Abstract
A safe and effective human immunodeficiency virus type 1 (HIV-1) vaccine is urgently needed, but remains elusive. While HIV-1 live-attenuated vaccine can provide potent protection as demonstrated in rhesus macaque-simian immunodeficiency virus model, the potential pathogenic consequences associated with the uncontrolled virus replication preclude such vaccine from clinical applications. We investigated a novel approach to address this problem by controlling live-attenuated HIV-1 replication through an unnatural genetic switch that was based on the amber suppression strategy. Here we report the construction of all-in-one live-attenuated HIV-1 mutants that contain genomic copy of the amber suppression system. This genetic modification resulted in viruses that were capable of multi-cycle replication in vitro and could be switched on and off using an unnatural amino acid as the cue. This stand-alone, replication-controllable attenuated HIV-1 virus represents an important step towards the generation of a safe and efficacious live-attenuated HIV-1 vaccine. The strategy reported in this work can be adopted for the development of other live-attenuated vaccines.
Keywords: HIV-1 vaccine, genetic code, unnatural amino acid, live-attenuated vaccine, virus engineering
Graphical Abstract
A safe and effective vaccine for human immunodeficiency virus type 1 (HIV-1) infection is urgently needed, but remains elusive.1 Among all the HIV-1 vaccine modalities developed thus far, an HIV-1 live-attenuated vaccine (LAV) that was constructed by the deletion of the nef gene (referred thereafter as conventional Δnef-LAV) conferred the best protection against HIV-1 acquisition as demonstrated in simian immunodeficiency virus (SIV)/rhesus macaque model.2–7 However, the SIV Δnef-LAV showed pathogenic potential in neonatal and even in adult macaques, which was likely the consequence of uncontrolled viral replication events.8, 9 Therefore, the current HIV-1 LAV, without a significant enhancement in safety, has been precluded from clinical applications.
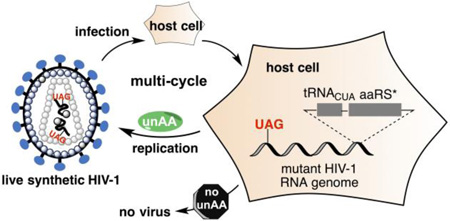
We recently applied an unnatural genetic switch10–13 to regulate HIV-1 replication in order to facilitate the development of a safe HIV-1 vaccine.14 The unnatural genetic switch consists of three essential components (Figure 1): (1) a special tRNA (tRNACUA) that can decode amber nonsense codon; (2) an engineered aminoacyl-tRNA synthetase (aaRS*) that specifically charges the tRNACUA with a desired unnatural amino acid (unAA); and (3) the desired unAA. By substitution of a regular codon with an amber codon in its encoding gene, the expression of this essential HIV-1 protein can be controlled in a biological system that expresses the first two components. In the presence of the unAA cue, the full-length essential HIV-1 protein can be synthesized, and HIV-1 is able to replicate (the ‘on’ state). In the absence of the unAA, the essential HIV-1 protein cannot be functionally expressed and the replication of HIV-1 is prevented (the ‘off’ state). Since the potential vaccination hosts do not have endogenous unAA, HIV-1 LAV (and by extension other LAV) can be turned on (with unAA) and off (without unAA) to mimic prime-boost vaccination strategy. Once the desirable protective immune responses have been achieved, the replication of HIV-1 LAV can be permanently turned off, which would overcome the safety barrier for the clinical uses of HIV-1 LAV. The unnatural genetic switch strategy is theoretically better than systems using inducible promoters15–23 in which basal replication of HIV-1 cannot be completely avoided. This is because the chance to read through one or more blank codons is much lower than the chance to initiate undesirable transcription. In addition to the intrinsically stringent control mechanism, the unAA-mediated genetic switch also benefits from the level of control it exerts. In our strategy, virus replication is controlled at the protein translation level and one read-through event leads to the synthesis of one copy of protein. In the other strategy, the control is at the DNA transcription level and one transcription event leads to the synthesis of one copy of mRNA, which serves as the template for the synthesis of numerous copies of protein.
Figure 1. Applying an unnatural genetic switch to the development of HIV-1 LAV.
Abbreviations: unAA, unnatural amino acid; aaRS*, an engineered aminoacyl-tRNA synthetase.
We successfully demonstrated that HIV-1 replication could be precisely turned on and off in 293T cells using the unnatural genetic switch when the tRNACUA-aaRS* pair was expressed from a separate plasmid.14 While HIV-1 mutants assembled in these experiments could functionally infect reporter cells (TZM-bl), they were only able to sustain one cycle of infection due to the lack of the tRNACUA-aaRS* pair in host cells. A similar approach was recently reported for the generation of live but replication-incompetent influenza vaccines.24 Unlike the influenza vaccines,24 replication-incompetent or transient replicating HIV-1 was insufficient to elicit potent protective immune responses in vaccination hosts.25 To this end, HIV-1 mutants that are capable of completing multi-cycle infection with stringent regulation are needed as LAV. To meet with this challenge, we now report the construction of all-in-one HIV-1 mutants that carry genomic copies of the tRNACUA-aaRS* pair through a synthetic biology approach. We successfully demonstrated that the new HIV-1 mutants were capable of completing multi-cycle of replication and infection. This work represents a significant step towards the generation of a safe and effective HIV-1 LAV through the novel unAA-mediated genetic switch strategy.
RESULTS AND DISCUSSION
Screening of genomic insertion sites for the tRNACUA-aaRS* pair
The organization of HIV-1 genome is complex and efficient. The HIV-1 genome has overlapping open reading frames that lead to nine primary translation products from unspliced, single- and multi-spliced mRNAs, from which a total of 15 proteins are made.26 As shown in Figure 2, the HIV-1 genome contains 5’-long terminal repeat (LTR), gag, pol, env, and 3’-LTR with 6 accessory genes (vif, vpu, vpr, tat, rev and nef) scattered around the env region. The gag gene encodes p17 matrix protein (MA), p24 capsid protein (CA), spacer peptide 1 (SP1), nucleocapsid protein (NC), spacer peptide 2 (SP2), and p6 polypeptide. The pol gene encodes protease (PR), reverse transcriptase (RT), and integrase (IN). The env gene encodes glycoprotein gp120 and gp41. In addition to the complex genome structure, the pre-RNA transcribed from the provirus undergoes alternative splicing of more than 40 different mRNAs. Therefore, it is critical to identify suitable sites for the insertion of the tRNACUA-aaRS* pair. Such insertion should not interfere with normal HIV-1 life cycle.
Figure 2. The HIV-1 genome organization.
