Abstract
Precision Medicine involves the delivery of a targeted, personalized treatment for a given patient. By harnessing the power of electronic health records (EHR), we are increasingly able to practice precision medicine to improve patient outcomes. In this article, we introduce the scientific community at large to important building blocks for personalized treatment, such as terminology standards that are the foundation of the EHR and allow for exchange of health information across systems. We briefly review different types of clinical decision support (CDS) and present the current state of CDS, which is already improving the care patients receive with genetic profile-based tailored recommendations regarding diagnostic and treatment plans. We also report on limitations of current systems, which are slowly beginning to integrate new genomic data into patient records but still present many challenges. Finally, we discuss future directions and how the EHR can evolve to increase the capacity of the healthcare system in delivering Precision Medicine at the point of care.
Graphical Abstract
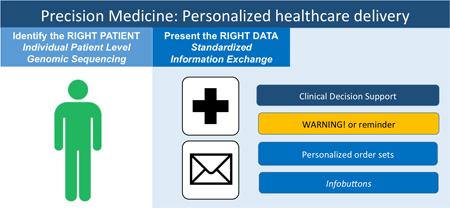
Building blocks of personalized healthcare delivery
PRECISION MEDICINE, ELECTRONIC HEALTH RECORDS, AND CLINICAL DECISION SUPPORT
In an era where the complexity of medicine grows exponentially, we face an acute need to develop systems that will integrate disparate data sources, provide real-time decision support, and enhance our ability to positively impact patient outcomes. The full spectrum of Precision Medicine spans the discovery of a patient-specific pattern of disease progression, determination of the precise therapy for that pattern, and the corresponding personalized delivery of care. EHRs are instrumental across this spectrum, but in this article we will focus on personalized healthcare delivery based on the rapidly evolving knowledge base brought about by advances in genomic medicine. Integration of implementation science with basic and translational sciences is essential to fully realize the potential of therapeutic discoveries. The landscape of the electronic health record has substantially evolved over the course of the past decade from basic adoption to sophisticated decision support. The evolution of complex terminologies that serve to electronically communicate shared data is a critical component. We explain here key terms and contextual applications of EHRs (Appendix 1). The following two example cases highlight current opportunities and challenges in personalized treatment given the current state of EHR systems.
Personalized treatment: example use cases
Case 1: N.M. is a 72 years old man with a history of atrial fibrillation and prior stroke who had been taking anticoagulant medication (warfarin) for 1 year. He presented with 7 days of runny nose, fever, and a new onset of pain in the left ear. On exam, he was noted to have otitis media, and his doctor was planning to start him on penicillin. He was aware of an increased risk of minor bleeding due to this drug combination (warfarin and penicillin), but felt that the risks were small. Three days later, the patient presented with sudden onset headache, weakness of the right arm, and difficulty with speech and vision. He was transported via ambulance to the hospital due to concern for stroke. On CT imaging of the brain, he was noted to have an acute stroke due to a subarachnoid hemorrhage, and underwent emergent neurosurgery. On further lab work, his INR was noted to be 8.4, significantly increased from a value of 2.1 three days before (the target INR range on warfarin is between 2 and 3). A potential explanation was that the interaction between warfarin and penicillin contributed to this increase in INR. As the patient was recovering in the ICU, his family brought in reports of a commercial genotyping assay that the patient had performed recently as part of a research study in another facility. That testing revealed the patient carried a specific CYP2C9 variant, which significantly increases the risk of life-threatening bleeding in patients taking warfarin and penicillin together.
This bad outcome could have been averted if the genetic testing result had been available via health information exchange between the two organizations, and a point-of-care clinical decision support (CDS) system was in place to alert the prescribing physician of this potentially significant drug-drug interaction.1
Case 2: C.P. is a 68 years old female diagnosed with breast cancer that was discovered by screening mammography. As with many breast cancer patients, C.P. has been asked to make a variety of treatment decisions. After much deliberation, she settled on lumpectomy and radiation. Her surgery went smoothly, as did radiation. She was found to have a 1 cm low-grade tumor and a negative lymph node biopsy. Her tumor was positive for estrogen and progesterone receptors. The next step along her treatment pathway would be chemotherapy. Like many patients her age, C.P. was hesitant about undergoing chemotherapy.2 Chemotherapy is one of the riskiest components of the breast cancer treatment, exposing patients to an increased risk of serious infections, cardiopulmonary complications and poorly tolerated side effects including nausea and hair loss.3 It has been shown to decrease risk of metastasis in many trials.4 Small tumors (<.5 cm) in patients with negative lymph nodes do not require chemotherapy but, given the size of C.P.’s tumor, there was no clear recommendation.5 Without a precision medicine approach, clinicians would attempt to integrate information about C.P.’s health status, personal preferences and data from large studies to decide about chemotherapy. Instead, C.P.’s oncologist employed a 21-gene recurrence score (RS) assay to predict the likelihood of distant metastasis and assess the benefit she would get from chemotherapy.6 C.P.’s score indicated high risk of recurrence and chemotherapy was recommended.
The RS has been shown to alter treatment recommendations for breast cancer patients, such as in this case.7 Its incorporation into the EHR is a clear example of CDS.
