Abstract
The mitochondria are dynamic organelles that regulate oxidative metabolism and mediate cellular redox homeostasis. Proteins within the mitochondria are exposed to large fluxes in the surrounding redox environment. In particular, cysteine residues within mitochondrial proteins sense and respond to these redox changes through oxidative modifications of the cysteine thiol group. These oxidative modifications result in a loss in cysteine reactivity, which can be monitored using cysteine-reactive chemical probes and quantitative mass spectrometry (MS). Analysis of cell lysates treated with cysteine-reactive probes enable the identification of hundreds of cysteine residues, however, the mitochondrial proteome is poorly represented (<10% of identified peptides), due to the low abundance of mitochondrial proteins and suppression of mitochondrial peptide MS signals by highly abundant cytosolic peptides. Here, we apply a mitochondrial isolation and purification protocol to substantially increase coverage of the mitochondrial cysteine proteome. Over 1,500 cysteine residues from ~450 mitochondrial proteins were identified, thereby enabling interrogation of an unprecedented number of mitochondrial cysteines. Specifically, these mitochondrial cysteines were ranked by reactivity to identify hyper-reactive cysteines with potential catalytic and regulatory functional roles. Furthermore, analyses of mitochondria exposed to nitrosative stress revealed previously uncharacterized sites of protein S-nitrosation on mitochondrial proteins. Together, the mitochondrial cysteine enrichment strategy presented herein enables detailed characterization of protein modifications that occur within the mitochondria during (patho)physiological fluxes in the redox environment.
Graphical abstract
Mitochondria are double-membrane organelles of α-proteobacterial ancestry and are deeply integrated into the metabolic and structural machinery of the eukaryotic cell. Far from being isolated sites of oxidative respiration and ATP production, mitochondria dynamically communicate with numerous subcellular organelles through close contact junctions that facilitate exchange of metabolites1–3. Mitochondria are composed of 4 distinct and functionally heterogeneous sub-compartments: (1) the outer mitochondrial membrane (OMM) that establishes organelle contacts and recruits mitochondrial fission and fusion machinery; (2) the intermembrane space (IMS) where oxidative protein folding events are initiated; (3) the inner mitochondrial membrane (IMM) that provides the site of electron transport, oxidative phosphorylation, and generation of the proton gradient; and, (4) the mitochondrial matrix, which harbors the mitochondrial genome and a variety of metabolic enzymes and oxidoreductases. The mitochondrial proteome, estimated to consist of ~1,500 proteins in humans4, is the subset of cellular proteins that is associated with the mitochondria under various cellular conditions. The MitoCarta5 database, which was generated through computational and experimental means, defines a list of mitochondrial proteins. The vast majority of the mitochondrial proteome is expressed from the nuclear genome, with only 13 proteins expressed by the mitochondrial genome. While some nuclear-encoded proteins show strict localization to the mitochondria, other proteins, such as PINK1 and Bax, transiently associate with the surface of the mitochondria under specific cellular conditions6–8. Many mitochondrial proteins have been demonstrated to be dysregulated in a myriad of human disease states, from neurodegenerative disorders9.10 and aging11,12 to cancer13,14.
The mitochondria are critical mediators of cellular redox homeostasis due to their role in the generation and dissipation of reactive oxygen/nitrogen species (ROS/RNS)15. These ROS/RNS that are generated can dynamically regulate mitochondrial proteins through oxidative post-translational modifications (PTMs) on cysteine residues, which affect protein localization16,17, catalytic activity18,19, and protein-protein interactions20. These cysteine PTMs include oxidation to sulfenic/sulfinic/sulfonic acids, S-nitrosation, S-glutathionylation, and disulfide-bond formation21,22. For example, oxidation of a cysteine residue on uncoupling protein 1 (UCP1) in adipocytes regulates thermogenic energy expenditure23, and S-nitrosation of a cysteine residue on complex I is cardioprotective24. Improvements in the technological platforms available to interrogate sites of cysteine oxidation in mitochondria are likely to reveal other functionally important sites of protein regulation.
Coupling cysteine-reactive chemical probes, such as an iodoacetamide-alkyne (IA), with quantitative mass spectrometry (MS) has enabled the global interrogation of the cysteine proteome to identify functional cysteines and sites of PTMs25–28. These global cysteine-profiling studies typically involve MS analysis of unfractionated cell lysates labeled with the IA probe and provide limited coverage of the mitochondrial proteome; for example, 131 cysteines from 94 mitochondrial proteins were identified out of a total of 1076 cysteines on 812 cellular proteins25. The poor mitochondrial coverage is likely due to the low abundance of mitochondrial proteins relative to highly abundant cytosolic proteins, resulting in suppression of mitochondrial peptide MS signals. To enhance the number of identified mitochondrial cysteines, Hamachi et al. report the use of a mitochondrial-targeting tetraethylrhodamine moiety to direct a cysteine-reactive chloroacetamide probe to the mitochondria of living cells29. MS analysis identified 290 cysteines from 196 mitochondrial proteins out of a total of 378 cysteines on 244 proteins. This represents an increase in the percentage of mitochondrial cysteines identified relative to all cysteines, however, the total number of mitochondrial cysteines is relatively low and represent a small fraction of the ~1500 proteins estimated to be localized to the mitochondria. This poor coverage is attributed to the selective localization of the tetraethylrhodamine probe in the mitochondrial matrix and inner membrane, rendering limited labeling of OMM and IMS proteins. Furthermore, the use of mitochondrial targeting probes is limited to polarized mitochondria and excludes analysis of depolarized mitochondria. Mitochondrial depolarization is a characteristic feature of mitochondrial dysfunction in a number of disease states, including neurodegenerative disorders30. Depolarization is associated with a number of cellular changes that could affect cysteine reactivity including translocation of cysteine containing proteins between subcellular compartments31, changes in mitochondrial redox balance and cysteine oxidation32,33, and/or changes in mitochondrial pH and the proton gradient34,35.
In global proteomic studies, improved coverage of the mitochondrial proteome was achieved through selective enrichment of mitochondrial proteins prior to MS-based analysis. These enrichment strategies include: (1) the isolation of purified mitochondria by centrifugation prior to MS analysis5; and (2) selective biotinylation of mitochondrial proteins by an engineered peroxidase (APEX) localized to various mitochondrial membranes36. In particular, the mitochondrial-isolation strategy identified >1000 mitochondrial proteins and was instrumental in generating the MitoCarta database. Here we coupled a similar mitochondrial isolation strategy with IA labeling to selectively enrich and identify reactive cysteines within the mitochondria. Importantly, in contrast to mitochondrial-targeted chemical probes, this approach will enable the study of multiple regions of the mitochondria, including OMM and IMS proteins, and is feasible for the study of reactive cysteines in depolarized mitochondria. Previously, biotinylated iodoacetamide and maleimide derivatives were applied to purified mitochondrial lysates, identifying 1255 cysteine residues on 809 proteins across the two probes37. Of these, only ~33% (~250 proteins) were annotated as mitochondrial. Replacing the bulky biotin reporter tag on these electrophiles with the terminal alkyne group in the IA probe enables labeling of intact mitochondria to better preserve native redox environments and protein-protein interactions that could affect cysteine reactivity and PTM state. We demonstrate that purification and isolation of mitochondria from HEK293T cells followed by IA labeling provides significantly improved coverage of mitochondrial cysteine residues. MS analysis afforded identification of ~1500 cysteine residues from ~450 proteins from all 4 mitochondrial sub-compartments. Using quantitative MS, these mitochondrial cysteines were ranked by reactivity and sensitivity to S-nitrosation, thereby providing a comprehensive inventory of functional, reactive and redox-sensitive cysteines within the mitochondria. These studies provide the foundation to globally interrogate the effects of (patho)physiological mitochondrial oxidative stress on protein function.