Abbreviations: MA, p17 matrix protein; CA, p24 capsid protein; NC, nucleocapsid protein; SP1, Spacer peptide 1; SP2, Spacer peptide 2; LTR, long terminal repeat; PR, protease; RT, reverse transcriptase; IN, integrase. The red arrows indicate the insertion sites of foreign genes.
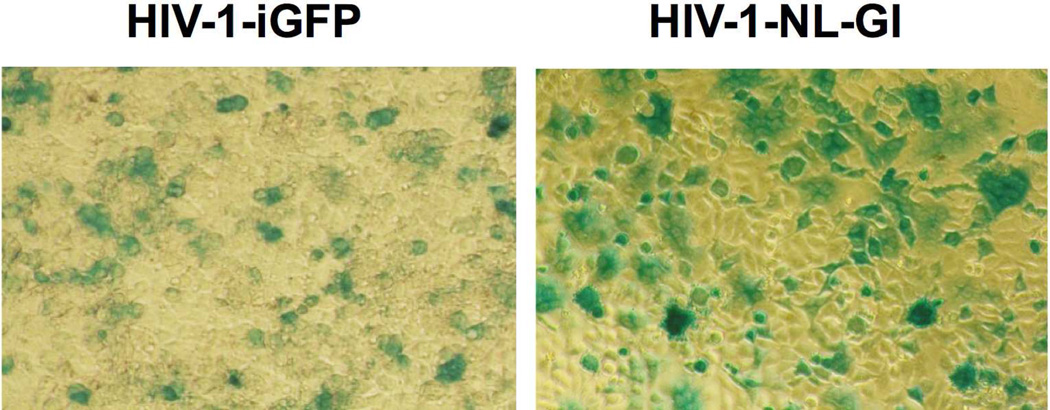
Multiple research groups have demonstrated the successful insertion of GFP-encoding gene (~ 720 bp) into two positions onto the HIV-1 genome. One position is between MA and CA (e.g., HIV-1-iGFP27, 28), the other one is to replace the 5’ end of the nef (e.g., HIV-1-NL-GI29, 30). We compared the effect of genomic insertion sites on the reproduction of infective virus using TZM-bl cells in which the expression of β-galactosidase is under the control of an HIV-1 LTR. Functional HIV-1 could infect TZM-bl cells and activate the expression of β-galactosidase, which catalyzes the formation of a blue-colored product from X-gal. As shown in Figure 3, both HIV-1-NL-GI29, 30 and HIV-1-iGFP27, 28 variants were functional, with the HIV-1-NL-GI mutant displaying relatively higher infectivity according to a qualitative analysis of the number of blue cells from multiple infection assays. According to the reported Δnef-LAV studies,2–7, 29, 30 the genomic structure around and within the nef gene showed high-level of plasticity. We therefore selected this region for the insertion of genes that encode the tRNACUA-aaRS* pair.
Figure 3. Infection assay of HIV-1-iGFP and HIV-1-NL-GI strains.
TZM-bl cells infected with viruses collected from 293T cell cultures.
Construction and examination of HIV-aaRS* mutants
A 4-azido-L-phenylalanine (AzF)-based genetic switch was chosen in this study. This switch has been successfully used in our previous efforts to control HIV-1 replication in vitro.14 We first focused on the insertion of AzFRS (1284 bp, 52% GC content), which encodes an aaRS variant that specifically charges amber suppressor tRNACUA with AzF, into HIV-1 genome and examined the replication and infectivity of the resulting HIV-1 mutant.
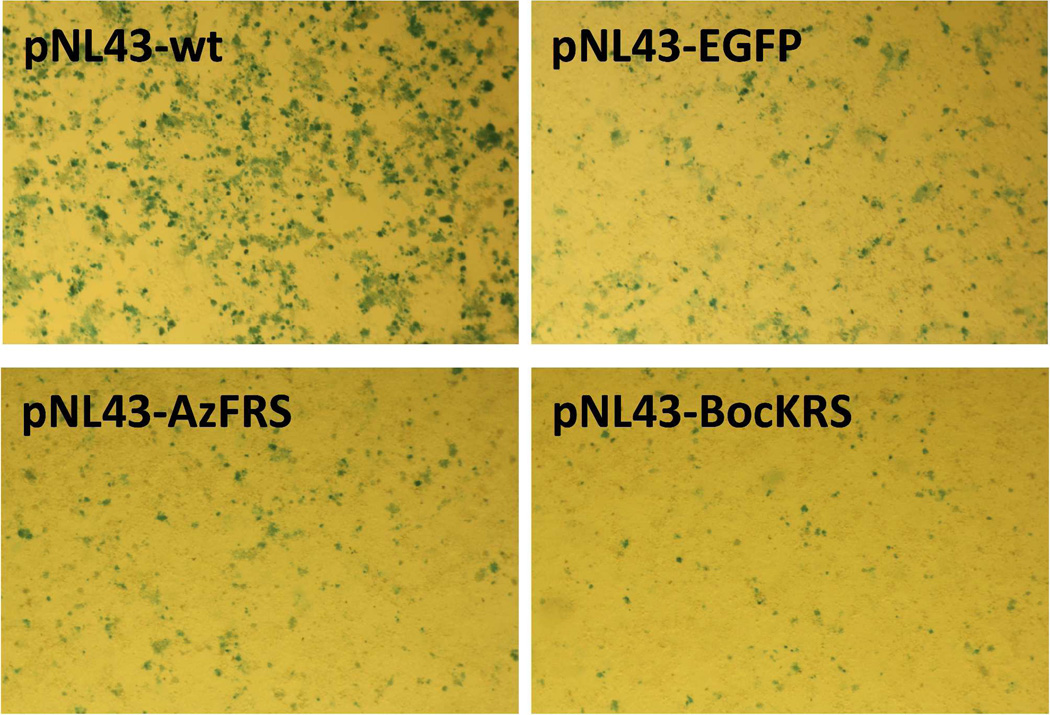
A widely used NL4-3 strain, which is the parental strain of HIV-1-NL-GI, was chosen as the starting virus for the generation of HIV-1 mutants. Two NL4-3 variants, NL43-AzFRS (encoded by plasmid pNL43-AzFRS) and NL43-EGFP (encoded by plasmid pNL43-EGFP), were constructed by directly inserting the AzFRS-encoding gene or the EGFP-encoding gene between the env and the nef genes of the NL4-3 genome. As shown in Figure 4, infective viruses were produced by both constructs, although in lower titers than that obtained from the wild-type NL4-3 construct (encoded by plasmid pNL43-wt). To further examine the effect of env/nef intergenic insertion, we constructed, NL43-BocKRS (encoded by plasmid pNL43-BocKRS), which contains BocKRS-encoding gene (1260 bp, 43% GC content) in the same region. BocKRS is a pyrrolysyl-tRNA synthetase (PyIRS) variant that specifically charges its cognate tRNACUA with Nε-(tert-butyloxy-carbonyl)-L-lysine (BocK). The BocKRS-tRNACUA pair together with BocK constitutes another type of unnatural genetic switch that can be potentially used in the generation of HIV-1 LAV. Again, significant but lower than NL43-wt infectivity was observed for the NL43-BocKRS mutant (Figure 4). Above results indicated that the intergenic region between env and nef of the NL4-3 genome could tolerate the insertion of a large piece of foreign DNA with a wide range of GC contents.