BUILDING BLOCKS: STANDARDIZING AND EXCHANGING DATA TO DRIVE CLINICAL DECISION SUPPORT
Adoption of EHR and order entry systems has continued to increase, spurred by federal incentives and mandates.8 These electronic systems facilitate increasingly effective CDS, defined by (healthit.gov) as systems or processes that “[provide] clinicians, staff, patients or other individuals with knowledge and person-specific information, intelligently filtered or presented at appropriate times, to enhance health and health care.”9
A number of accurate data sources are required to drive appropriate opportunities for CDS. As illustrated in the two cases above, a substantial amount of data needs to be present at the time of care delivery in order to optimize opportunities for improved outcomes and treatment. The respective data sources include different types, such as diagnoses, ordered medications, completed procedures, and completed lab tests. Appendix 1 provides a glossary of key terms related to the use and sharing of clinical data as contextualized by the preceding cases.
Data Models and Terminology Standards
Each type of data has an associated terminology that enables the vocabulary to be operationalized within the context of the EHR. These terminology systems have unique data formatting, coding, domain coverages, and hierarchical relationships between a specific instantiation, such as amoxicillin capsule 250 mg, and a concept, such as penicillin. Terminology standards have the ability to influence design and utilization of the respective data. For example, a medication such as warfarin (Case 1) in the National Drug Code (NDC) file will include not only the medication itself, but also the type and strength of the product (e.g., 5 mg tablet) and the quantity included in the product (e.g., 100 tablets).10, 11
In order to facilitate common data formatting and structure that enables effective communication between healthcare organizations, the use of standardized medical terminologies is critical. Current opportunities and challenges with the use and exchange of health data are listed in Table 1. Often, new data types have complexity that is not adequately represented or matured in the standard vocabulary of its domain. In addition, non-automated processes such as specimen interpretation by a pathologist or image interpretation by a radiologist are dictated and archived as free text in an unstructured, non-standardized format. Relevant data need to be identified and parsed from the unstructured narrative texts using natural language processing (NLP) techniques so that they can be used by a CDS tool.
Table 1.
Data Sources for EHR relevant to drive Clinical Decision Support
| Type of Information |
Standardization | Opportunities | Challenges |
|---|---|---|---|
| Laboratory | LOINC, HGVS, HL7 FHIR value sets |
Clinical laboratory tests have a mature standardization capabilities via LOINC LOINC and HL7 genomics groups have started developing standards for genetic tests - that enable standardized discrete coding of some genetic test information |
|
| Medication | RxNorm, NDC | Clinical drug names have been standardized using these codes Dictionaries provide the opportunity to include manufacturer, dosing, and route information |
|
| Diagnosis | ICD 9, ICD 10, SNOMED-CT |
Most institutions adopt ICD system to support both active problem lists and encounter diagnoses Diagnosis names are interrelated; meaning that terms encoded with other one terminology such as SNOMED-CT, can be converted to ICD through cross-mapping established between the two systems |
|
| Radiology | RadLex, SNOMED-CT DICOM |
Standards to capture the key findings and metadata about the radiologic studies exist |
|
| Pathology | SNOMED-CT HL7 (anatomic pathology) |
Standards to capture the key findings and metadata about the pathology test exist NAACCR is interested in adopting standard for cancer pathology reporting |
|
| Clinical Evidence & Outcomes |
OMOP CDM and all terminology systems listed above |
EHR data stored in a clinical data warehouse serves a powerful knowledge resource OMOP CDM is recognized as a de facto standard and adopted by many institutions |
|
| Procedures | Terms to represent clinical procedures |
Standardized terms that define common clinical procedures and their associated charges |
|
Causality and association are often inferred from context. Some EHR systems do not accurately code the time of many events. Additionally, many variables lack intensity qualifiers. For instance, most symptoms are coded as present or absent and are not scaled (pain is an exception).
Precision Medicine is developing a new vocabulary related to genetic conditions, which has yet to be standardized in the EHR. Genetic test results should follow relevant data standards, such as LOINC, HL7 Genomics, HGVS, etc., that contain information about test findings and potential risk; yet, this a challenge since these standards are not adopted by all laboratories. The rapid evolution of tests makes this challenging for the field of genetics, posing challenges for discrete data retrieval of this information in the EHR. Precision medicine also relies on other types of data that were not traditionally recorded in EHRs. Patient reported outcomes (PROs) are still early in standardization and the reporting is highly variant according to race, ethnicity, and literacy. PROs are vital to enhance our understanding of the value of healthcare to its primary “customers”. Patient preferences, for example, are not universally standardized today but this is necessary because they are intimately connected with the definition of “value”.
The process of translating clinical information across terminologies is called “data harmonization”. Mapping between terminology systems makes this possible, but increases complexity and the risk of introducing errors. Yet, this mapping across standard terminologies is essential for clinical decision support, quality measurement, research, and information exchange across healthcare systems.