RESULTS AND DISCUSSION
Mitochondrial enrichment
The poor coverage of the mitochondrial proteome in whole-cell lysate analyses for cysteine reactivity25,38 is attributed to the relatively low abundance of mitochondrial proteins and suppression by highly abundant cytoskeletal proteins. Removal of abundant proteins in the cytosol and other organelles by mitochondrial isolation (Figure 1A) was anticipated to afford greater coverage of mitochondrial cysteines. A mitochondrial isolation strategy previously employed in a shotgun-proteomic method by Pagliarini et al. to catalogue the entire mitochondrial proteome5, was adapted to generate crude and purified mitochondrial preparations (Figure 1B). Briefly, HEK293T cells were gently lysed by homogenization (to preserve the mitochondrial membrane structure) and a ‘crude’ mitochondrial preparation (Mito-C) was obtained by differential centrifugation39. A step-wise percoll gradient centrifugation subsequently afforded a ‘pure’ mitochondrial preparation (Mito-P)5.
Figure 1.
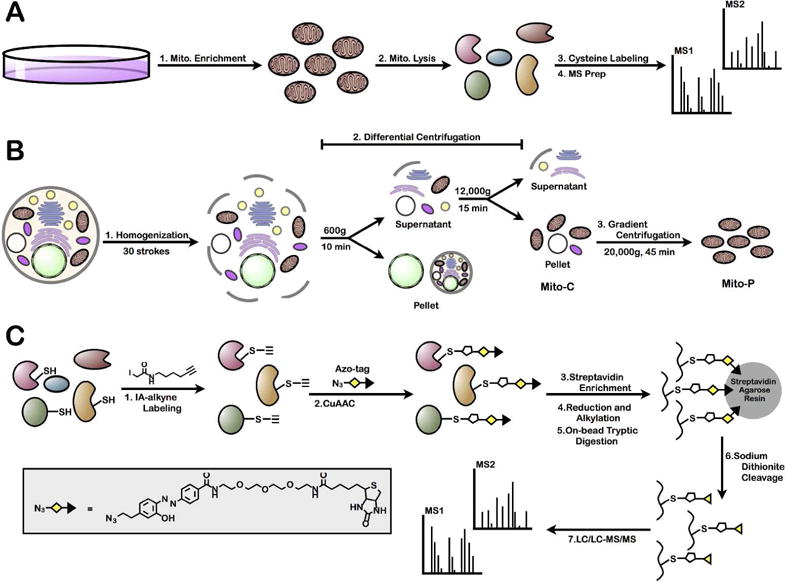
MS platform for enrichment and identification of mitochondrial cysteine residues. (A) General strategy to identify mitochondrial cysteine residues through the purification of mitochondria and MS identification of cysteine-containing peptides. (B) Schematic representation of a two-step mitochondrial purification strategy for the generation of either ‘crude’ (Mito-C) or ‘pure’ (Mito-P) mitochondrial preparations. (C) Cysteine-targeted MS workflow for the identification of IA-labeled cysteine residues.
Western-blot analysis of Mito-C and Mito-P preparations demonstrated mitochondrial enrichment compared to a whole-cell (WC) lysate (Figure 2A). The relative levels of a cytosolic protein (GAPDH; glyceraldehyde-3-phosphate dehydrogenase), an ER protein (Calreticulin), and a mitochondrial protein (Complex V inhibitor protein) were monitored for each preparation. The WC sample exhibited signals for proteins from all three subcellular compartments. The Mito-C preparation displayed significant loss of the cytosolic protein signals relative to WC, but still maintained ER protein signals. In contrast, the Mito-P preparation displayed only mitochondrial signals, demonstrating a high degree of purity. IA labeling of reactive cysteines in WC, Mito-C and Mito-P preparations was assessed by in-gel fluorescence upon copper-catalyzed azide-alkyne cycloaddition (CuAAC)-mediated conjugation of a fluorophore to IA-labeled proteins. In-gel fluorescence analysis of IA-labeled Mito-C and Mito-P preparations (Supplemental Figure 1) displayed differential banding patterns compared to the WC sample, demonstrating that a unique subset of cysteine-containing proteins were being modified upon mitochondrial enrichment.
Figure 2.
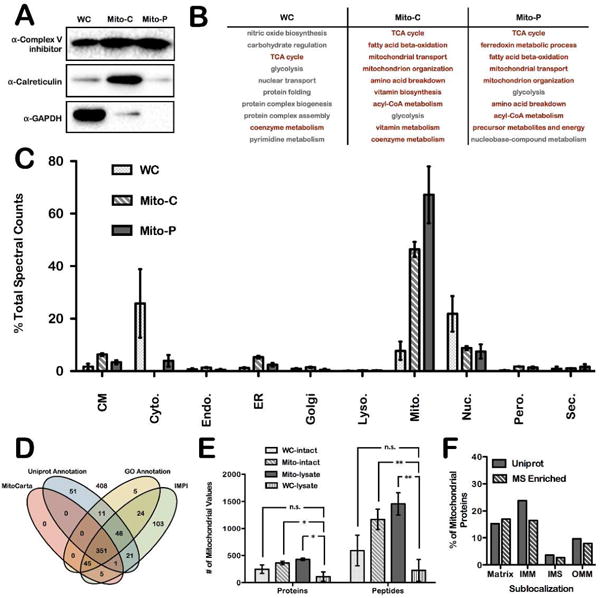
MS-based proteomic analysis of reactive cysteine-containing proteins from enriched and purified mitochondria. (A) Western-blot analysis of whole cell (WC), ‘crude’ mitochondrial (Mito-C), and ‘pure’ mitochondrial (Mito-P) lysates against GAPDH, calreticulin, and Complex V inhibitor protein. (B) PANTHER analysis of the 10 most overrepresented GO Biological Processes for proteins identified in each biological preparation. Pathways highlighted in red indicate a mitochondrial process. (C) Uniprot subcellular localization annotations of proteins identified by MS analysis, plotted as the spectral counts as a percent of total. Data represent the mean ± s.d. of three biological replicates. Organelles included in the analysis are cell membrane (CM), cytoskeleton (Cyto.), endosomes (Endo.), endoplasmic reticulum (ER), golgi apparatus (Golgi), lysosomes (Lyso.), mitochondria (Mito.), nucleus (Nuc.), peroxisomes (Pero.), and secreted (Sec.). (D) Visualization of filtered enriched proteins identified by LC/LC-MS/MS in Mito-C and Mito-P preparations. Mitochondrial localization was determined for these proteins from the overlap of 4 different mitochondrial protein databases; Uniprot, GO, IMPI, and MitoCarta. (E) Absolute number of mitochondrial proteins and peptides identified under four different IA-labeling conditions; WC-intact, Mito-intact, Mito-lysates and WC-lysates. Data represent the mean ± s.d. of three biological replicates; *P < 0.05, paired Student’s t-test (two-tailed). (F) Mitochondrial sublocalization analysis of proteins identified in the Mito-intact sample; localizations are denoted as matrix, IMM (inner mitochondrial membrane), IMS (Intermembrane space), or OMM (outer mitochondrial membrane).
