is present in affected but not unaffected family members.
Squares, males; circles, females; open shapes, unaffected; filled shapes, affected; diagonal strike, deceased. The numbers beneath the icons are individual identification numbers and correspond to the respective chromatograms. The heterozygous variant in females shown as overlapping peaks in the chromatograms is denoted by brackets.
Table 1.
Clinical features of Alport family 68
| Subject ID | Pedigree | Sex | Diagnosis |
|---|---|---|---|
| 3223 | I.1 | Female | Alport. Hematuria, proteinuria Skin Biopsy: Segmental loss of α5 staining |
| 3226 | II.1 | Female | Alport. Hematuria, proteinuria before age 10 Progressed to CKD 3 by age 55 Skin Biopsy: Segmental loss of α5 staining |
| 3231 | II.2 | Male | Unaffected |
| 3 | II.3 | Male | Alport. Hematuria, proteinuria before age 10 ESRD age 14. Transplant at age 15+17 |
| 3224 | III.1 | Female | Alport. Hematuria |
| 3225 | III.2 | Female | Alport. Hematuria |
| 3227 | III.3 | Male | Alport. hematuria, proteinuria age 5 Skin Biopsy age 7: Absence of α5 staining Wilsons disease |
| 3222 | III.4 | Male | Alport. Hematuria, proteinuria age 5 Kidney biopsy age 7: Thin GBM 90–100nm Skin Biopsy: Absence of α5 staining |
| 3232 | III.4 | Female | Unaffected Skin biopsy: Normal α5 staining |
CKD–Chronic kidney disease; ESRD–End-stage renal disease; GBM–Glomerular basement membrane
Mother (3226) and uncle (3) of the proband (3222) developed persistent hematuria in the first decade of life. The uncle (3) progressed to end-stage renal disease (ESRD) by the age of 14 years. At the time of submission the mother (3226) had chronic kidney disease (CKD) stage 3 and the maternal grandmother (3223) had preserved renal function. The proband (3222) and his brother (3227) developed hematuria and proteinuria by 5 years of age and have been on ACEi/ARB therapy since ages 8 and 7, respectively. At the time of manuscript submission both showed preserved renal function (serum creatinines of 1.0 and 0.8 mg/dL, estimated GFRs > 60 ml/min) at the ages of 21 and 19 years, respectively.
The proband was diagnosed with Alport syndrome after a kidney biopsy showing thin glomerular basement membranes (GBM) of 90–100 nm thickness with areas of lamina densa lamellation and a skin biopsy showing absence of collagen α5(IV) staining. All of the affected family members (except 3224 and 3225) had a skin biopsy to confirm their Alport syndrome diagnosis, with absence of α5(IV) staining in the male biopsies and segmental loss of α5(IV) staining in the female biopsies. Two female first cousins (3224 and 3225) of the proband were also diagnosed with Alport syndrome during childhood after the finding of hematuria in the setting of a family history of Alport Syndrome.
Germline genetic analysis
Patient 3227 had custom targeted next-generation sequencing performed. This revealed a novel splice region variant, c.1780-6T>G, in COL4A5 intron 24. No other potentially pathogenic variants in any of the collagen IV genes associated with Alport syndrome (COL4A3, COL4A4 and COL4A5) were found. Sanger sequencing confirmed that the c.1780-6T>G variant segregated with disease (Fig. 1). This variant was not found in the unaffected sibling (3232) of the proband and was not found in the married-in father (3231) of the proband. One heterozygous variant in complement gene CFI, c.782G>A, was found. This has a minor allele frequency of <1 % but has been previously described (rs112534524), and studies by Nilsson et al. found that this variant is not likely to be pathogenic [11]. No other variants in the genes included in the panel were found.
In silico splicing analysis
The c.1780-6T>G variant in COL4A5 was predicted to have widely variable impact on splicing, depending upon the specific in silico algorithm used. At one extreme, Human Splicing Finder detected no significant impact on splicing. MaxEntScan::score3ss predicted 13.2 % reduced splicing efficiency. NetGene2 Server predicted a 75 % drop in confidence that splicing would occur when the variant was present. The inconsistencies in these predictions precluded defining the variant as pathogenic.