Figure 4. Relative replication efficiency of four NL4-3 variants.
TZM-bl cells were infected with viruses that were collected from 293T cells transfected with indicated plasmids.
Construction and examination of HIV-tRNA-AzFRS mutants
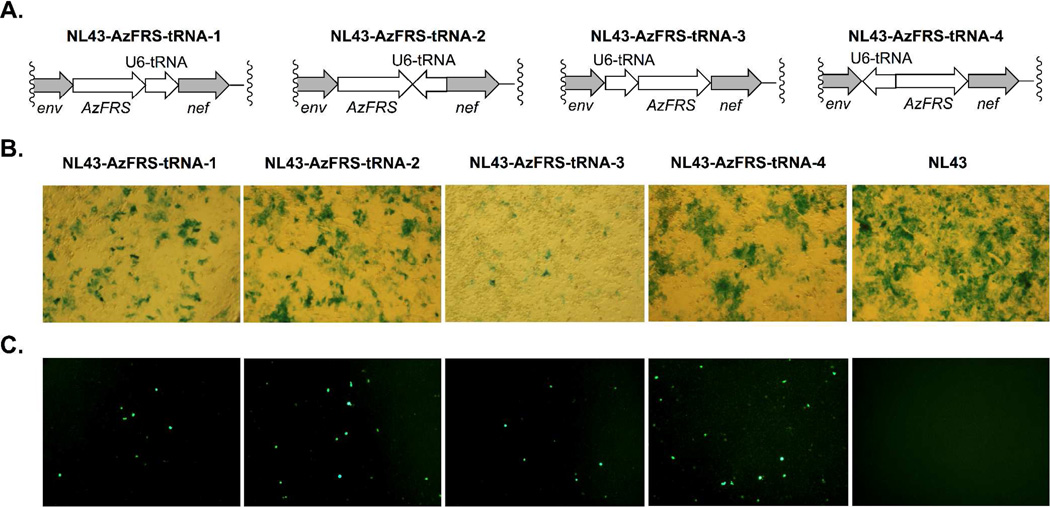
To complete the unnatural genetic switch, tRNACUA needs to be included in the HIV-1 genome for the decoding of amber nonsense codons. A tRNACUA transcription cassette was constructed by installing a human U6 (pol III) promoter immediately upstream of the tRNACUA-encoding gene. Four different HIV-1 constructs were generated where the position and the orientation of the U6-tRNACUA cassette were varied. The schematic drawing of these constructs are shown in Figure 5A. One pair of the constructs, NL43-AzFRS-tRNA-1 (encoded by plasmid pNL43-AzFRS-tRNA-1) and NL43-AzFRS-tRNA-2 (encoded by plasmid pNL43-AzFRS-tRNA-2), contain the U6-tRNACUA cassette between the AzFRS and the nef genes either in the same (NL43-AzFRS-tRNA-1) or in the opposite (NL43-AzFRS-tRNA-2) transcription direction as that of AzFRS. The other pair of constructs contain the U6-tRNACUA cassette between the env and the AzFRS genes either in the same (NL43-AzFRS-tRNA-3; encoded by plasmid pNL43-AzFRS-tRNA-3) or in the opposite (NL43-AzFRS-tRNA-4; encoded by plasmid pNL43-AzFRS-tRNA-4) transcription direction as that of AzFRS. These constructs are designed to fully explore the potential of the env/nef intergenic region to the tolerance of foreign DNA insertion. In the meantime, by sampling both insertion directions, we examined potential polar effects caused by transcription and/or translation of neighboring genes. Next we conducted in vitro assays and assessed the infectivity of the four HIV-1 mutants. As shown in Figure 5B, insertion of the U6-tRNACUA cassette into the AzFRS/env region resulted in two mutants (NL43-AzFRS-tRNA-1 and NL43-AzFRS-tRNA-2) that displayed infectivity, while the second insertion site only led to mutant NL43-AzFRS-tRNA-4 that showed infectivity.
Figure 5. Construction and examination of HIV-tRNA-AzFRS mutants.
(A) The four HIV-tRNA-AzFRS constructs with different positions and orientations of tRNA; (B) Infectivity assay of the four HIV-tRNA-AzFRS mutants and the parental NL43 virus; (C) Functional assay of the inserted tRNACUA-AzFRS pair of the four HIV-tRNA-AzFRS mutants and the parental NL43 virus in the presence of AzF.
Next, we examined whether the tRNACUA-AzFRS pair could be correctly transcribed, processed, and expressed as functional components in mammalian host cells. This was achieved by an assay that entails the co-transfection of an HIV-1 mutant of interest together with a plasmid (pEGFP) that contains an EGFP gene with an amber mutation at a permissive site (position Tyr40). Functional expression of the tRNACUA-AzFRS pair should completes the genetic switch, which would enable the decoding of the amber codon and the subsequent expression of full-length EGFP in the presence of AzF cue. Fluorescence analysis of 293T cells showed that tRNACUA-AzFRS was functional in all four HIV-1 constructs (Figure 5C). The relative fluorescence intensities were estimated using the ImageJ software (Figure S1).
Construction and multi-cycle replication of HIV-UAG-tRNA-AzFRS variants
As the NL43-AzFRS-tRNA-2 mutant displayed one of the best combinations of infectivity and amber suppression activity in the presence of 1 mM AzF, this mutant was chosen for subsequent investigations on the control of multi-cycle replication of HIV-1. Two new mutants were derived from NL43-AzFRS-tRNA-2 by substituting amber codon for either the codon of Trp36 in p17 (NL43-Trp36-AzFRS-tRNA-2; encoded by plasmid pNL43-Trp36-AzFRS-tRNA-2) or the codon of Tyr59 in HIV-1 protease (NL43-Tyr59-AzFRS-tRNA-2; encoded by plasmid pNL43-Tyr59-AzFRS-tRNA-2). Neither of the two mutants contained AzF in HIV-1 envelope proteins, which are the major immunogen for developing HIV-1 vaccine.