Cysteine enrichment and MS analysis
Isolated WC, Mito-C and Mito-P proteomes were subjected to cysteine enrichment and subsequent MS analysis. Cysteine enrichment was achieved by the following workflow40 (Figure 1C): (1) cysteine labeling with IA-alkyne; (2) CuAAC to append a chemically cleavable biotin-azide tag (Azo tag); (3–5) on-bead streptavidin enrichment, reduction, alkylation, and trypsin digestion; and, (6) chemical cleavage of the Azo-tag to release IA-labeled peptides from the bead. The resulting IA-labeled peptide samples were analyzed by LC/LC-MS/MS, whereby fragmentation (MS2) spectra were used to identify IA-labeled peptides and cysteine residues. MS data for three biological replicates of each preparation were collected, and filtered to obtain a list of proteins identified with average spectral counts ≥ 2 (Supplemental Table 1). WC, Mito-C and Mito-P preparations afforded 953, 1142, and 759 protein identifications, respectively. For each of the three preparations, Panther overrepresentation analysis41 provided a measure of GO biological process enrichment (Figure 2B, Supplemental Tables 2–4). The top ten most overrepresented processes from WC samples are mostly cytosolic processes (e.g. glycolysis and glycogen metabolism). In contrast, for both Mito-C and Mito-P samples, primarily mitochondrial associated processes were overrepresented (e.g. beta-oxidation, amino acid breakdown, and mitochondrial transport and organization).
The subcellular distribution of identified proteins was examined by mining the Uniprot database for subcellular localization (e.g. cell membrane, cytoskeleton, endosome, ER, golgi, lysosome, nucleus, peroxisome, or secreted). The percentage of spectral counts attributed to proteins from each subcellular localization was determined for each preparation (Figure 2C). In WC samples, spectral counts from mitochondrial proteins represent only ~10% of total, while in Mito-C and Mito-P preparations they represent ~50% and ~70%, respectively. Spectral counts from both cytoskeletal and nuclear proteins significantly decrease with increasing mitochondrial purity, and spectral counts from the endomembrane system and peroxisomes increase slightly in the Mito-C preparation, but decrease upon further mitochondrial purification. Analysis of identified peptides and proteins reflected the same trend as spectral counts, whereby as mitochondrial purity increased, mitochondrial peptides and proteins contributed to a greater percentage of the total identified (Supplemental Figure 2A). The total number of mitochondrial proteins and peptides significantly increased in the Mito-C preparation (~450 proteins and 1500 peptides) as compared to WC samples (~100 proteins and 200 peptides). However, there was no significant difference in the total number of mitochondrial proteins and peptides identified in the Mito-C (428 proteins and 1455 peptides) and Mito-P (397 proteins and 1332 peptides) samples (Supplemental Figure 2B). Therefore, the additional purification step provides no increased benefit for identifying mitochondrial proteins and peptides, likely indicating that the multidimensional separation method (LC/LC) used prior to MS identification is sufficient to overcome any suppression of mitochondrial signals resulting from non-mitochondrial impurities in the crude preparations.
In addition to Uniprot annotations of protein subcellular localization42,43, there are a number of other databases of mitochondrial localized proteins, including GO annotations44, the IMPI database45,46, and the MitoCarta (a list of 1158 human mitochondrial proteins determined by MS, fluorescence, and computational predictive algorithms5,47). Of the proteins found to be enriched in the Mito-C and Mito-P preparation, 504 were annotated to be mitochondrial in at least two of the databases (Figure 2D, Supplemental Table 5). Interestingly, every protein listed in the MitoCarta database was identified in at least one other database, while the other databases had a number of unique mitochondrial identifications.
IA labeling in intact mitochondria
To preserve the native environment surrounding mitochondrial proteins during the cysteine-labeling step, IA labeling of intact cells and intact mitochondria were compared to IA labeling in mitochondrial lysates. Four conditions were investigated: (1) treatment of intact cells with IA, followed by subsequent mitochondrial isolation, lysis and MS analysis (WC-intact); (2) treatment of intact mitochondria with IA, followed by mitochondrial lysis and MS analysis (Mito-intact); (3) treatment of mitochondrial lysates with IA, followed by MS analysis, as a positive control (Mito-lysate); and, (4) treatment of whole-cell lysates with IA-alkyne, without mitochondrial enrichment, as a negative control (WC-lysates) (Supplemental Figure 3A). WC-intact and Mito-intact best preserve the native environment of mitochondrial proteins during the IA labeling step. The goal was to compare protein identifications in WC-intact and Mito-intact samples and identify which of these two conditions provided coverage similar to that observed in the positive control (Mito-lysate).
In-gel fluorescence analysis showed similar IA-labeling profiles for WC-intact, Mito- intact and Mito-lysates (Supplemental Figure 3B), confirming mitochondrial enrichment. MS analysis was then performed on each of the four preparations and subcellular location of each identified protein was annotated (Supplemental Table 6). As expected, WC-lysates provided the lowest number of mitochondrial protein identifications (114 proteins) (Figure 2E). WC-intact, Mito-intact and Mito-lysates generated 329, 406 and 485 mitochondrial protein identifications, respectively (Supplemental Figure 3C, Supplemental Table 7). Therefore, labeling of intact mitochondria with the IA probe (Mito-intact) provides optimal coverage of mitochondrial proteins, whilst providing minimal perturbation of the local mitochondrial environment during IA labeling.
To determine the distribution of the identified proteins amongst the mitochondrial sub-compartments (OMM, IMS, IMM, and matrix), the Uniprot database was used to assign the location of the 406 proteins identified in the Mito-intact experiment. Importantly, proteins in all four mitochondrial locations were identified (Figure 2F). Interestingly, the percentage of proteins identified from each sub-compartment closely matched the percentage of total annotated proteins in each location, suggesting that minimal enrichment of individual mitochondrial sub-compartments is occurring. This is in contrast to previous studies utilizing a mitochondria-targeted cysteine probe, which showed predominant identification of matrix proteins, with poor coverage of other sub-compartments29.