In vitro splicing analysis
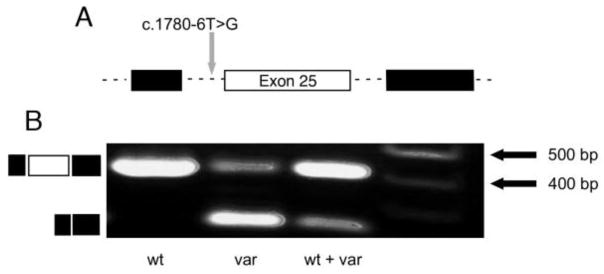
Functional assessment of the WT and c.1780-6T>G splice region variant was performed in cultured cells using the pCAS2.1 splicing reporter minigene assay (see Materials and Methods). The WT and VAR plasmids were transfected into HEK293T cells both separately and together. RNA was prepared from the cells and subjected to RT-PCR using amplification primers targeting the vectors’ exonic sequences flanking the COL4A5 insert (Fig. 2A). Agarose gel electrophoresis (Fig. 2B) showed that cells transfected with the WT vector yielded the expected 404 bp amplicon containing the vector’s endogenous exons and the COL4A5 exon 25 between them, with the intronic sequences spliced out. In contrast, cells transfected with the VAR vector yielded a major amplicon of 235 bp lacking COL4A5 exon 25 and a minor amplicon of 404 bp. Cells transfected with both WT and VAR vectors also yielded amplicons of 404 bp and 235 bp, but the 404 bp band was brighter than the 235 bp band (Fig. 2B, wt + var). The 404 bp band contained COL4A5 exon 25 (169 bp) flanked by the exonic sequences intrinsic to the pCAS2.1 splicing vector (exons 2 and 3 of C1NH); the 235 bp band represents the spliced pCAS2.1 exons without COL4A5 exon 25 (Supplemental Figure 1). Thus, the c.1780-6T>G variant caused frequent skipping of exon 25 in HEK293T cells, indicating that it is pathogenic.
Figure 2. In vitro analysis of the splice region variant’s function in HEK293T cells reveals frequent skipping of exon 25.
(A) The diagram illustrates the relevant intronic (dashed lines) and exonic (boxes) regions of the splicing reporter minigene vector. Black boxes represent C1NH exons 2 and 3, and the open box represents exon 25 of COL4A5. The c.1780-6T>G splice region variant is indicated. (B) RT-PCR products obtained from cells transfected with wild type (wt) vector only, variant (var) vector only, or wt and var vectors together. Product sizes are indicated based on size markers in lane 4. Diagrams to the left represent the normally spliced transcript containing exon 25 (top) and the abnormally spliced transcript resulting from the c.1780-6T>G variant (bottom).
Analysis of collagen IV expression in hair follicles
Because the epidermal basement membrane is continuous with the hair follicle basement membrane, we reasoned that analysis of collagen IV in plucked hair follicles could be a new method to help diagnose Alport syndrome noninvasively. Hair follicles plucked from affected and married-in members of Family 68 and from controls were stained for collagen α2(IV) and α5(IV) proteins by direct immunofluorescence. There was reduced and abnormal α5(IV) staining in the follicles of affected individuals (Fig. 3B, C) compared to control follicles (Fig. 3A) and follicles from a married-in family member (Fig. 3D). We noted that hairs plucked from affected individuals were less likely to have basement membrane attached to the hair root compared to unaffected individuals and controls.
Figure 3. Immunofluorescence staining of plucked hair follicle basement membranes from a control and from Alport family members.
FITC-conjugated antibodies to collagen α5(IV) (green) and Texas Red-conjugated antibody to collagen α2(IV) (red) were used. A, Control follicle, COL4A5WT/WT. B, 3223 follicle, COL4A5c.1780-6T>G/WT. C, 3227 follicle, COL4A5c.1780-6T>G/Y. D, 3231 follicle, COL4A5WT/Y. COL4A2 staining shows where basement membrane is present. COL4A5 was abundant in the control (A) and unaffected family member (D) follicle basement membranes but absent (B) or weak/segmental (C) in those with the variant.