Health Information exchange
The Health Information Technology for Economics and Clinical Health (HITECH) Act of 2009 was proposed to promote interoperable health information.12, 13 Meaningful Use of EHR is an incentive program put forth by the Medicare and Medicaid programs in response to HITECH to promote effective use of EHR systems.14, 15 The receipt of the incentive payment requires hospitals and providers to prove their “meaningful use” of EHR by satisfying a set of requirements such as recording patient information in a structured format, ordering medications using a computerized order entry system, and sharing test results with patients using personal health records tethered to the EHR system. Health information exchange (HIE) initiatives aim at realizing timely and appropriate level of access to the patient level of health information stored in the EHR by healthcare providers through a secure means to exchanging health data among healthcare organizations.16 Having complete information about disease progression and treatment data at the point of care helps healthcare providers make better treatment decisions and achieve better patient outcomes. Utilizing information collected from different healthcare systems is an important step towards this goal. The Nationwide Health Information Network (NwHIN) specifies data and messaging standards, services, and policies required realizing secure exchange of health information on Internet.17
HIE covers three types of data exchange: (1) Directed exchange that occurs between healthcare providers to complete the planned healthcare services such as sending and receiving laboratory test orders and results, exchanging patient referral documents, etc. (2) Query-based exchange that occurs when a healthcare provider delivers unplanned services and requires accessing necessary health information about the patient. For example, when an emergency room physician needs to access patient’s disease history, current medications, allergies, etc. (3) Consumer-mediated exchange that lets patients control their health information. In this model, patients grant access to their health information to healthcare providers.18
However, establishing a sustainable HIE is not a trivial task; there are a number of technical and non-technical barriers that need to be addressed first. For example, lack of business incentives, specifically concerns on losing patients to other hospitals by making their health data available anywhere, has long been recognized as a factor that makes some healthcare systems hesitant to embrace HIEs.8, 19 Patients and providers sometimes opt out from HIEs due to privacy concerns.20 Other recognized challenges are poor data standardization,19 inefficient processes of sorting through overloaded unselective information of a patient,21 and difficulties in understanding the shared data in the absence of context when detailed clinical notes are withheld due to privacy concerns.22
DELIVERING PERSONALIZED CARE WITH CDS SYSTEMS
CDS systems help providers and patients answer certain types of questions in the course of care, such as what the most likely diagnosis is, what tests are most appropriate to arrive at the diagnosis, and what treatment would be best. In addition, CDS can help optimize the effectiveness and efficiency of care delivery, and can highlight when patients’ conditions do not follow expected trajectories. (Appendix 2)
Effective CDS can be constructed in various ways. Some tools, such as Infobuttons, provide individuals ready access to relevant clinical guidelines when actively sought out or “pulled.”23 Other systems, including alerts, reminders, and event detectors, will proactively “push” information to individuals with varying levels of interruption and urgency. Certain CDS tools can straddle the “push” or “pull” approaches depending on how well they can be integrated into the EHR. For example, risk calculators or differential diagnosis generators can be automatically engaged if all the required data elements are available; otherwise, they can be made available for individuals to enter the data manually. Another class of CDS systems is designed to provide clinicians guidance on the appropriate evidence-based care for certain disease states, and to reduce variation in delivery of that care. These systems include order sets, care pathways, and documentation templates. Finally, data summarization and visualization tools address the growing issue of “information overload” facing patients and clinicians and provide CDS by displaying a filtered version of clinical data in a manner better aligned with human cognition and decision-making.
Early leaders in CDS have provided valuable lessons and best practices to maximize the impact of these systems.24, 25 One framework developed by Osheroff and colleagues is known as the “five rights”—that effective CDS requires that the right information be provided to the right person in the right format and communication channel at the right time in the workflow.24 This framework highlights many of the challenges faced when designing CDS systems for use in clinical settings. Often, such systems have failed to demonstrate tangible improvements when the five rights are not appropriately addressed; even if they are, the rigidity of the support tools or the phenomenon of alert fatigue may limit the effectiveness of CDS.26 Another significant challenge is the ongoing maintenance of the knowledge base underlying CDS systems, as new clinical research informing these tools is being constantly generated.
There are some published examples of CDS solutions which overcome these challenges. Two of the most successful and widely used applications of computerized clinical decision support have been evidence-based order sets and alerts (for minimizing medication alerts, especially of drug-drug interactions and drug dose adjustment). For example, Ballard and colleagues observed that implementation of a standardized heart failure order set resulted in reduced inpatient mortality (odds ratio [OR], 0.49; 95% confidence intervals [CI], 0.28–0.88), and improved compliance with core measures (OR, 1.51; 95% CI, 1.08–2.12).27 Similarly, with the use of a smart order set on evidence-based, risk-appropriate venous thromboembolism prophylaxis, Zeidan and colleagues observed a significant decline in 90-d risk of venous thromboembolism after hospital discharge (pre- vs. post: 2.5% vs. 0.7%, p=0.002) and complete elimination of preventable harm (1.1% vs. 0%, p<0.001), paralleling an increase in prescription of risk-appropriate venous thromboembolism prophylaxis (65.6% vs. 90.1%, p<0.001).28 To evaluate the impact of clinical decision support on medication prescription patterns in patients with kidney disease (who often require dose adjustment or drug discontinuation of certain medications based on dynamic renal function), Awdishu and colleagues designed a cluster-randomized trial comparing clinical decision support through real-time alerts generated based on dynamic and integrated monitoring of renal function vs. usual workflow in patients with kidney disease. Over a course of 1 year, 4068 alerts were generated recommending either dose adjustment (n=827) or drug discontinuation (n=3241). The investigators observed that physicians randomized to CDS were significantly more likely to make drug adjustments, as compared to physicians in usual workflow (17.0% vs. 5.7%, p<0.001).29
In our examples, Case 1 showed that the CYP2C9 variant could explain why the co-administration of both penicillin and warfarin resulted in the higher than intended level of anticoagulation and the subsequent development of a hemorrhagic stroke. It illustrates several requirements for effective CDS. First, the CYP2C9 test occurred at an outside hospital – mechanisms for health information exchange between the hospitals would have been needed to bring the test information to the clinician at the point of care. Second, the provider had to agree that this variant implied an increased risk for a dangerous interaction between these two medications. An alert would have needed to be in place to notify the clinician at the time penicillin was ordered, and to outline the risks of giving penicillin and warfarin together for this patient. Such a CDS tool could have guided the clinician toward better treatment by suggesting an alternative medication.