Identifying hyper-reactive mitochondrial cysteines
Analysis of time-or concentration-dependent IA labeling by quantitative MS has enabled the ranking of cysteines by reactivity and the identification of a hyper-reactive subset enriched in functional cysteines25. However, mitochondrial cysteines were poorly represented in these global cysteine-reactivity profiling studies. Identifying hyper-reactive mitochondrial cysteines will reveal novel functional sites involved in catalysis, metal binding and redox regulation. To rank mitochondrial cysteines by reactivity, lysates from crude mitochondrial preparations were treated with either 10 μM or 100 μM IA, prior to CuAAC-mediation conjugation to either heavy or light isotopically-labeled, chemically-cleavable Azo-tags, respectively33 (Figure 3A). The light and heavy samples were combined, enriched on streptavidin beads, and subjected to on-bead trypsin digestion and sodium dithionite treatment. Four replicate samples were analyzed by LC/LC-MS/MS and light to heavy (L/H) ratios for all identified peptides were calculated (Supplemental Table 8). The L/H ratio is a measure of cysteine reactivity, as it reflects the degree of cysteine labeling between the 10 μM and 100 μM IA-treated samples. A hyperreactive cysteine will saturate labeling at low IA concentrations, and display a ratio of ~1, whereas less reactive cysteine residues will show ratios >>1. L/H ratios were obtained for 522 cysteine-containing peptides from mitochondrial proteins (Figure 3B, Supplemental Table 9). The distribution of ratios for mitochondrial peptides does not differ noticeably from non-mitochondrial peptides (Supplemental Figure 4), suggesting that mitochondrial cysteine residues show similar reactivity profiles compared to cysteine residues from other subcellular locations.
Figure 3.
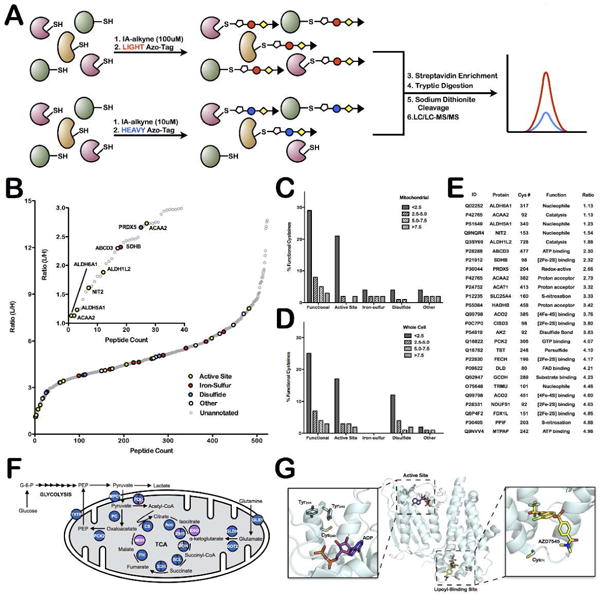
Quantitative-MS analysis of mitochondrial proteins identifies hyper-reactive and functional cysteine residues. (A) Schematic representation of workflow to rank cysteines by reactivity. (B) L/H ratio plot for all mitochondrial cysteine-containing peptides. Functional cysteines are displayed in bold, colored circles (based on function) while unannotated cysteines are displayed as light, gray circles. Inset shows those cysteine residues with ratios less than 3 (ie. hyper-reactive cysteine residues). (C and D) Analysis of the relationship between functional annotation and reactivity in (C) current mitochondrial dataset and (D) previously published whole-cell lysate data from Weerapana et al. Nature 201025. Data are divided into generally functional, active-site, iron-sulfur ligating, disulfide-bond forming, or other and into reactivity bins; L/H ratio less than 2.5, between 2.5 and 5.0, between 5.0 and 7.5, and greater than 7.5. (E) List of identified functional mitochondrial reactive cysteine residues. Only cysteine residues with L/H ratios below 5 are included. (F) Schematic of the TCA cycle proteins that are identified in mitochondrial enriched samples (blue circles), a handful are also found in whole cell samples (purple circles) or not at all (white circles). Detailed information on identified cysteine-labeled TCA-cycle peptides is presented in Supplemental Table 11. (G) Structure of the ATP and lipoyl binding sites of pyruvate dehydrogenase kinase 2 (PDK2) (PDB = 2Q8F).
The reactivity of mitochondrial cysteines identified in previous whole-cell lysate analyses were mined to determine if this subset is biased toward cysteines with elevated reactivity (Supplemental Figure 5, Supplemental Table 9)25. While many highly reactive mitochondrial cysteine residues were identified in whole cell lysates (with comparable L/H ratios), ~75% of the hyper-reactive cysteine residues (L/H ratios < 2.5) were novel identifications in this current dataset. Therefore, mitochondrial isolation prior to IA labeling serves to enrich for cysteine residues across the reactivity spectrum.
The human Uniprot database was mined for functional annotation of the identified mitochondrial cysteines, including active sites, disulfide bonds, modifications, substrate and metal binding, as well as disease variants and conflicting residues (Figure 3B). To examine the correlation between reactivity and functionality of mitochondrial cysteines, identified residues were divided into four ratio bins (<2.5, 2.5–5.0, 5.0–7.5, >7.5) and the percentage of annotated functional cysteines indicated for each bin (Figure 3C). Interestingly, ~25% of cysteine residues with ratios below 2.5 are functionally annotated, while less than 10% of cysteines in the other three ratio bins are annotated. These results are similar to previous studies with whole-cell lysates25 (Figure 3D). The annotated hyperreactive cysteine residues identified include active-site residues from 3-ketoacyl-CoA thiolase (ACAA2), omega-amidase (NIT2), and three aldehyde dehydrogenases (methylmalonate-semialdehyde dehydrogenase (ALDH6A1); succinate-semialdehyde dehydrogenase (ALDH5A1); and 10-formyltetrahydrofolate dehydrogenase (ALDH1L2)). Additionally, a highly reactive redox-active cysteine from peroxiredoxin 5 (PRDX5) and a [2Fe-2S] ligating cysteine from succinate dehydrogenase (SDHB) were also identified, demonstrating the diverse functional roles of reactive cysteines (Figure 3E). Interestingly, while iron-sulfur (Fe-S) ligating cysteine residues do not show a strong correlation with ratio, they are significantly enriched overall in the mitochondrial dataset compared to the previous whole-cell analysis25, in which no annotated Fe-S ligands were identified (Figure 3C and D). This result could be a consequence of the large number of Fe-S proteins in the mitochondria, including Complexes I and II, and several Fe-S maturation proteins48,49.
In addition to cysteine function, cysteine conservation was also examined by performing BLAST searches against 5 other organisms; mouse, Drosophila, C. elegans, baker’s yeast, and Arabidopsis. Importantly, there was little correlation between reactivity and conservation (Supplemental Figure 6), as previously shown for all cellular cysteines25. Interestingly, all functionally annotated, hyper-reactive cysteine residues (ratio < 3) were 100% conserved, with the exception of the redox-active cysteine on PRDX5. This observation suggests that highly reactive and conserved cysteines are likely to be functional. In fact, even though unannotated in Uniprot, examination of the literature provided evidence that the conserved and reactive Cys219 on Cox11 is a copper Cu(I) binding ligand50,51, and based on homology, Cys77 of MTO1 is an active-site residue52. Similarly, a number of other reactive and conserved cysteine residues were assigned function based on either literature or homology (Supplemental Table 10), confirming that reactivity and conservation is an extremely strong indicator of functionality. These studies therefore provide the first analysis of cysteine reactivity in the mitochondria and reveal several highly reactive cysteines that could serve diverse functions on mitochondrial proteins.