Discussion
This study describes a novel COL4A5 variant in an Alport Syndrome family in which both males and females are affected. Males tended to have more severe disease than females, suggesting the classic X-linked pattern of inheritance. This disease severity difference between the sexes is demonstrated by individual 3 who progressed to ESRD by the age of 14 versus his affected sister (individual 3226) and mother (3223) who had not progressed to severe renal failure by the ages of 55 and 75, respectively.
All of the affected members of this family who were tested carried the c.1780-6T>G splice region variant in COL4A5, but the unaffected sibling and married-in father of the proband did not. Thus, this variant segregates with disease and is highly penetrant. Initial examination of this variant using online in silico tools suggested the variant might be pathogenic, but results from different algorithms were inconsistent. This is frequently the case for variants that are subsequently shown to be pathogenic using in vitro or in vivo models. For this reason we tested the functional consequences of this variant using a simple in vitro splicing reporter minigene assay. The c.1780-6T>G variant caused frequent yet apparently incomplete exon skipping (Fig. 2 and Supplementary Figure 1). RT-PCR data from the variant vector showed both skipped exon 25 product and retained exon 25 product; however, the predominant product lacked exon 25. When equal amounts of wild type and variant vector were co-expressed, the retained exon 25 message was the predominant transcript detected.
A particularly interesting finding in this study is that the variant vector can express some normally spliced transcript, at least in transfected HEK293T cells. Although other groups have reported using RNA extracted from hair follicles to assay COL4A5 expression and/or mRNA splicing by RT-PCR [21–23], we were unable to obtain enough intact RNA to determine the extent to which normal splicing of exon 25 occurs in vivo. However, if a low frequency of exon 25 retention does occur in vivo, it could partly explain the differences in disease severity among the affected males in this family (e.g., individual 3 compared to individuals 3222 and 3227), and it may also partly explain differences in phenotype between affected females (e.g., individual 3223 who has preserved renal function and her daughter, individual 3226, who has CKD stage 3).
In this regard, a recent article on females and Alport syndrome highlights the increased risk of disease, variable phenotype-genotype correlations in females with X-linked Alport mutations, and the importance of Lyonization, the random inactivation of one X chromosome [24]. This adds further complexity to the diagnosis and management of Alport syndrome in females with COL4A5 mutations. The extent of inactivation of the normal X chromosome in the affected females, in addition to the extent of COL4A5 exon 25 retention, may further increase the degree of phenotypic variation between affected females. Analysis of X inactivation in this family was not performed. The presence of unidentified modifier gene variants in this family may also explain variation in phenotype within this and within other families with Alport syndrome.
As COL4A5 exon 25 is 169 base pairs in length, its loss causes a frameshift and truncation of the α5(IV) protein. Gross et al. have previously reported that males with X-linked AS caused by a frame shift mutation progress to ESRD at a mean age of 19.8+/−5.7 years [25]. On the other hand, mild renal disease in a male, despite carrying a splice region variant that causes a truncation, has also been described before. Nozu et al. demonstrated the presence of wild type and abnormal transcripts in the kidney of an affected male with a splice region variant in COL4A5 that causes a truncation [21]. Our findings also suggest that incomplete splicing abnormalities might occur in the kidney and lead to a variable phenotype.
The incomplete skipping of exon 25 shown in this study would be expected to lead to reduced and variable protein deposition in tissues where COL4A5 is expressed. As COL4A5 is expressed in the skin, we examined the basement membranes of plucked hair follicles. Both α2 (in [α1(IV)]2α2(IV)) and α5 (in [α5(IV)]2α6(IV)) proteins are normally present in hair follicles [26]. Using the above described novel and non-invasive method we found that α5(IV) protein expression was reduced in the basement membranes of hair follicles of affected family members compared to controls. We also found that hair plucked from affected family members frequently lacked basement membrane compared to hairs plucked from controls (data not shown). This finding may be explained by the discontinuity of the lamina densa of skin basement membranes in AS patients [27].
Demonstration of a reduction or loss of α5(IV) expression in hair follicle basement membranes is a potentially useful and non-invasive method of diagnosis in AS. In this regard it is important to note that alternative splicing of COL4A5 RNA has been shown to generate two mRNAs (t1 and t2) and that hair follicles contain only t1, which lacks two small exons of 9 bp each. To our knowledge, no mutations have been described in these exons, but a mutation impacting these exons could cause kidney disease without altering expression of COL4A5 in hair follicles. With this caveat in mind, this α5(IV) protein assay is novel and has the potential to be a clinically meaningful screening tool for Alport syndrome. It would be especially valuable in determining the functional consequence of variants that would otherwise be described as ‘variants of uncertain significance’. Further studies on a large cohort of Alport families representing the full spectrum of mutation types is needed to determine the clinical utility of this hair follicle assay.