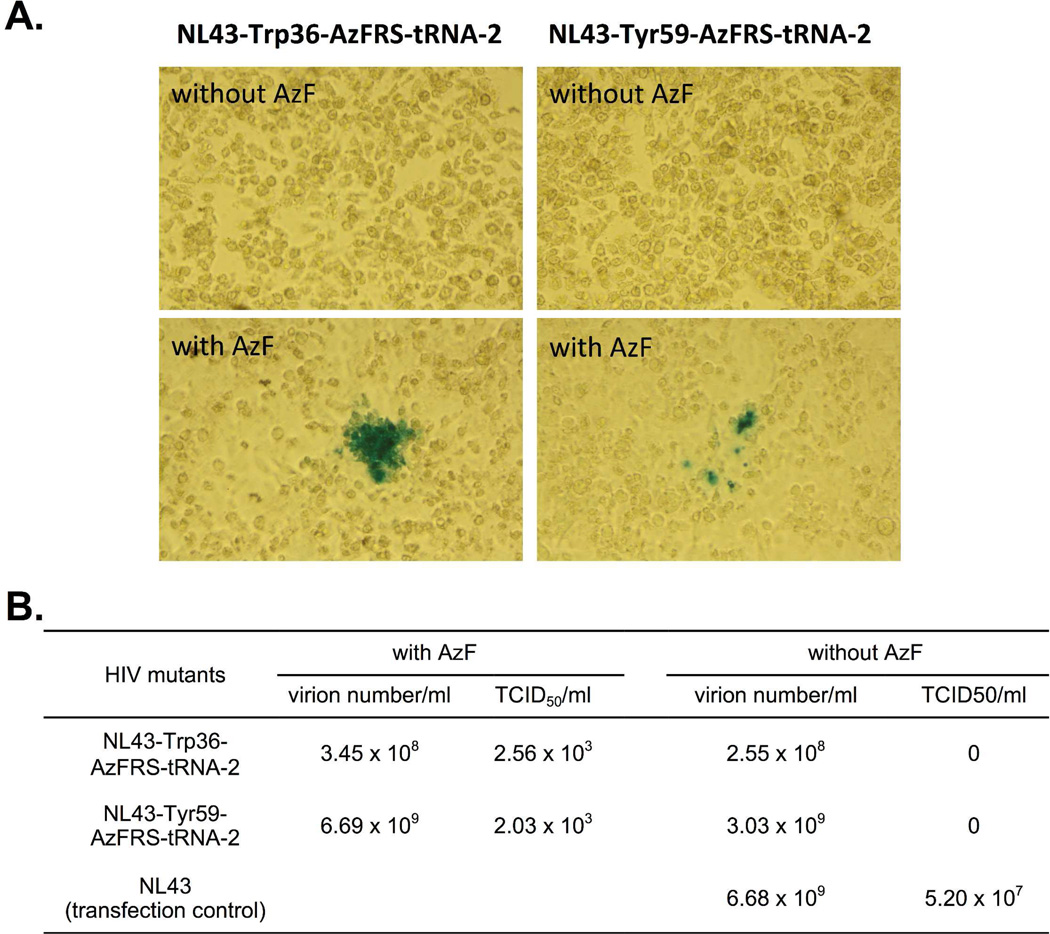
As shown in Figure 6, both mutants could undergo functional replication to form infective virus in the presence of AzF, whereas, no infective virus was obtained in the absence of AzF. Above results represent the first ever demonstration that the replication of an HIV-1 mutant can be controlled by an unAA (i.e. AzF) without the need of exogenously added tRNACUA-aaRS* pair. According to the TCID50 measurements (Figure 6B), both of the NL43-Trp36-AzFRS-tRNA-2 and the NL43-Tyr59-AzFRS-tRNA-2 mutant displayed significantly lower infectivity than that of the parental NL43 virus. On the other hand, the overall viral particle numbers of the two mutants are either slightly lower than or similar to that of the parental NL43 virus (Figure 6B). These results indicate that the modification of viral genome and the genetic incorporation of AzF do not significantly affect the expression of viral proteins. However, truncated p17 or protease (if protein translation stops at the amber mutation site) may interfere with the generation of infectious virions.
Figure 6. Infectivity assay of the NL43-Trp36-AzFRS-tRNA-2 and the NL43-Tyr59-AzFRS-tRNA-2 mutant in the presence and in the absence of AzF.
(A) Cell images of the infectivity assay; (B) TCID50 values and virion numbers. TZM-bl cells were infected with viruses that were collected from 293T cell cultures.
Next, the multi-cycle replications of the NL43-Trp36-AzFRS-tRNA-2 and NL43-Tyr59-AzFRS-tRNA-2 mutants were examined. First, the two mutant viruses were produced in 293T cells in the presence of 1 mM AzF. While this concentration of amino acid is generally employed in many unnatural amino acid mutagenesis studies, lower AzF concentrations, e.g., 0.1 or 0.25 mM, were sufficient for protein expression (Figure S6). In fact, no significant difference in protein expression levels was observed when 0.1 and 1.0 mM AzF was used, respectively (Figure S6). Our results are consistent with a previously reported study, where an even lower concentration (0.01 mM) of unnatural amino acid was used.31 Therefore, the concentration of amino acid can be adjusted according to the requirement of the experiment and is not a limiting factor.