Case 2 highlights the potential ways by which CDS systems can improve the quality, safety and efficiency of the care delivered by our health care system. A risk assessment tool, such as OncotypeDx,30 indicated the risk of cancer recurrence and clarified the risk-benefit tradeoff of undergoing chemotherapy. To get to that point, her clinician needed to be aware of the appropriateness of this test for her particular scenario as well as be able to accurately interpret the test results. Different CDS systems (Table 2) might make this process more reliable, including: 1) order sets or care pathways that guide clinicians caring for similar patients down a step-wise decision-making process that includes this specific tumor genome panel; 2) alerts or reminders for the clinician to order the tumor genome panel, if appropriate; and 3) Infobuttons that provide the patient and clinician access to current guidelines and test interpretation.
Table 2.
Types of clinical decision support. Adapted from Greenes1.
| CDS type | Prerequisites | Strengths | Weaknesses | Example(s) |
|---|---|---|---|---|
| Links
to references InfoButtons |
Ability to link patient context with specific information sources |
Facilitates access to current best evidence |
Only engaged proactively by provider or patient (pull-mode) |
KnowledgeLink31 |
| Diagnosis (Dx) generators |
Accurate identification of clinical features of patient’s condition |
Facilitates
Bayesian reasoning May improve effectiveness and efficiency of diagnostic testing strategy |
Dx algorithms may rely on unstructured data (needs manual entry or NLP) Have been difficult to integrate into clinical workflow |
DxPlain32, Isabel33, VisualDx34 |
| Probability calculators/Risk scoring systems |
Access to components of model in computable format |
Provides individualized predictions for prognosis or risk of events |
Often difficult to obtain confidence intervals of predictions for individuals. |
Yale New Haven Readmission Risk Score35 |
| Alerts, reminders and rule-based event detection |
Development of reliably computable definition of event or condition |
Can bring important or urgent scenarios to attention of decision- makers |
Patients and providers can become desensitized to alerts (alert fatigue). |
Alerts for medications in kidney disease36 and temporary catheter use.37 |
| Evidence-based ordersets, care pathways and protocols |
Careful review and understanding of ideal process |
Facilitates evidence- based care May reduce variation of care |
Labor-intensive
to create May increase steps to placing orders |
Ordersets for heart failure38 and venous thromboembolis m prophylaxis.39 |
| Documentation templates |
General agreement of ideal documentation |
Can improve efficiency and completeness of documentation Can prompt consideration of diagnoses, treatments or care coordination |
In some circumstances, may increase time of documentation May be difficult to document nuances of clinical scenario |
Templates for documentation for breast cancer tumor board40 and palliative care encounters.41 |
| Data visualization and summarization techniques |
Reliable methods to categorize and condense underlying data elements |
Highlights key information for decision-making Reduces cognitive load Improves ability to make causal inferences from disparate data |
Labor-intensive to create |
Lifelines42 and problem- oriented display of health records.43 |
Simple CDS as described in the example cases is currently available. However, as the use of genetic tests based on whole genome sequences becomes more common, the evidence base will have to evolve to support their use in guiding clinical decisions. Since practicing clinicians will likely find it difficult to keep their knowledge up to date in this area, it is critical that interpretation and recommendations be done by more sophisticated CDS that operates “behind the scenes”. CDS systems will play a large role in Precision Medicine by incorporating the evidence base into clinical practice. However, maintaining the underlying knowledge base driving such systems will require significant effort.
THE ROAD AHEAD
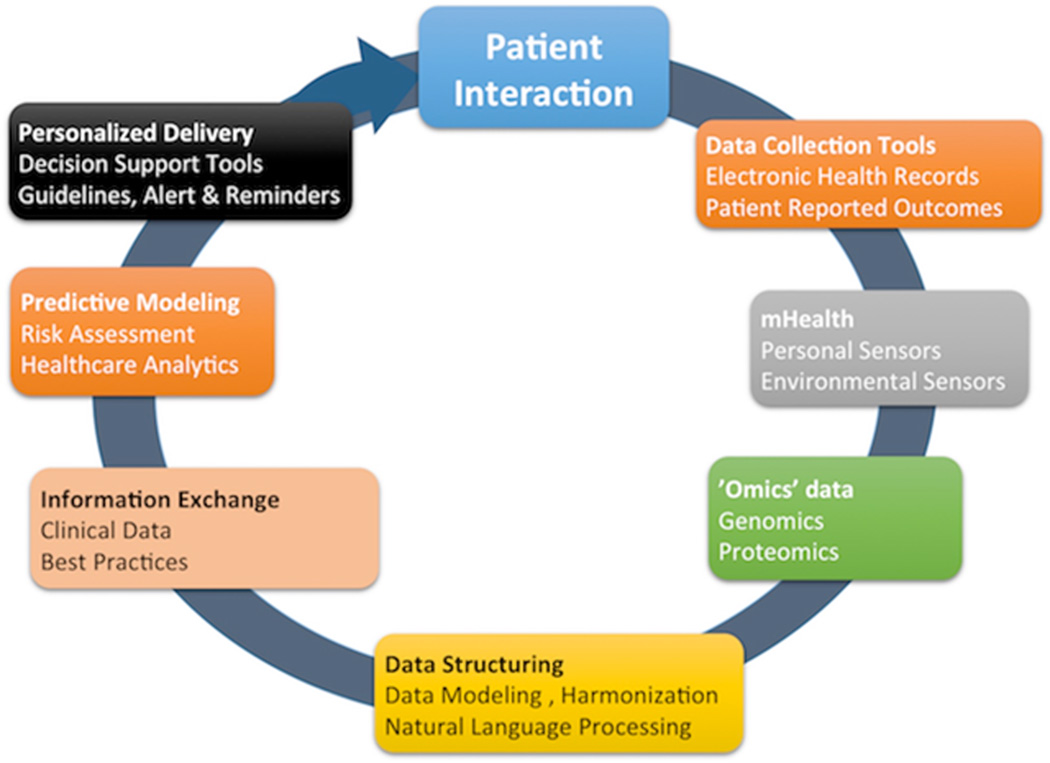
Realizing the goals envisioned that electronic health records will enable us to provide better, safer, and more effective care is an active pursuit for our healthcare systems. Through data integration and real-time decision support, we have the capacity to alter patient outcomes and drive value-driven care. Precision Medicine impresses the need to rapidly adopt an evolving knowledge base brought about by advances in genomic medicine and making it actionable by putting it at the hands of clinicians who can intervene. To get there, we need vision, a culture of sharing, commitment to standardized terminologies, and iterative learning. Our knowledge on the associations among gene, disease, and the effectiveness of various therapeutic approaches is still quite limited.44–48 Making new discoveries at the molecular or cell level is still much needed thus often the emphasis of articles describing Precision Medicine but it is actually one component in a vast spectrum of activities that are necessary to make it happen in practice. At the end of this spectrum lies implementation through guidance of actions by patients, caregivers, and healthcare providers. We focused this article at this less prominently but equally important component of Precision Medicine to help the scientific community at large understand why it is critical to “close the loop” (Figure 1).