Reduction and variable expression of α5(IV) protein in glomeruli could not be examined here due to lack of specimens. However, our findings in the basement membrane of hair follicles suggest that there is likely reduced and/or variable α3α4α5(IV) protein expression in the GBM. Reduced but not absent α5(IV) protein in the GBM of males with X-linked AS has been described before. Only one of the α5(IV)-positive patients described by Hashimura et al. had a splice site mutation; however, we know from Nozu et al. that splice region variants can lead to expression of both normal and abnormal transcripts in the kidneys of male patients with X-linked AS [21, 28].
The amount of normal COL4A5 transcript expressed in vivo by the c.1780-6T>G variant may vary significantly between cell types. For example, keratinocytes may express no normal transcript, and thus would explain the skin biopsy results in affected family members showing loss of α5(IV) protein. It is interesting to speculate that the relevant cells in the inner ear that normally express COL4A5 are able to properly splice exon 25 to some extent, resulting in the preserved hearing function observed in this family.
In summary, this study reports a novel splice region variant in intron 24 of COL4A5, c.1780-6T>G, in a family with X-linked Alport syndrome. We used an in vitro assay to show that this variant is a pathogenic mutation that causes frequent but incomplete skipping of exon 25. This mutation likely leads to expression of mostly abnormal but also some normal transcripts that may partially explain the variable intra-family phenotypes seen in individuals carrying the mutation. Furthermore, we describe a novel non-invasive method to characterize α5(IV) protein deposition in the basement membranes of plucked hair follicles that may be an alternative method of diagnosis for Alport syndrome.
Supplementary Material
Acknowledgments
We thank Diane Salamon in the Division of Nephrology’s Kidney Translational Research Core for processing human specimens, Gloriosa Go for technical assistance, and Dr. Mario Tosi (Inserm U1079, Rouen, France) for generously providing the pCAS2.1 splicing vector. J.H.M. was supported by NIH grants R56DK100593, R01DK078314, and R01DK058366 and by the Alport Syndrome Foundation/Pedersen Family/Kidney Foundation of Canada Alport Syndrome Research Funding Program. S.D.F. was supported by T32DK007126.
Footnotes
Disclosures
JHM has received grants from Hoffmann-La Roche, Basel and RGDI3, Inc., Boston; has provided consultation to Third Rock Ventures, Boston; and has received licensing fees from Eli Lilly, Indianapolis and Genentech, South San Francisco.
Ethics statement
Human studies were conducted according to protocols approved by the Washington University Human Research Protection Office. Informed consent was obtained from all individual participants included in the study.
References
- 1.Kashtan CE. Alport syndrome. An inherited disorder of renal, ocular, and cochlear basement membranes. Medicine (Baltimore) 1999;78:338–360. doi: 10.1097/00005792-199909000-00005. [DOI] [PubMed] [Google Scholar]
- 2.Tryggvason K, Zhou J, Hostikka SL, Shows TB. Molecular genetics of Alport syndrome. Kidney Int. 1993;43:38–44. doi: 10.1038/ki.1993.8. [DOI] [PubMed] [Google Scholar]
- 3.Renieri A, Meroni M, Sessa A, Battini G, Serbelloni P, Torri Tarelli L, Seri M, Galli L, De Marchi M. Variability of clinical phenotype in a large Alport family with Gly 1143 Ser change of collagen alpha 5(IV)-chain. Nephron. 1994;67:444–449. doi: 10.1159/000188020. [DOI] [PubMed] [Google Scholar]