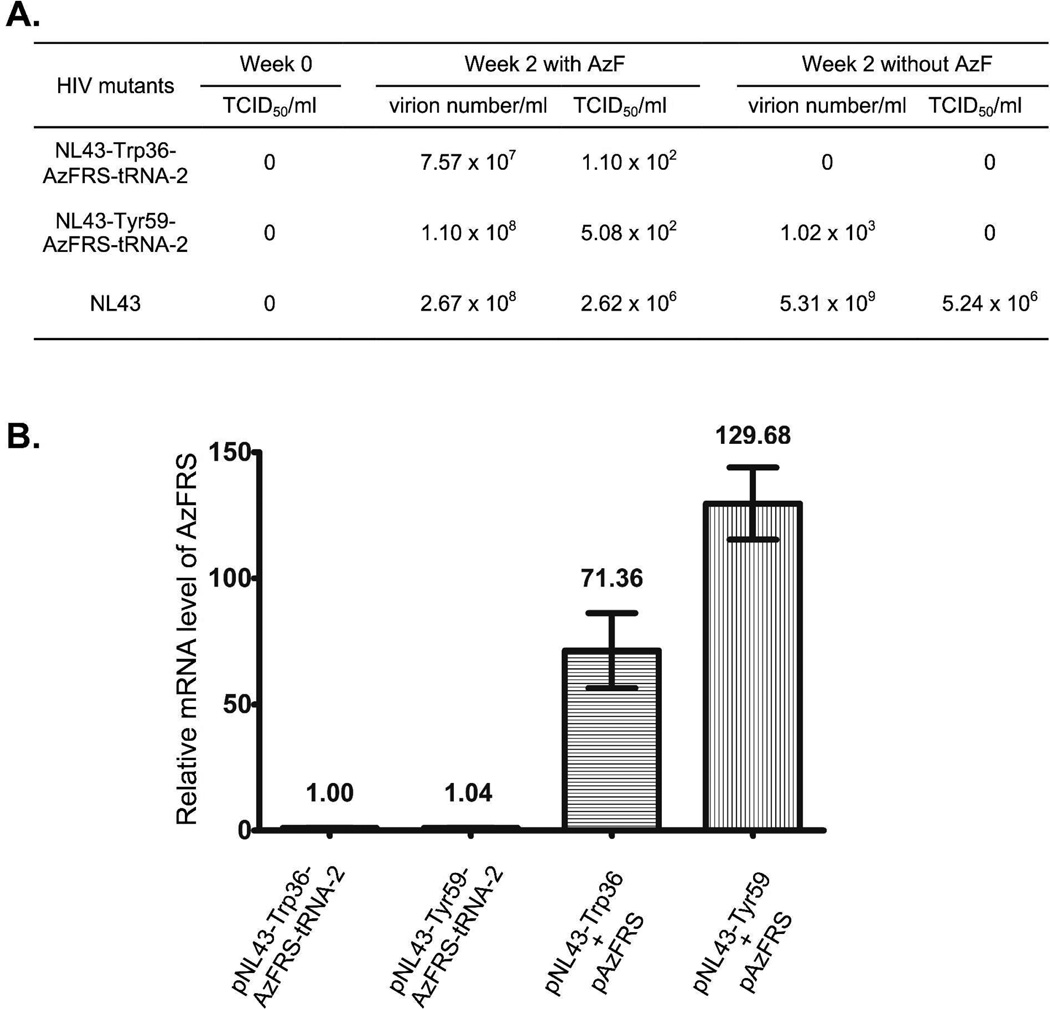
After 48 hours of cultivation, viruses were harvested and used to infect SupT1 cells that are CD4 positive and can support the entire life cycle of HIV-1. One day post infection, SupT1 cells were washed twice to remove any virions that remained in the culture media. Provirus-containing SupT1 cells were subsequently cultivated either in the presence or in the absence of AzF. As shown in Figure 7A, HIV-1 replications were only observed in the presence of AzF. Higher virus titer was detected after two weeks of cultivation (Figure 7A), which indicates that multi-cycle replications of the NL43-Trp36-AzFRS-tRNA-2 and the NL43-Tyr59-AzFRS-tRNA-2 mutant were achieved. Between the two, the NL43-Tyr59-AzFRS-tRNA-2 mutant displayed better replication capacity than that of the NL43-Trp36-AzFRS-tRNA-2 mutant. As a negative control, when the infected SupT1 cells were cultivated in the absence of AzF, no infective virus was detected two weeks after the initial infection. In comparison to the parental NL4-3 virus, the infectivity of the NL43-Trp36-AzFRS-tRNA-2 and the NL43-Tyr59-AzFRS-tRNA-2 mutant was weaker according to TCID50 measurements (Figure 7A). To assess the effects of AzF on the infectivity of HIV-1, TCID50 values of the parental NL4-3 virus were measured both in the presence and in the absence of AzF. The NL4-3 virus displayed similar but slightly lower infectivity in the presence of AzF (Figure 7A).
Figure 7. Evaluation of the infectivity of the NL43-Trp36-AzFRS-tRNA-2 and the NL43-Tyr59-AzFRS-tRNA-2 mutants.
(A) Multi-cycle replications of the NL43-Trp36-AzFRS-tRNA-2, the NL43-Tyr59-AzFRS-tRNA-2, or the parental NL43 variants. TCID50, tissue culture infectious dose 50; (B) Relative expression levels of AzFRS. The AzFRS mRNA level that was transcribed from the pNL43-Trp36-AzFRS-tRNA-2 plasmid was set as 1.
We have also measured the transcription level of aaRS gene in 293T cells after the transfection of mutant HIV-1 genome. As shown in Figure 7B, the levels of AzFRS mRNA transcribed from pNL43-Trp36-AzFRS-tRNA-2 and pNL43-Tyr59-AzFRS-tRNA-2 constructs were significantly lower than that transcribed from plasmid pAzFRS. One plausible explanation is that much more copies of pAzFRS plasmid (could be more than 100 copies) were present in the transfected cells, which led to higher expression level of the AzFRS gene (Figure 7B). As only one copy of AzFRS gene can likely be inserted into the HIV genome, we sought to increase the AzFRS expression by using a strong CMV promoter. Two new HIV-1 constructs, pNL43-Trp36-CMV-AzFRS-tRNA-2 and pNL43-Tyr59-CMV-AzFRS-tRNA-2, were constructed. However, the use of CMV promoter did not improve the infectivity of HIV-1 mutants (Figure S2).