Enrichment of cysteine residues on mitochondrial metabolic proteins
Two different mitochondrial pathways found to be enriched in a Panther analysis of the proteins identified in Mito-C and Mito-P preparations, the TCA cycle (Figure 3F, Supplemental Table 11), and the electron-transport chain/oxidative phosphorylation (ETC)/OxPhos complexes (Supplemental Figure 7, Supplemental Table 12), were further evaluated for protein coverage. In both cases, a significant number of proteins from each of these pathways was only identified in enriched mitochondrial samples. In total, proteins in the TCA cycle (including the pyruvate dehydrogenase complex) contain ~285 cysteine residues, of which 139 were identified. Similarly, 96 out of 267 cysteines were identified from subunits of the ETC complexes, with higher coverage observed for the soluble subunits of these complexes (e.g. 44% identification for NDUFS1-8/NDUFV1-3 and 66% identification for SHDA/B). These data highlight the comprehensive detection of cysteine residues from important mitochondrial metabolic pathways.
Reactive and unannotated cysteine residues from highly conserved TCA-cycle proteins were examined as potential novel functional sites. Pyruvate dehydrogenase kinase 1 (PDK1) contained one highly conserved reactive cysteine residue, Cys240, as well as a second less conserved reactive cysteine residue, Cys71. The homologous cysteine residues were also identified as reactive in PDK3 and PDK2, respectively. PDK is responsible for phosphorylating and inactivating pyruvate dehydrogenase (PDH), and is itself regulated by phosphorylation at a number of tyrosine residues53. Both cysteine residues reside proximal to ligand-binding sites of PDK (Figure 3G); Cys240 is positioned above the ATP-binding site, adjacent to two tyrosine residues, whose phosphorylation is known to regulate PDK activity. Similarly, Cys71 sits near the lipoyl binding pocket, which controls PDK binding to PDH. Cys71 was identified as ligandable in a recent screen of cysteine-reactive small-molecule fragments54. In general, ligandable cysteine residues were found across the spectrum of reactivity for mitochondrial cysteines (Supplemental Figure 8, Supplemental Table 9). The high reactivity, partial conservation, ligandability, and proximity to functional sites within PDK support the potential for these cysteines to be important for PDK function.
Of particular interest are the iron-sulfur (Fe-S) ligating cysteine residues that were enriched in our mitochondrial datasets. Fe-S clusters are synthesized in the mitochondria, with an input of iron, sulfur, and reducing equivalents on the scaffold protein ISCU48,49 (Figure 4A). A number of chaperones and Fe-S cluster-binding proteins of poorly defined function build up and deliver clusters of different nuclearities to the wide variety of mitochondrial Fe-S binding proteins. The large majority of Fe-S clusters are ligated by protein cysteine residues, and of the 33 known mitochondrial iron-sulfur clusters with cysteine ligation, at least one ligating cysteine residue was identified from 22 of these clusters (Figure 4B, Supplemental Table 13), representing a 70% coverage of the mitochondrial Fe-S proteome. Full occupancy of Fe-S clusters likely prevents cysteine labeling by the IA probe. However, Fe-S clusters are known to be highly sensitive to oxidative stress55 and it is likely that a subpopulation of protein would be cluster deficient and susceptible to labeling by the IA probe. The expansive coverage of the Fe-S proteome allows for adaptation of this cysteine-enrichment strategy to monitor Fe-S cluster occupancy in the mitochondria.
Figure 4.
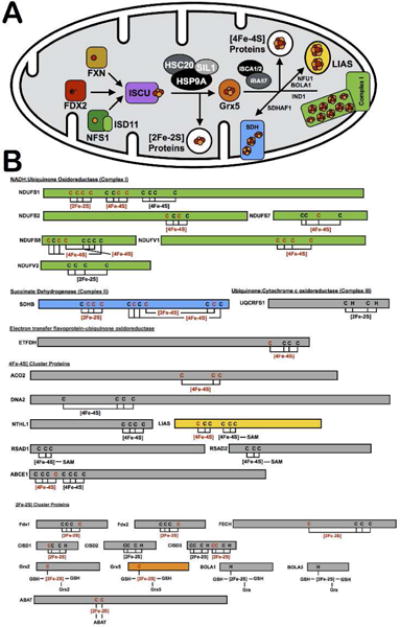
Cysteines involved in Fe-S clusters. (A) Schematic of the proteins involved in the Fe-S cluster biogenesis pathway, including cluster generation on the scaffold protein ISCU, proposed cluster transfer to Grx5, and delivery to target mitochondrial proteins, such as complex I and lipoyl synthase (LIAS). (B) List of all known mitochondrial Fe-S proteins, with their ligating residues depicted. Proteins are color coded to match the diagram in 4A. Cysteine residues and Fe-S clusters are highlighted in red if they were identified in the mitochondrial datasets. Detailed information on identified mitochondrial iron-sulfur proteins is presented in Supplemental Table 13.
Sensitivity of mitochondrial cysteines to transnitrosation by GSNO
To demonstrate the utility of this mitochondrial enrichment platform to identify sites of cysteine PTMs, S-nitrosation by the transnitrosating agent S-nitrosoglutathione (GSNO) was monitored. Intact mitochondria were treated with varying concentrations of GSNO (0, 10, 100, 1000 μM) prior to IA treatment. It is known that GSNO can permeate membranes56, and is therefore likely to access proteins within the different sub-compartments of intact mitochondria. After lysis of mitochondria, CuAAC was performed with heavy (untreated control) and light (GSNO treated) Azo-tags (Figure 5A). Samples were analyzed by LC/LC-MS/MS and heavy to light (H/L) ratios for peptides were calculated (Supplemental Table 14). The H/L ratio is a measure of cysteine GSNO sensitivity, with higher ratios representative of a greater percentage of cysteine S-nitrosation, while a ratio of ~1 represents virtually no S-nitrosation. Filtering of the data for mitochondrial localization, afforded 576 cysteine-containing peptides with H/L ratios at all four GSNO concentrations tested (Supplemental Table 15). The 576 mitochondrial cysteine residues displayed a wide range of sensitivities (Figure 5B), with highly GSNO-sensitive cysteine residues like Cys45 of GPD2 displaying increasing levels of S-nitrosation with GSNO concentration. In contrast, many cysteines, such as Cys71 of PDK1 displayed only modest GSNO-mediated S-nitrosation, and Cys458 of HADHB showing complete insensitivity to GSNO. Importantly, Cys458 of HADHB is the active-site proton acceptor, demonstrating that cysteine functionality and GSNO sensitivity are not directly correlated. Additionally, correlation of the GSNO sensitivity of mitochondrial cysteines to their ligandability54 was poor, suggesting that GSNO targets a selective subset of cysteine residues independent of the presence of a proximal ligandable pocket (Supplemental Table 15). A list of the 22 most GSNO-sensitive mitochondrial cysteine residues was generated; defined by ratios above 1.5 at 10 μM GSNO, above 3 at 100 μM GSNO, and above 5 at 1 mM GSNO (Figure 5C, Supplemental Table 16). Correlation with the dbSNO database of previously annotated and characterized cysteine SNO sites57 showed poor overlap (Supplemental Figure 9). However, the dbSNO database provided poor coverage of mitochondrial proteins in general, where 149 of the 1250 listed human nitrosation events are found on mitochondrial proteins, likely due to the majority of these analyses being performed on whole-cell and tissue proteomes without mitochondrial fractionation. Furthermore, our platform enables elucidation of the extent of cysteine S-nitrosation, which is not accounted for in the majority of entries included in the dbSNO database. Of note, the transcription factor, p53, contains three IA-labeled cysteine residues, one of which, Cys182, shows significant GSNO sensitivity (Figure 5D, Supplemental Figure 10). This residue is located on the DNA binding face of the protein and near but not involved in zinc binding (Figure 5E). Cys141 and Cys124 on p53 were found to be GSNO-insensitive, yet all three cysteine residues are annotated as sites of cysteine nitrosation in the dbSNO database (Figure 5F). Our data suggest that Cys182 is a high-affinity site of cysteine S-nitrosation, while the other two sites are only nitrosated to a very small degree.