- 4.Tsiakkis D, Pieri M, Koupepidou P, Demosthenous P, Panayidou K, Deltas C. Genotype-phenotype correlation in X-linked Alport syndrome patients carrying missense mutations in the collagenous domain of COL4A5. Clin Genet. 2012;82:297–299. doi: 10.1111/j.1399-0004.2012.01849.x. [DOI] [PubMed] [Google Scholar]
- 5.Pierides A, Voskarides K, Kkolou M, Hadjigavriel M, Deltas C. X-linked, COL4A5 hypomorphic Alport mutations such as G624D and P628L may only exhibit thin basement membrane nephropathy with microhematuria and late onset kidney failure. Hippokratia. 2013;17:207–213. [PMC free article] [PubMed] [Google Scholar]
- 6.Demosthenous P, Voskarides K, Stylianou K, Hadjigavriel M, Arsali M, Patsias C, Georgaki E, Zirogiannis P, Stavrou C, Daphnis E, Pierides A, Deltas C. X-linked Alport syndrome in Hellenic families: phenotypic heterogeneity and mutations near interruptions of the collagen domain in COL4A5. Clin Genet. 2012;81:240–248. doi: 10.1111/j.1399-0004.2011.01647.x. [DOI] [PubMed] [Google Scholar]
- 7.Jais JP, Knebelmann B, Giatras I, De Marchi M, Rizzoni G, Renieri A, Weber M, Gross O, Netzer KO, Flinter F, Pirson Y, Verellen C, Wieslander J, Persson U, Tryggvason K, Martin P, Hertz JM, Schroder C, Sanak M, Krejcova S, Carvalho MF, Saus J, Antignac C, Smeets H, Gubler MC. X-linked Alport syndrome: natural history in 195 families and genotype-phenotype correlations in males. J Am Soc Nephrol. 2000;11:649–657. doi: 10.1681/ASN.V114649. [DOI] [PubMed] [Google Scholar]
- 8.Jais JP, Knebelmann B, Giatras I, De Marchi M, Rizzoni G, Renieri A, Weber M, Gross O, Netzer KO, Flinter F, Pirson Y, Dahan K, Wieslander J, Persson U, Tryggvason K, Martin P, Hertz JM, Schroder C, Sanak M, Carvalho MF, Saus J, Antignac C, Smeets H, Gubler MC. X-linked Alport syndrome: natural history and genotype-phenotype correlations in girls and women belonging to 195 families: a “European Community Alport Syndrome Concerted Action” study. J Am Soc Nephrol. 2003;14:2603–2610. doi: 10.1097/01.asn.0000090034.71205.74. [DOI] [PubMed] [Google Scholar]
- 9.Hertz JM, Thomassen M, Storey H, Flinter F. Clinical utility gene card for: Alport syndrome. Eur J Hum Genet. 2012;20(6) doi: 10.1038/ejhg.2011.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larsen CP, Durfee T, Wilson JD, Beggs ML. A Custom Targeted Next-Generation Sequencing Gene Panel for the Diagnosis of Genetic Nephropathies. Am J Kidney Dis. 2016;67:992–993. doi: 10.1053/j.ajkd.2015.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nilsson SC, Karpman D, Vaziri-Sani F, Kristoffersson AC, Salomon R, Provot F, Fremeaux-Bacchi V, Trouw LA, Blom AM. A mutation in factor I that is associated with atypical hemolytic uremic syndrome does not affect the function of factor I in complement regulation. Mol Immunol. 2007;44:1835–1844. doi: 10.1016/j.molimm.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 12.Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 14.Brunak S, Engelbrecht J, Knudsen S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J Mol Biol. 1991;220:49–65. doi: 10.1016/0022-2836(91)90380-o. [DOI] [PubMed] [Google Scholar]
- 15.Schwartz S, Hall E, Ast G. SROOGLE: webserver for integrative, user-friendly visualization of splicing signals. Nucleic Acids Res. 2009;37:W189–192. doi: 10.1093/nar/gkp320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vreeswijk MP, van der Klift HM. Analysis and interpretation of RNA splicing alterations in genes involved in genetic disorders. Methods Mol Biol. 2012;867:49–63. doi: 10.1007/978-1-61779-767-5_4. [DOI] [PubMed] [Google Scholar]