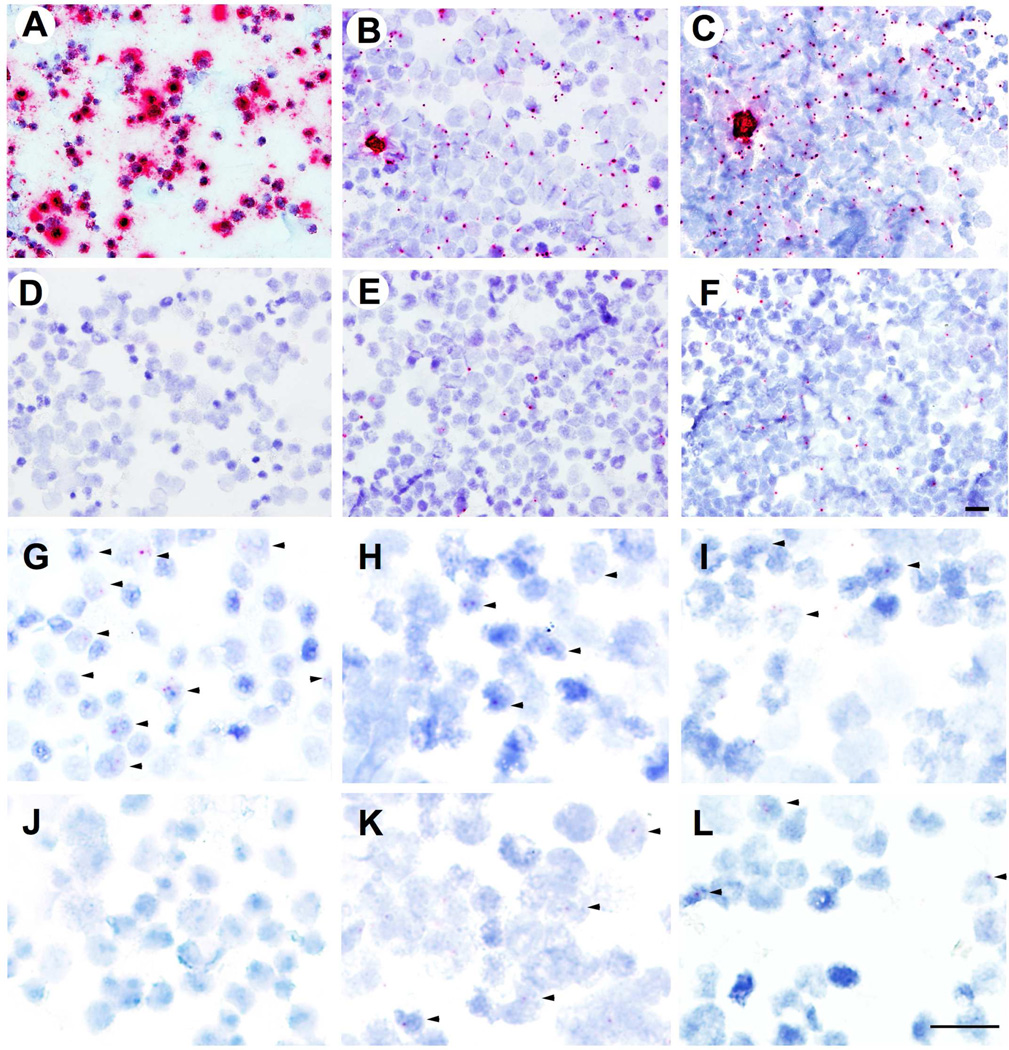
Next, we examined HIV-1 viral RNA (vRNA) and DNA (vDNA) in cells that were infected with the NL43-Tyr59-AzFRS-tRNA-2 and the NL43-Tyr59-AzFRS-tRNA-2 mutants using RNAscope in situ hybridization (ISH; Figures 8A–8F) and DNAscope ISH (Figures 8G–8L), respectively. Viral transcriptions were detectable both in the presence and in the absence of AzF (Figures 8B, 8C, 8E, and 8F). However, higher level of viral transcriptions was observed in the presence of AzF (Figures 8B, 8C), which indicated a multi-cycle replication. As a negative control (Figure 8D), no viral transcription was detected with uninfected Sup-T1 cells. As a positive control, ACH2 cells with PMA stimulation (Figure 8A) showed robust signal of viral transcriptions. The DNAscope ISH studies were performed to detect the vDNA from both of the NL43-Tyr59-AzFRS-tRNA-2 and the NL43-Tyr59-AzFRS-tRNA-2 mutants. The data clearly showed that the integration of proviral DNA could be observed in the target cells both in the presence and in the absence of AzF (Figures 8H, 8I, 8K, and 8L).
Figure 8. Viral RNA (vRNA) and DNA (vDNA) detection using RNAscope and DNAscope in situ hybridization (ISH).
A–F, in situ detection of HIV-1 vRNA. HIV-1 vRNA was detected using RNAscope ISH with V-HIV Clade B anti-sense probes and RNAscope 2.0 HD red reagent kit. (A) Positive control, ACH2 cells stimulated with PMA; (B) Sup-T1 cells infected with NL43-Trp36-AzFRS-tRNA-2 and in the presence of AzF; (C) Sup-T1 cells infected with NL43-Tyr59-AzFRS-tRNA-2 and in the presence of AzF; (D) Negative control, Sup-T1 cells without HIV-1 infection; (E) Sup-T1 cells infected with NL43-Trp36-AzFRS-tRNA-2 and in the absence of AzF; (F) Sup-T1 cells infected with NL43-Tyr59-AzFRS-tRNA-2 and in the absence of AzF. G–L, in situ detection of HIV-1 vDNA. HIV-1 vDNA was detected using DNAscope ISH with V-HIV Clade B sense probes and RNAscope 2.0 HD red reagent kit. Arrows indicate the cells harboring HIV-1 vDNA (red dots). (G) Positive control, ACH2 cells; (H) Sup-T1 cells infected with NL43-Trp36-AzFRS-tRNA-2 and in the presence of AzF; (I) Sup-T1 cells infected with NL43-Tyr59-AzFRS-tRNA-2 and in the presence of AzF; (J) Negative control, uninfected Sup-T1 cells; (K) Sup-T1 cells infected with NL43-Trp36-AzFRS-tRNA-2 and in the absence of AzF; (L) Sup-T1 cells infected with NL43-Tyr59-AzFRS-tRNA-2 and in the absence of AzF. Scale bar, 20 microns.
Conclusions
In our proof-of-concept experiment, we demonstrated that the unAA-mediated genetic switch could be used to turn on and off multi-cycle HIV-1 replication through the addition and the clearance of a small-molecule cue. This approach can in theory be extended as the control of other live-attenuated vaccines. Our ability to control the timing and the duration of attenuated HIV-1 replication represents an important step towards the generation of a safe and efficient live-attenuated HIV-1 vaccine. As demonstrated in the SIV/rhesus macaque model of HIV-1 infection, live-attenuated Δnef-SIV vaccines could be pathogenic in neonatal macaques. Our approach has the potential to address this safety concern. Once the protective immunity is elicited, the replication of the attenuated HIV-1 can be permanently turned off by stopping the administration of a desirable unAA. Our approach to address the safety concerns of HIV-1 LAV is different from a few previously reported strategies in which the vaccine strains were further attenuated through additional deletions or mutations in accessory genes or regulatory elements of SIV as demonstrated in the macaque model. While the safety of these vaccines was improved, their efficacy was significantly decreased or lost due to the reduction of virus replication and viral immunogen levels. In contrast, our approach does not require the further deletion of other viral genes. The only differences between our HIV-1 mutant and the wild-type HIV-1 are the replacement of one (or two to three) naturally occurring amino acid residues with their close unnatural amino acid analogs and the inclusion of a genomic copy of the tRNACUA-aaRS encoding gene. Neither of the modification in theory significantly interferes with the immunological properties of the engineered HIV-1 vaccine. Another advantage of our vaccine approach is the temporal control of HIV-1 replication to mimic prime and boost vaccination for the elicitation of protective immune responses. If a booster vaccination is required to mount a recall immune response, virus replication can be transiently turned on and then off again at later time.