Figure 5.
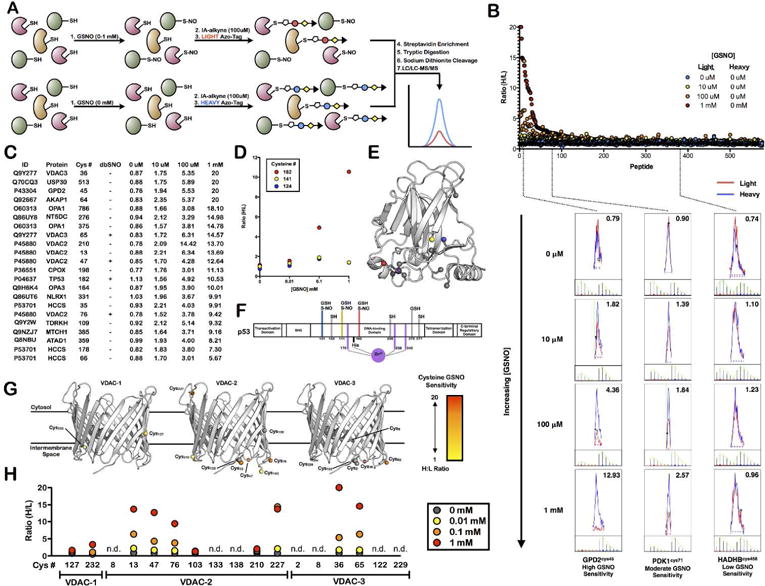
Quantitative MS analysis identifies mitochondrial cysteine residues sensitive to GSNO-mediated transnitrosation. (A) Schematic of quantitative proteomic workflow. (B) H/L ratio plot for all mitochondrial cysteine-containing peptides with valid ratios in all four GSNO treatments. Extracted ion chromatograms and isotopic envelopes are displayed for three cysteine-containing peptides (GPD2 – high GSNO sensitivity; PDK1 – moderate GSNO sensitivity; and HADHB – low GSNO sensitivity) at all four GSNO concentrations. (C) List of GSNO-sensitive mitochondrial cysteine residues. (D–F) Identification of a single GSNO-sensitive cysteine on p53. (D) Plot of GSNO sensitivity for three identified cysteine residues of p53. (E) Crystal structure of the DNA binding domain of p53, cysteine residues are highlighted (PDB = 5LGY). (F) Protein schematic of p53 showing previously annotated cysteine post-translational modifications. (G,H) Identification of a number of GSNO-sensitive cysteine residues on human VDACs. (G) Structural analysis of all cysteine residues of VDAC-1-3. Cysteine residues from VDAC-2 and VDAC-3 were mapped onto the crystal structure of VDAC-1 (PDB = 2JK4). (H) GSNO sensitivity plots for all VDAC-1-3 cysteine residues (n.d. indicates incomplete or no ratio data for that residue).
Our platform allows for the investigation of outer mitochondrial membrane proteins, compared to chemically targeted probes that are selective for matrix proteins. Analysis of the sub-mitochondrial localization of the GSNO-sensitive cysteines shows little to no preference for location (Supplemental Figure 11), confirming the ability of GSNO to diffuse into all compartments of the mitochondria. Some of the most GSNO-sensitive cysteine residues are found on voltage-dependent anion channels (VDACs) 1–3 that reside in the outer mitochondrial membrane and regulate the diffusion of small hydrophilic molecules58. VDAC1 contains only two cysteine residues, while VDAC2 and VDAC3 have 9 and 6, respectively, which reside on the intermembrane faces of the proteins (Figure 5G). These residues have been shown to be sensitive to oxidation and may play a role in regulating pore function59. The two cysteine residues of VDAC1 appear to be relatively insensitive to GSNO-mediated transnitrosation, while four cysteine residues in VDAC2 are highly sensitive to GSNO (Figure 5H). These four residues are also conserved in VDAC3 (Supplemental Figure 12), but only two of these residues Cys36 and Cys65 from VDAC3 were identified in our data, both with high GSNO-mediated transnitrosation sensitivity. These data provide a global assessment of oxidative/nitrosative cysteine PTMs on outer mitochondrial membrane proteins.
In conclusion, we report the development of a platform to study reactive and functional cysteines within the mitochondria. In contrast to previous studies utilizing mitochondrial targeted cysteine probes, which rely on the polarization of the mitochondrial inner membrane for enrichment, and are limited to the study of mitochondrial matrix proteins, we have achieved significant enrichment of the mitochondrial proteome by performing an isolation and enrichment procedure. Using our platform of mitochondrial enrichment and subsequent cysteine-targeted MS analysis, we successfully identified ~1,500 reactive cysteine residues on ~450 mitochondrial proteins. This represents a significant improvement in mitochondrial cysteine identifications compared to previous whole-cell studies25,29. Cysteine reactivity was quantified for ~500 residues, and it was demonstrated that hyper-reactivity was correlated and predictive of cysteine functionality. Additionally, 22 highly GSNO-sensitive cysteine residues were identified, including those on the outer mitochondrial membrane VDAC proteins. The benefits of this cysteine-enrichment platform include the ability to label all compartments of the mitochondria equally, confirmed by our observation of significant labeling of outer membrane and intermembrane-space proteins. As appreciation of the dynamic nature of the mitochondria grows, it is clear that a number of important cellular signaling and communication processes occur at the surface of the mitochondria and include the recruitment of transiently associated mitochondrial proteins. This platform has the ability to interrogate these processes and identify functional cysteine residues that may play roles in regulating mitochondrial dynamics, organelle-organelle contacts, and apoptosis and autophagy. Additionally, chemically directed probes would be unable to label depolarized mitochondria efficiently, which may preclude the use of these probes in the study of a number of mitochondrial-associated disease states. Future work will focus on the quantitative comparison of mitochondria in various (patho)physiological states, to identify cysteine residues involved in cell signaling and redox homeostasis within the mitochondria. It has been previously demonstrated that unique subsets of cysteine residues may be labeled by electrophiles that function via a SN2 nucleophilic displacement versus Michael-addition mechanism37. Therefore, future studies that multiplex diverse cysteine-reactive electrophiles could provide even greater coverage of the mitochondrial reactive cysteine proteome.