Figure 1.
Key components of Precision Medicine
Acknowledgments
HK and LOM are partially supported by CDRN-1306-04819 from PCORI, U24AI117966, R01HG008802, and UL1TR001442 from NIH. REK is partially funded by K22LM011435. SS and RM are supported by T15LM011271 from NIH.
Appendix 1
Glossary of Key Terms including Reference and Case Context
| Abbreviation | Full name | Reference | Case Context |
|---|---|---|---|
| CDM49–52 | Common Data Model |
http://mini-sentinel.org/data_activities/distributed_db_and_data/default.aspx | Standardized data
storage Many Common Data Models exist for various use cases |
| OMOP53 | Observational Medical Outcomes Partnership |
http://www.ohdsi.org/data-standardization/the-common-data-model/ | Standardized formatting of data |
| LOINC | Logical Observation Identifiers Names and Codes |
http://loinc.org | Lab formatting and code for the INR test |
| NDC | National Drug Code |
http://www.fda.gov/Drugs/InformationOnDrugs/ucm142438.htm | Code to support identification of
Warfarin or chemotherapy medication |
| RxNorm | Normalized names for clinical drug (Rx) |
https://www.nlm.nih.gov/research/umls/rxnorm/ | Normalized names to support warfarin
in system |
| ICD | International Classification of Diseases |
http://www.who.int/classifications/icd/en/ | Classification of specific disease
such as intermittent atrial fibrillation |
| SNOMED-CT | Systematized Nomenclature of Medicine – Clinical Terms |
http://www.ihtsdo.org/snomed-ct | Groupings of clinical terms that
enable aggregation of concepts such as breast cancer |
| RadLex | Radiology Lexicon |
http://www.radlex.org | Terms for identification of CT imaging
or mammography |
| DICOM | Digital
Imaging and Communication in Medicine |
http://dicom.nema.org | Standard format for digital
imaging transmittal |
| HGVS | Human Genome Variation Society code |
http://www.hgvs.org | Standardized name and syntax
for describing genetic variations |
| NACCR | The
North American Association of Central Cancer Registries |
http://www.naaccr.org | Centralized repository of cancer registries |
| CPT | Current Procedural Terminology |
http://www.ama-assn.org/ama/pub/physician-resources/solutions-managing-your-practice/ coding-billing-insurance/cpt/about-cpt.page? |
Standardized terms that describe
procedure such as lumpectomy, R breast with axillary dissection |
| IMO | Intelligent Medical Objects |
https://www.e-imo.com | Intelligent Medical Objects is a
privately held company specializing in developing, managing and licensing medical vocabularies and terminology maps |
| PROMIS | Patient
– Reported Outcomes Measurement Information System |
http://www.healthmeasures.net/explore-measurement-systems/promis | Validated sets of measures designed
assess physical, emotional, social aspects of health |
| Exchange of Health Information | |||
| HIE54 | Health Information Exchange |
https://www.healthit.gov/providers-professionals/health-information-exchange/what-hie | Shared EMR based results between
health systems related to hospitalization from hemorrhage |
| Communication Standards | |||
| HL7 | Health Level 7 | http://www.hl7.org | Formatting that enables import of a
file about genetic test into the EHR |
| C-CDA55 | Consolidated Clinical Document Architecture |
http://www.hl7.org/implement/standards/product_brief.cfm?product_id=258 | File formatting for data exchange |
| FHIR | Fast
Healthcare Interoperability Resource |
http://www.hl7.org/fhir/?ref=learnmore | Specifications that support complex
record exchange between organizations |
Appendix 2
Purposes and methodologies of clinical decision support. Adapted from Greenes1
| Purpose | Potential methodologies |
|---|---|
| Answering questions | Links to
references InfoButtons |
| Making decisions | |
| • Diagnosis | Differential Diagnosis
generators Probability calculators Alerts and reminders Documentation templates |
| • Test selection | Evidence-based
ordersets Alerts and reminders Documentation templates |
| • Choice of treatment | Evidence-based
ordersets Documentation templates |
| • Prognosis | Predictive
modeling Risk scoring systems |
| Optimizing workflow | Care pathways and
protocols Evidence-based ordersets Alerts and reminders Documentation templates |
| Monitoring actions | Alerts and
reminders Rule-based event detection |
| Focusing attention
and enhancing visualization |
Data visualization
and summarization techniques |
Footnotes
The authors do not have any conflict of interests to report on this work.