The present study represents a significant advance over our previously reported proof-of-concept study,14 in which HIV-1 mutant can only complete a single infection cycle due to the dependency on exogenously provided tRNACUA-aaRS pair. Although a virus that can execute only a single round of replication may be potentially used as a vaccine,32–35 it is unlikely to provide necessary protective immunity against HIV-1 due to transient replication.25 To solve this problem, we have constructed HIV-1 mutants that are capable of multi-cycle replication and can be turned on/off by the addition and the clearance of an unAA. This is achieved by the modification of HIV-1 genome to include a complete set of unnatural genetic switch that consists of amber nonsense codons in essential genes and a full set of nonsense codon suppression system. Although HIV-1 has a complex genome structure and undergoes intricate alternative splicing, we successfully identified a permissive site on the HIV-1 genome for the incorporation of a gene cassette that encodes the tRNACUA-aaRS* pair. While the current constructs are capable of multi-cycle replication, there is still need for further optimization. As shown in Figure 7A, the replication and infectivity of the current HIV-1 mutants are much lower than those of the wild-type virus. While the attenuated replication and infectivity may be desirable for a vaccine, these HIV-1 mutants with weakened replication may not be able to mount an optimal immune response. Subsequent optimization is needed to further increase the replication capacity of these HIV-1 mutants. For example, we can fine-tune the promoter and terminator sequences so that the expression level of AzFRS can be improved. In addition, protein engineering approach could be used to improve the kinetic properties of AzFRS in order to achieve sufficient catalytic turnovers even at relatively low protein concentrations.
As HIV-1 lacks a proofreading reverse transcriptase, it remains possible that the HIV-1 can regain functional replication by mutating an amber codon back to a sense codon. In an actual vaccine, multiple amber nonsense codons will be introduced into several essential genes of HIV-1 to further tighten the control on viral replication. Based on our calculation,14 the possibility of mutating all amber codons back to sense codons is approximately 1%, 0.01%, and 0.0001%, respectively, in the human life span (100 years) when one, two, and three blank codons are used. The other concern of our strategy is the genetic stability of the inserted tRNACUA-aaRS pair, whose function could be eventually lost through deletion or accumulation of detrimental mutations. While the loss of tRNACUA-aaRS* function was not observed in our experiments (2–3 weeks period) according to the RT-PCR (Figure S3) and DNA sequencing (Figures S4, S5) experiments, this may happen in a longer time span. However, the chance of losing tRNACUA-aaRS* function in all viruses within the vaccination window could be low. In a very long term, the loss of tRNACUA-aaRS* function will be beneficial to the safety aspect of the vaccine, since the replication of the virus is dependent on the function of the tRNACUA-aaRS* pair.
In summary, we have demonstrated in our proof-of-concept experiment that multi-cycle replication of attenuated HIV-1 could be controlled by using an unAA-mediated genetic switch based on the amber codon suppression strategy. This work represents a significant step towards the generation of a safe and effective live-attenuated HIV-1 vaccine. The approach/tools that were developed in this study can be potentially applied to the development of other live attenuated vaccines.
METHODS
Materials and General Methods
AzF was purchased from Bachem. Primers were ordered from Sigma. Restriction enzymes, Antarctic phosphatase (AP) and T4 DNA ligase were purchased from New England Biolabs. KOD hot start DNA polymerase was purchased from EMD Millipore. Standard molecular biology techniques36 were used throughout. Site-directed mutagenesis was carried out using overlapping PCR. E. coli GeneHogs were used for routine cloning and DNA propagation for none-HIV plasmids. E. coli MAX Efficiency Stbl2 (Thermo Fisher Scientific Inc) was used for routine cloning and DNA propagation for HIV plasmids. All solutions were prepared in deionized water further treated by Barnstead Nanopure® ultrapure water purification system (Thermo Fisher Scientific Inc). Antibiotics were added where appropriate to following final concentrations: ampicillin, 100 mg L−1; kanamycin, 50 mg L−1; tetracycline, 12.5 mg L−1.
Construction of HIV-1 mutants
To facilitate the insertion of foreign genes into HIV-1 genome, two unique restriction enzyme cutting sites, ClaI and XmaI, were introduced into pNL43-wt to yield pNL43-ClaI-XmaI. The two cutting sites were installed between env and nef by overlapping PCR using primers P7, P14, P15, and P16 (see primer list in supplementary Table S1). The AzFRS-encoding gene was PCR amplified using primers P12 and P13 and inserted into pNL43-ClaI-XmaI between ClaI and XmaI sites to give pNL43-AzFRS. AzFRS gene is in the same transcription direction as that of other HIV-1 genes. The transcription of tRNACUA is under the control of a human U6 promoter. The U6-tRNACUA cassette was PCR amplified and inserted into ClaI or XmaI site of pNL43-AzFRS to yield four different constructs, pNL43-AzFRS-tRNA-x (x = 1, 2, 3, and 4). Amber mutations of interest at position Trp36 of p17 and position Tyr59 of HIV-1 protease were introduced into pNL43-AzFRS-tRNA-2 using overlapping PCR.
Transfection and generation of HIV-1
293T cells were grown in a medium containing DMEM, 10% FBS, and 2 mM L-glutamine at 37 °C in a humidified atmosphere of 5% CO2. When reached 60–70% confluency, cells (in 2 mL medium in a 6-well plate) were transfected with 3 µg of appropriate plasmid(s) using 10 µL lipofectamine 2000 by following manufacture’s procedures. After 6 hours of incubation, the culture medium was carefully removed and replaced with 2 mL of fresh medium. AzF was subsequently added to a final concentration of 1 mM. After 48 hours of cultivation, culture supernatant was collected separately from each well. The FBS concentration in the harvested culture medium was adjusted to 20%. The culture medium was then filtered through a 0.45-micron filter. Viruses in the supernatant were used directly or aliquoted into sterile screw-cap vials and stored at −150°C.
Qualitative infection assay
The infectivity of HIV-1 variants was evaluated using TZM-bl cells and X-Gal Staining Kit (Genlantis). Briefly, virion-containing samples, mixed with 10% DMEM growth medium and DEAE-dextran at a final concentration of 40 mg/mL, were added into each well of a 6-well flat bottom plate that contains TZM-bl cells. After 48 hours of cultivation at 37 °C in a humidified atmosphere of 5% CO2, TZM-bl cells were washed, fixed, and stained using X-gal solution for 2 hours at 37 °C. Infectivity was evaluated by counting X-gal-positive cells under a light microscope.