METHODS
See the Supporting Information for details about mitochondrial isolation, mass spectrometry sample preparation, and data analysis.
Supplementary Material
Acknowledgments
This work was funded by NIH grant 1R01GM117004 to E. W.
Footnotes
Supporting Information
Detailed experimental procedures and supplementary figures and tables. The Supporting Information is available free of charge on the ACS Publications website at DOI:
Notes
The authors declare no competing financial interests.
References
- 1.Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148(6):1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashrafi G, Schwarz TL. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013;20(1):31–42. doi: 10.1038/cdd.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wai T, Langer T. Mitochondrial Dynamics and Metabolic Regulation. Trends Endocrin Met. 2016;27(2):105–117. doi: 10.1016/j.tem.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Lopez MF, Kristal BS, Chernokalskaya E, Lazarev A, Shestopalov AI, Bogdanova A, Robinson M. High-throughput profiling of the mitochondrial proteome using affinity fractionation and automation. Electrophoresis. 2000;21(16):3427–3440. doi: 10.1002/1522-2683(20001001)21:16<3427::AID-ELPS3427>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 5.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134(1):112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, Sou YS, Saiki S, Kawajiri S, Sato F, Kimura M. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189(2):211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Losón OC, Song Z, Chen H, Chan DC. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol Biol Cell. 2013;24(5):659–667. doi: 10.1091/mbc.E12-10-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soriano ME, Scorrano L. Traveling Bax and forth from mitochondria to control apoptosis. Cell. 2011;145(1):15–17. doi: 10.1016/j.cell.2011.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Itoh K, Nakamura K, Iijima M, Sesaki H. Mitochondrial dynamics in neurodegeneration. Trends Cell Biol. 2013;23(2):64–71. doi: 10.1016/j.tcb.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramonet D, Perier C, Recasens A, Dehay B, Bové J, Costa V, Scorrano L, Vila M. Optic atrophy 1 mediates mitochondria remodeling and dopaminergic neurodegeneration linked to complex I deficiency. Cell Death Differ. 2013;20(1):77–85. doi: 10.1038/cdd.2012.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120(4):483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Hekimi S. Mitochondrial dysfunction and longevity in animals: Untangling the knot. Science. 2015;350(6265):1204–1207. doi: 10.1126/science.aac4357. [DOI] [PubMed] [Google Scholar]
- 13.Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12(10):685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saxena N, Maio N, Crooks DR, Ricketts CJ, Yang Y, Wei MH, Fan TW, Lane AN, Sourbier C, Singh A, Killian JK. SDHB-Deficient Cancers: The Role of Mutations That Impair Iron Sulfur Cluster Delivery. J Natl Cancer I. 2016;108(1):287. doi: 10.1093/jnci/djv287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bak DW, Weerapana E. Cysteine-mediated redox signalling in the mitochondria. Mol BioSyst. 2015;11(3):678–697. doi: 10.1039/c4mb00571f. [DOI] [PubMed] [Google Scholar]
- 16.Canet-Avilés RM, Wilson MA, Miller DW, Ahmad R, McLendon C, Bandyopadhyay S, Baptista MJ, Ringe D, Petsko GA, Cookson MR. The Parkinson’s disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc Natl Acad Sci. 2004;101(24):9103–9108. doi: 10.1073/pnas.0402959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wabnitz GH, Goursot C, Jahraus B, Kirchgessner H, Hellwig A, Klemke M, Konstandin MH, Samstag Y. Mitochondrial translocation of oxidized cofilin induces caspase-independent necrotic-like programmed cell death of T cells. Cell Death Disease. 2010;1(7):58. doi: 10.1038/cddis.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ozawa K, Komatsubara AT, Nishimura Y, Sawada T, Kawafune H, Tsumoto H, Tsuji Y, Zhao J, Kyotani Y, Tanaka T, Takahashi R. S-nitrosylation regulates mitochondrial quality control via activation of parkin. Scientific Reports. 2013;3:2202. doi: 10.1038/srep02202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang KS, Kang SW, Woo HA, Hwang SC, Chae HZ, Kim K, Rhee SG. Inactivation of human peroxiredoxin I during catalysis as the result of the oxidation of the catalytic site cysteine to cysteine-sulfinic acid. J Biol Chem. 2002;277(41):38029–38036. doi: 10.1074/jbc.M206626200. [DOI] [PubMed] [Google Scholar]
- 20.McSTAY GP, Clarke SJ, Halestrap AP. Role of critical thiol groups on the matrix surface of the adenine nucleotide translocase in the mechanism of the mitochondrial permeability transition pore. Biochem J. 2002;367(2):541–548. doi: 10.1042/BJ20011672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paulsen CE, Carroll KS. Cysteine-mediated redox signaling: chemistry, biology, and tools for discovery. Chem Rev. 2013;113(7):4633–4679. doi: 10.1021/cr300163e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Couvertier SM, Zhou Y, Weerapana E. Chemical-proteomic strategies to investigate cysteine posttranslational modifications. BBA-Proeins Proteom. 2014;1844(12):2315–2330. doi: 10.1016/j.bbapap.2014.09.024. [DOI] [PubMed] [Google Scholar]
- 23.Chouchani ET, Kazak L, Jedrychowski MP, Lu GZ, Erickson BK, Szpyt J, Pierce KA, Laznik-Bogoslavski D, Vetrivelan R, Clish CB, Robinson AJ. Mitochondrial ROS regulate thermogenic energy expenditure and sulfenylation of UCP1. Nature. 2016;532:112–116. doi: 10.1038/nature17399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chouchani ET, Methner C, Nadtochiy SM, Logan A, Pell VR, Ding S, James AM, Cochemé HM, Reinhold J, Lilley KS, Partridge L. Cardioprotection by S-nitrosation of a cysteine switch on mitochondrial complex I. Nat Med. 2013;19(6):753–759. doi: 10.1038/nm.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, Bachovchin DA, Mowen K, Baker D, Cravatt BF. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468(7325):7901795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang C, Weerapana E, Blewett MM, Cravatt BF. A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles. Nat Methods. 2014;11(1):79–85. doi: 10.1038/nmeth.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pace NJ, Weerapana E. A competitive chemical-proteomic platform to identify zinc-binding cysteines. ACS Chem Biol. 2013;9(1):258–265. doi: 10.1021/cb400622q. [DOI] [PubMed] [Google Scholar]