Contributor Information
Amy Sitapati, UC San Diego, Department of Medicine, Health System Department of Biomedical Informatics, asitapati@ucsd.edu.
Hyeoneui Kim, UC San Diego, Department of Medicine, Health System Department of Biomedical Informatics, Hyk038@ucsd.edu.
Barbara Berkovich, UC San Diego Health System, bberkovich@ucsd.edu.
Rebecca Marmor, UC San Diego, Department of Medicine, Health System Department of Biomedical Informatics, marmor@ucsd.edu.
Siddharth Singh, UC San Diego, Department of Medicine, Health System Department of Biomedical Informatics, sis040@ucsd.edu.
Robert El-Kareh, UC San Diego, Department of Medicine, Health System Department of Biomedical Informatics, relkareh@ucsd.edu.
Brian Clay, UC San Diego, Department of Medicine, Health System Department of Biomedical Informatics, bclay@ucsd.edu.
Lucila Ohno-Machado, UC San Diego, Department of Medicine, Health System Department of Biomedical Informatics, machado@ucsd.edu.
References
- 1.Greenes RA. Features of Computer-Based Clinical Decision Support. In: Greenes RA, editor. Clinical Decision Support. 2014. [Google Scholar]
- 2.Hamelinck VC, Bastiaannet E, Pieterse AH, de Glas NA, Portielje JE, Merkus JW, et al. A Prospective Comparison of Younger and Older Patients' Preferences for Adjuvant Chemotherapy and Hormonal Therapy in Early Breast Cancer. Clinical breast cancer. 2016 doi: 10.1016/j.clbc.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Hassett MJ, O'Malley AJ, Pakes JR, Newhouse JP, Earle CC. Frequency and cost of chemotherapy-related serious adverse effects in a population sample of women with breast cancer. Journal of the National Cancer Institute. 2006;98(16):1108–1117. doi: 10.1093/jnci/djj305. [DOI] [PubMed] [Google Scholar]
- 4.Early Breast Cancer Trialists' Collaborative Group (EBCTCG) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. The Lancet. 2005;365(9472):1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 5.National Comprehensive Cancer Network (NCCN) NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) Breast Cancer [Internet]. Version 1. 2016:1–191. [2016]. Available from: https://www.nccn.org/professionals/physician_gls/f_guidelines.asp. [Google Scholar]
- 6.Paik S, Tang G, Shak S, Kim C, Baker J, Kim W, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor–positive breast cancer. Journal of clinical oncology. 2006;24(23):3726–3734. doi: 10.1200/JCO.2005.04.7985. [DOI] [PubMed] [Google Scholar]
- 7.Joh JE, Esposito NN, Kiluk JV, Laronga C, Lee MC, Loftus L, et al. The effect of oncotype DX recurrence score on treatment recommendations for patients with estrogen receptor–positive early stage breast cancer and correlation with estimation of recurrence risk by breast cancer specialists. The oncologist. 2011;16(11):1520–1526. doi: 10.1634/theoncologist.2011-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adler-Milstein J, DesRoches CM, Kralovec P, Foster G, Worzala C, Charles D, et al. Electronic health record adoption in US hospitals: progress continues, but challenges persist. Health Affairs. 2015 doi: 10.1377/hlthaff.2015.0992. 10.1377/hlthaff. 2015.0992. [DOI] [PubMed] [Google Scholar]
- 9. HealthIT.gov. Clinical Decision Support (CDS) [June 28, 2016];2016 Available from: https://www.healthit.gov/policy-researchers-implementers/clinical-decision-support-cds.