- 17.Bonnet C, Krieger S, Vezain M, Rousselin A, Tournier I, Martins A, Berthet P, Chevrier A, Dugast C, Layet V, Rossi A, Lidereau R, Frébourg T, Hardouin A, Tosi M. Screening BRCA1 and BRCA2 unclassified variants for splicing mutations using reverse transcription PCR on patient RNA and an ex vivo assay based on a splicing reporter minigene. J Med Genet. 2008;45:438–446. doi: 10.1136/jmg.2007.056895. [DOI] [PubMed] [Google Scholar]
- 18.Gaildrat P, Killian A, Martins A, Tournier I, Frébourg T, Tosi M. Use of splicing reporter minigene assay to evaluate the effect on splicing of unclassified genetic variants. Methods Mol Biol. 2010;653:249–257. doi: 10.1007/978-1-60761-759-4_15. [DOI] [PubMed] [Google Scholar]
- 19.Kagawa M, Kishiro Y, Naito I, Nemoto T, Nakanishi H, Ninomiya Y, Sado Y. Epitope-defined monoclonal antibodies against type-IV collagen for diagnosis of Alport’s syndrome. Nephrol Dial Transplant. 1997;12:1238–1241. doi: 10.1093/ndt/12.6.1238. [DOI] [PubMed] [Google Scholar]
- 20.Borza DB, Bondar O, Ninomiya Y, Sado Y, Naito I, Todd P, Hudson BG. The NC1 domain of collagen IV encodes a novel network composed of the alpha 1, alpha 2, alpha 5, and alpha 6 chains in smooth muscle basement membranes. J Biol Chem. 2001;276:28532–28540. doi: 10.1074/jbc.M103690200. [DOI] [PubMed] [Google Scholar]
- 21.Nozu K, Vorechovsky I, Kaito H, Fu XJ, Nakanishi K, Hashimura Y, Hashimoto F, Kamei K, Ito S, Kaku Y, Imasawa T, Ushijima K, Shimizu J, Makita Y, Konomoto T, Yoshikawa N, Iijima K. X-linked Alport syndrome caused by splicing mutations in COL4A5. Clin J Am Soc Nephrol. 2014;9:1958–1964. doi: 10.2215/CJN.04140414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.King K, Flinter FA, Green PM. Hair roots as the ideal source of mRNA for genetic testing. J Med Genet. 2001;38:E20. doi: 10.1136/jmg.38.6.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tazon-Vega B, Ars E, Burset M, Santin S, Ruiz P, Fernandez-Llama P, Ballarin J, Torra R. Genetic testing for X-linked Alport syndrome by direct sequencing of COL4A5 cDNA from hair root RNA samples. Am J Kidney Dis. 2007;50:257e251–214. doi: 10.1053/j.ajkd.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 24.Savige J, Colville D, Rheault M, Gear S, Lennon R, Lagas S, Finlay M, Flinter F. Alport Syndrome in Women and Girls. Clin J Am Soc Nephrol. 2016;11:1713–1720. doi: 10.2215/CJN.00580116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gross O, Netzer KO, Lambrecht R, Seibold S, Weber M. Meta-analysis of genotype-phenotype correlation in X-linked Alport syndrome: impact on clinical counselling. Nephrol Dial Transplant. 2002;17:1218–1227. doi: 10.1093/ndt/17.7.1218. [DOI] [PubMed] [Google Scholar]
- 26.Hasegawa H, Naito I, Nakano K, Momota R, Nishida K, Taguchi T, Sado Y, Ninomiya Y, Ohtsuka A. The distributions of type IV collagen alpha chains in basement membranes of human epidermis and skin appendages. Arch Histol Cytol. 2007;70:255–265. doi: 10.1679/aohc.70.255. [DOI] [PubMed] [Google Scholar]
- 27.Kuroki A, Ito J, Yokochi A, Kato N, Sugisaki T, Sueki H, Akizawa T. Diagnosing Alport syndrome using electron microscopy of the skin. Kidney Int. 2008;73:364–365. doi: 10.1038/sj.ki.5002682. [DOI] [PubMed] [Google Scholar]
- 28.Hashimura Y, Nozu K, Kaito H, Nakanishi K, Fu XJ, Ohtsubo H, Hashimoto F, Oka M, Ninchoji T, Ishimori S, Morisada N, Matsunoshita N, Kamiyoshi N, Yoshikawa N, Iijima K. Milder clinical aspects of X-linked Alport syndrome in men positive for the collagen IV alpha5 chain. Kidney Int. 2014;85:1208–1213. doi: 10.1038/ki.2013.479. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.