- 28.Zhou Y, Wynia-Smith SL, Couvertier SM, Kalous KS, Marletta MA, Smith BC, Weerapana E. Chemoproteomic Strategy to Quantitatively Monitor Transnitrosation Uncovers Functionally Relevant S-Nitrosation Sites on Cathepsin D and HADH2. Cell Chem Biol. 2016;23(6):727–737. doi: 10.1016/j.chembiol.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yasueda Y, Tamura T, Fujisawa A, Kuwata K, Tsukiji S, Kiyonaka S, Hamachi I. A set of organelle-localizable reactive molecules for mitochondrial chemical proteomics in living cells and brain tissues. J Am Chem Soc. 2016;138(24):7592–7602. doi: 10.1021/jacs.6b02254. [DOI] [PubMed] [Google Scholar]
- 30.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443(7113):787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 31.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183(5):795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levraut J, Iwase H, Shao ZH, Vanden Hoek TL, Schumacker PT. Cell death during ischemia: relationship to mitochondrial depolarization and ROS generation. Am J Physiol Heart Circ Physiol. 2003;53(2):H549. doi: 10.1152/ajpheart.00708.2002. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Nartiss Y, Steipe B, McQuibban GA, Kim PK. ROS-induced mitochondrial depolarization initiates PARK2/PARKIN-dependent mitochondrial degradation by autophagy. Autophagy. 2012;8(10):1462–1476. doi: 10.4161/auto.21211. [DOI] [PubMed] [Google Scholar]
- 34.Qian T, Nieminen AL, Herman B, Lemasters JJ. Mitochondrial permeability transition in pH-dependent reperfusion injury to rat hepatocytes. Am J Physiol Cell Physiol. 1997;273(6):C1783–C1792. doi: 10.1152/ajpcell.1997.273.6.C1783. [DOI] [PubMed] [Google Scholar]
- 35.Santo-Domingo J, Giacomello M, Poburko D, Scorrano L, Demaurex N. OPA1 promotes pH flashes that spread between contiguous mitochondria without matrix protein exchange. EMBO J. 2013;32(13):1927–1940. doi: 10.1038/emboj.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rhee HW, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, Ting AY. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339(6125):1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong HL, Liebler DC. Mitochondrial protein targets of thiol-reactive electrophiles. Chem Res Toxicol. 2008;21(4):796–804. doi: 10.1021/tx700433m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abo M, Weerapana E. A Caged Electrophilic Probe for Global Analysis of Cysteine Reactivity in Living Cells. J Am Chem Soc. 2015;137(22):7087–7090. doi: 10.1021/jacs.5b04350. [DOI] [PubMed] [Google Scholar]
- 39.Frezza C, Cipolat S, Scorrano L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured filroblasts. Nat Protoc. 2007;2(2):287–295. doi: 10.1038/nprot.2006.478. [DOI] [PubMed] [Google Scholar]
- 40.Qian Y, Martell J, Pace NJ, Ballard TE, Johnson DS, Weerapana E. An Isotopically Tagged Azobenzene-Based Cleavable Linker for Quantitative Proteomics. ChemBioChem. 2013;14(12):1410–1414. doi: 10.1002/cbic.201300396. [DOI] [PubMed] [Google Scholar]
- 41.Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016;44(D1):D336–D342. doi: 10.1093/nar/gkv1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.UniProt, C. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith AC, Robinson AJ. MitoMiner, an integrated database for the storage and analysis of mitochondrial proteomics data. Mol Cell Proteom. 2009;8(6):1324–1337. doi: 10.1074/mcp.M800373-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith AC, Blackshaw JA, Robinson AJ. MitoMiner: a data warehouse for mitochondrial proteomics data. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr1101. gkr1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calvo SE, Clauser KR, Mootha VK. MitoCarta2. 0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv1003. gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stehling O, Lill R. The role of mitochondria in cellular iron–sulfur protein biogenesis: mechanisms, connected processes, and diseases. Cold Spring Harbor Perspectives in Biology. 2013;5(8):a011312. doi: 10.1101/cshperspect.a011312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stehling O, Wilbrecht C, Lill R. Mitochondrial iron–sulfur protein biogenesis and human disease. Biochimie. 2014;100:61–77. doi: 10.1016/j.biochi.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 50.Banci L, Bertini I, Cantini F, Ciofi-Baffoni S, Gonnelli L, Mangani S. Solution structure of Cox11: a novel type of β-immunoglobulin-like fold involved in CuB site formation of cytochrome c oxidase. J Biol Chem. 2004 doi: 10.1074/jbc.M403655200. [DOI] [PubMed] [Google Scholar]
- 51.Carr HS, George GN, Winge DR. Yeast Cox11, a protein essential for cytochrome c oxidase assembly, is a Cu (I)-binding protein. J Biol Chem. 2002;277(34):31237–31242. doi: 10.1074/jbc.M204854200. [DOI] [PubMed] [Google Scholar]
- 52.Osawa T, Ito K, Inanaga H, Nureki O, Tomita K, Numata T. Conserved cysteine residues of GidA are essential for biogenesis of 5-carboxymethylaminomethyluridine at tRNA anticodon. Structure. 2009;17(5):713–724. doi: 10.1016/j.str.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 53.Hitosugi T, Fan J, Chung TW, Lythgoe K, Wang X, Xie J, Ge Q, Gu TL, Polakiewicz RD, Roesel JL, Chen GZ. Tyrosine phosphorylation of mitochondrial pyruvate dehydrogenase kinase 1 is important for cancer metabolism. Mol Cell. 2011;44(6):864–877. doi: 10.1016/j.molcel.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Backus KM, Correia BE, Lum KM, Forli S, Horning BD, González-Páez GE, Chatterjee S, Lanning BR, Teijaro JR, Olson AJ, Wolan DW. Proteome-wide covalent ligand discovery in native biological systems. Nature. 2016;534(7608):570–574. doi: 10.1038/nature18002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beinert H, Holm RH, Münck E. Iron-sulfur clusters: nature’s modular, multipurpose structures. Science. 1997;277(5326):653–659. doi: 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]
- 56.Romero JM, Bizzozero OA. Extracellular S-nitrosoglutathione, but not S-nitrosocysteine or N2O3, mediates protein S-nitrosation in rat spinal cord slices. J Neurochem. 2006;99(4):1299–1310. doi: 10.1111/j.1471-4159.2006.04180.x. [DOI] [PubMed] [Google Scholar]
- 57.Chen YJ, Lu CT, Su MG, Huang KY, Ching WC, Yang HH, Liao YC, Chen YJ, Lee TY. dbSNO 2.0: a resource for exploring structural environment, functional and disease association and regulatory network of protein S-nitrosylation. Nucleic Acids Res. 2015;43(D1):D503–D511. doi: 10.1093/nar/gku1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Colombini M. VDAC structure, selectivity, and dynamics. BBA-Biomembranes. 2012;1818(6):1457–1465. doi: 10.1016/j.bbamem.2011.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Pinto V, Reina S, Gupta A, Messina A, Mahalakshmi R. Role of cysteines in mammalian VDAC isoforms’ function. BBA-Bioenergetics. 2016;1857(8):1219–1227. doi: 10.1016/j.bbabio.2016.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