- 10.de Keizer NF, Abu-Hanna A, Zwetsloot-Schonk J. Understanding terminological systems I: terminology and typology. Methods of information in medicine. 2000;39(1):16–21. [PubMed] [Google Scholar]
- 11.Saitwal H, Qing D, Jones S, Bernstam EV, Chute CG, Johnson TR. Cross-terminology mapping challenges: a demonstration using medication terminological systems. Journal of biomedical informatics. 2012;45(4):613–625. doi: 10.1016/j.jbi.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. HealthIT.gov. Title XIII-Health Information Technology. 2009 [Google Scholar]
- 13.Waldren SKD, Mitchell J. “Will the feds really buy me an EHR?” and other commonly asked questions about the HITECH Act. Fam Pract Manag. 2009;16(4):19–23. [PubMed] [Google Scholar]
- 14.Easterly E. Medicaid Meaningful Use Incentives. The Journal of the Arkansas Medical Society. 2016;112(11):204–205. [PubMed] [Google Scholar]
- 15.Resnick C, Meara J, Peltzman M, Gilley M. Meaningful use: A program in transition. Bulletin of the American College of Surgeons. 2016;101(3):10. [PubMed] [Google Scholar]
- 16.Vest JR, Gamm LD. Health information exchange: persistent challenges and new strategies. Journal of the American Medical Informatics Association. 2010;17(3):288–294. doi: 10.1136/jamia.2010.003673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaynor M, Lenert L, Wilson KD, Bradner S. Why common carrier and network neutrality principles apply to the Nationwide Health Information Network (NWHIN) Journal of the American Medical Informatics Association. 2014;21(1):2–7. doi: 10.1136/amiajnl-2013-001719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams C, Mostashari F, Mertz K, Hogin E, Atwal P. From the Office of the National Coordinator: the strategy for advancing the exchange of health information. Health affairs. 2012;31(3):527–536. doi: 10.1377/hlthaff.2011.1314. [DOI] [PubMed] [Google Scholar]
- 19.Thorn SA, Carter MA, Bailey JE. Emergency physicians' perspectives on their use of health information exchange. Annals of emergency medicine. 2014;63(3):329–337. doi: 10.1016/j.annemergmed.2013.09.024. [DOI] [PubMed] [Google Scholar]
- 20.Mac McCullough J, Zimmerman FJ, Bell DS, Rodriguez HP. Electronic health information exchange in underserved settings: examining initiatives in small physician practices & community health centers. BMC health services research. 2014;14(1):1. doi: 10.1186/1472-6963-14-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richardson J, Malhotra S, Kaushal R Investigators wtH. A case report in health information exchange for inter-organizational patient transfers. Applied clinical informatics. 2014;5(3):642–650. doi: 10.4338/ACI-2014-02-CR-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hyppönen H, Reponen J, Lääveri T, Kaipio J. User experiences with different regional health information exchange systems in Finland. international journal of medical informatics. 2014;83(1):1–18. doi: 10.1016/j.ijmedinf.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Cimino J, editor. Infobuttons: anticipatory passive decision support; AMIA Annual Symposium proceedings/AMIA Symposium AMIA Symposium; 2007. [PubMed] [Google Scholar]
- 24.Osheroff M, Jerome A, Teich M, FHIMSS JM, Levick M, Saldana M, et al. Improving outcomes with clinical decision support: an implementer's guide. 2012 [Google Scholar]
- 25.Bates DW, Kuperman GJ, Wang S, Gandhi T, Kittler A, Volk L, et al. Ten commandments for effective clinical decision support: making the practice of evidence-based medicine a reality. Journal of the American Medical Informatics Association. 2003;10(6):523–530. doi: 10.1197/jamia.M1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ash JS, Sittig DF, Campbell EM, Guappone KP, Dykstra RH, editors. AMIA. 2007. Some unintended consequences of clinical decision support systems. [PMC free article] [PubMed] [Google Scholar]
- 27.Ballard DJ, Ogola G, Fleming NS, Stauffer BD, Leonard BM, Khetan R, et al. Impact of a standardized heart failure order set on mortality, readmission, and quality and costs of care. International Journal for Quality in Health Care. 2010;22(6):437–444. doi: 10.1093/intqhc/mzq051. [DOI] [PubMed] [Google Scholar]
- 28.Zeidan AM, Streiff MB, Lau BD, Ahmed SR, Kraus PS, Hobson DB, et al. Impact of a venous thromboembolism prophylaxis “smart order set”: improved compliance, fewer events. American journal of hematology. 2013;88(7):545–549. doi: 10.1002/ajh.23450. [DOI] [PubMed] [Google Scholar]
- 29.Awdishu L, Coates CR, Lyddane A, Tran K, Daniels CE, Lee J, et al. The impact of real-time alerting on appropriate prescribing in kidney disease: a cluster randomized controlled trial. Journal of the American Medical Informatics Association. 2016;23(3):609–616. doi: 10.1093/jamia/ocv159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. New England Journal of Medicine. 2004;351(27):2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 31.Maviglia SM, Yoon CS, Bates DW, Kuperman G. KnowledgeLink: impact of context-sensitive information retrieval on clinicians' information needs. Journal of the American Medical Informatics Association : JAMIA. 2006;13(1):67–73. doi: 10.1197/jamia.M1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elkin PL, Liebow M, Bauer BA, Chaliki S, Wahner-Roedler D, Bundrick J, et al. The introduction of a diagnostic decision support system (DXplain) into the workflow of a teaching hospital service can decrease the cost of service for diagnostically challenging Diagnostic Related Groups (DRGs) International journal of medical informatics. 2010;79(11):772–777. doi: 10.1016/j.ijmedinf.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.John RM, Hall E, Bakken S. Use of the isabel decision support system to improve diagnostic accuracy of pediatric nurse practitioner and family nurse practitioner students. NI 2012: 11th International Congress on Nursing Informatics, June 23–27, 2012, Montreal, Canada International Congress in Nursing Informatics. 2012;2012:194. [PMC free article] [PubMed] [Google Scholar]
- 34.Vardell E, Bou-Crick C. VisualDx: a visual diagnostic decision support tool. Medical reference services quarterly. 2012;31(4):414–424. doi: 10.1080/02763869.2012.724287. [DOI] [PubMed] [Google Scholar]
- 35.Lindenauer PK, Normand SL, Drye EE, Lin Z, Goodrich K, Desai MM, et al. Development, validation, and results of a measure of 30-day readmission following hospitalization for pneumonia. Journal of hospital medicine. 2011;6(3):142–150. doi: 10.1002/jhm.890. [DOI] [PubMed] [Google Scholar]
- 36.Awdishu L, Coates CR, Lyddane A, Tran K, Daniels CE, Lee J, et al. The impact of real-time alerting on appropriate prescribing in kidney disease: a cluster randomized controlled trial. Journal of the American Medical Informatics Association : JAMIA. 2016;23(3):609–616. doi: 10.1093/jamia/ocv159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rattanaumpawan P, Teeratorn N, Thamlikitkul V. A Cluster-Randomized Controlled Trial of the Catheter Reminder and Evaluation Program. Infection control and hospital epidemiology. 2016;37(2):231–233. doi: 10.1017/ice.2015.262. [DOI] [PubMed] [Google Scholar]
- 38.Ballard DJ, Ogola G, Fleming NS, Stauffer BD, Leonard BM, Khetan R, et al. Impact of a standardized heart failure order set on mortality, readmission, and quality and costs of care. International journal for quality in health care : journal of the International Society for Quality in Health Care/ISQua. 2010;22(6):437–444. doi: 10.1093/intqhc/mzq051. [DOI] [PubMed] [Google Scholar]
- 39.Zeidan AM, Streiff MB, Lau BD, Ahmed SR, Kraus PS, Hobson DB, et al. Impact of a venous thromboembolism prophylaxis "smart order set": Improved compliance, fewer events. American journal of hematology. 2013;88(7):545–549. doi: 10.1002/ajh.23450. [DOI] [PubMed] [Google Scholar]
- 40.Farrugia DJ, Fischer TD, Delitto D, Spiguel LR, Shaw CM. Improved Breast Cancer Care Quality Metrics After Implementation of a Standardized Tumor Board Documentation Template. Journal of oncology practice/American Society of Clinical Oncology. 2015;11(5):421–423. doi: 10.1200/JOP.2015.003988. [DOI] [PubMed] [Google Scholar]
- 41.Lamba S, Berlin A, Goett R, Ponce CB, Holland B, Walther S, et al. Assessing Emotional Suffering in Palliative Care: Use of a Structured Note Template to Improve Documentation. Journal of pain and symptom management. 2016;52(1):1–7. doi: 10.1016/j.jpainsymman.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 42.Plaisant C, Mushlin R, Snyder A, Li J, Heller D, Shneiderman B. LifeLines: using visualization to enhance navigation and analysis of patient records. Proceedings/AMIA Annual Symposium AMIA Symposium. 1998:76–80. [PMC free article] [PubMed] [Google Scholar]
- 43.Bashyam V, Hsu W, Watt E, Bui AA, Kangarloo H, Taira RK. Problem-centric organization and visualization of patient imaging and clinical data. Radiographics : a review publication of the Radiological Society of North America, Inc. 2009;29(2):331–343. doi: 10.1148/rg.292085098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ashley EA. Towards precision medicine. Nat Rev Genet. 2016;17(9):507–522. doi: 10.1038/nrg.2016.86. [DOI] [PubMed] [Google Scholar]
- 45.Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nature Reviews Genetics. 2011;12(11):745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 46.Camicia R, Winkler HC, Hassa PO. Novel drug targets for personalized precision medicine in relapsed/refractory diffuse large B-cell lymphoma: a comprehensive review. Molecular cancer. 2015;14(1):1. doi: 10.1186/s12943-015-0474-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tenenbaum JD. Translational Bioinformatics: Past, Present, and Future. Genomics, proteomics & bioinformatics. 2016;14(1):31–41. doi: 10.1016/j.gpb.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Collins FS, Varmus H. A new initiative on precision medicine. New England Journal of Medicine. 2015;372(9):793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hripcsak G, Duke JD, Shah NH, Reich CG, Huser V, Schuemie MJ, et al. Observational Health Data Sciences and Informatics (OHDSI): opportunities for observational researchers. Studies in health technology and informatics. 2015;216:574. [PMC free article] [PubMed] [Google Scholar]
- 50.FitzHenry F, Resnic F, Robbins S, Denton J, Nookala L, Meeker D, et al. Creating a common data model for comparative effectiveness with the observational medical outcomes partnership. Applied clinical informatics. 2015;6(3):536–547. doi: 10.4338/ACI-2014-12-CR-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reisinger SJ, Ryan PB, O'Hara DJ, Powell GE, Painter JL, Pattishall EN, et al. Development and evaluation of a common data model enabling active drug safety surveillance using disparate healthcare databases. J Am Med Inform Assoc. 2010;17(6):652–662. doi: 10.1136/jamia.2009.002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ogunyemi OI, Meeker D, Kim H-E, Ashish N, Farzaneh S, Boxwala A. Identifying Appropriate Reference Data Models for Comparative Effectiveness Research (CER) Studies Based on Data from Clinical Information Systems. Medical Care. 2013;51:S45–S52. doi: 10.1097/MLR.0b013e31829b1e0b. [DOI] [PubMed] [Google Scholar]
- 53.Observational Health Data Sciences and Informatics. [July 29, 2016];OMOP Common Data Model V5.0.1. 2014 Available from: http://www.ohdsi.org. [Google Scholar]
- 54.Williams C, Mostashari F, Mertz K, Hogin E, Atwal P. From the Office of the National Coordinator: the strategy for advancing the exchange of health information. Health Aff (Millwood) 2012;31(3):527–536. doi: 10.1377/hlthaff.2011.1314. [DOI] [PubMed] [Google Scholar]
- 55.D'Amore JD, Mandel JC, Kreda DA, Swain A, Koromia GA, Sundareswaran S, et al. Are Meaningful Use Stage 2 certified EHRs ready for interoperability? Findings from the SMART C-CDA Collaborative. Journal of the American Medical Informatics Association. 2014;21(6):1060–1068. doi: 10.1136/amiajnl-2014-002883. [DOI] [PMC free article] [PubMed] [Google Scholar]