Virus ultracentrifugation
293T cells in T175 flasks were transfected with plasmids (60 µg) of interest using lipofectamine 2000 (120 µL; Life Technologies). After 48 hours of cultivation, media was collected from each flask and separately filtered through a 0.45-micron filter. The filtrate (35 mL) from each flask was loaded into Ultra-Clear™ Tube (Beckman coulter) for ultracentrifugation. Virus ultracentrifugation was conducted with Optima L-100X ultracentrifuge and SW 32 Ti rotor (Beckman coulter) at 25,000 rpm and 4°C for 90 min. Supernatant was discarded. The pellet in each tube was resuspended with 1 mL of fresh medium and aliquoted into 200 µL portions in sterile screw-cap vials and stored at −150°C.
Multi-cycle infection assay
Concentrated viruses (200 µL) were added to T25 flask containing 1×106 Sup-T1 cells. After 24 hours of incubation, cells were collected by centrifugation. Cell pellets were resuspended into 5 mL of fresh medium and collected again by centrifugation. Next, cell pellets were resuspended into fresh 5 mL of culture medium either with or without 1 mM AzF. The status of cells was monitored everyday using microscope. After 2 weeks of cultivation, viruses were collected by ultracentrifugation and TCID50 values were measured.
Tissue culture infectious dose 50 (TCID50)
Infectious titers of all viruses were determined by standard TCID50 method using X-gal staining assay with TZM-bl cells. Briefly, four-fold serial dilutions of viruses were performed in quadruplicate (96-well flat bottom plate) in the X-gal staining assay. The TCID50 values were calculated by Spearman-Karber formula according to the negative end-point and dilution folds. The TZM-bl cell-only control and the wild-type virus control were included in all TCID50 measurements. In addition, samples that were derived from viral assembly in the absence of AzF were used as negative controls.
Fluorescence spectroscopy and cell imaging
The fluorescent images and bright-field images were obtained by a Nikon ECLIPSE TE3000 microscope and an EVOS FL Auto Imaging System with DIC, respectively. The samples were excited at 488 nm to acquire EGFP fluorescence images at 530/25 nm.
Quantification of viral load
Viral RNAs were extracted from plasma using QIAamp Viral RNA Mini kit (Qiagen). Plasma viral load (pVL) was determined by qRT-PCR using C1000 Thermal Cycler and CFX96 Real-Time system (Bio-Rad) with TaqMan Fast Virus 1-Step Master Mix (Life technologies), primers V1 and V2, and probe V3. The detection limit of plasma HIV-1 RNA was 200 copies/mL and was determined through repeating end point detection from serial dilution of the AcroMetrix HIV-1 Panel (Life technologies).
Analysis of AzFRS transcription using quantitative PCR
After transfection and 48 hours of cultivation, 293T cells were collected. The total RNA from cells was extracted with RNeasy Plus Mini Kit (Qiagen). The quantity and purity of RNA samples were determined by NanoDrop 2000 (Thermo Scientific). The cDNA was synthesized using an Oligo DT primer (IDT) and Superscript III reverse transcriptase (Life Technologies) on C1000 Thermal Cycler (Bio-Rad). The qPCR were conducted on the CFX96 Real-Time detection system (Bio-Rad) using a hot start (95°C for 5 min) and 40 amplification cycles (95°C for 15 s, 60°C for 30 s). For amplification and detection of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) control, primers Q1 and Q2, and probe Q3 were used (Table S3). For amplification and detection of AzFRS gene, primers Q4 and Q5, and probe Q6 were used (Table S3).
Viral transcription and provirus detection using RNAscope and DNAscope in situ hybridization
Viral RNA and DNA were detected using single-copy sensitivity DNAscope and RNAscope in situ hybridization (ISH) techniques.37 Briefly, cells were collected after 72 h of cultivation with AzF, spotted on glass-slide after centrifugation, and fixed with 4% PFA for 1 h. HIV-1 viral RNA was detected with V-HIV Clade B anti-sense probes and RNAscope 2.0 HD red reagent kit (Advanced Cell Diagnostics, CA). HIV-1 viral DNA was detected with V-HIV Clade B sense probes and RNAscope 2.0 HD red reagent kit (Advanced Cell Diagnostics, CA). All experiments were conducted according to manufacturer’s instruction.
Amplification and sequencing for aaRS-tRNACUA insertion
Cells infected with wild-type NL43 virus, the NL43-Trp36-AzFRS-tRNA-2 mutant, or the pNL43-Tyr59-AzFRS-tRNA-2 mutant were collected after 2 weeks of cultivation in the presence of AzF. Viral RNA was extracted from plasma using QIAamp Viral RNA Mini kit (Qiagen). The cDNA synthesis, amplification, and sequencing methods followed published procedures.38 RNA quality was verified using Nanodrop (Thermo Scientific). The cDNA synthesis from isolated RNA was conducted by reverse transcription using Superscript III reagents (Invitrogen) with gene-specific antisense primer (5’-AAGATCTACAGCTGCCTT-3’). This mixture was heated to 65 °C for 5 minutes followed by incubation on ice for 2 minutes. A master mix of the following was then added: 5× First-Strand Buffer, dithiothreitol, RNaseOUT Recombinant RNase Inhibitor (40 units/µl), SuperScriptIII RT (200 units/µl), and RNAse free water. The mixture was incubated at 50 °C for 60 minutes and 70°C for 15 minutes followed by the addition of 1 µl RNase H (5U/µl) and incubated at 37C for 20 minutes. Then the cDNA was stored at −20°C until amplification. PCR amplification was performed using High Fidelity Platinum Taq polymerase (Invitrogen) in a 20 µl reaction with forward primer 5’-TTATAGAAGTATTACAAGCAGCTTATAG-3’ and reverse primer 5’-AAGATCTACAGCTGCCTT-3’ under the following conditions: 94°C for 5 minutes, 36 cycles of 94°C for 30 seconds, 53°C for 30 seconds, and 68°C for 2 minutes, and an extension at 68°C for 5 minutes. The amplicons were checked by 0.8% agarose gel stained with ethidium bromide. All PCR reactions were performed with procedural safeguards against sample contamination, including pre-aliquot of all reagents, use of dedicated equipment, and physical separation of sample processing from pre- and post-PCR amplification steps. The amplicons of the full-length insertion were sequenced at Sequetech (Mountain View, CA) using the Sanger method.
Acknowledgments
This work is supported by the New Faculty Startup Fund to J. Guo from the Chemistry Department of University of Nebraska – Lincoln and by Grant 1R01AI111862 (to J. Guo, Q. Li, and W. Niu) from DHHS-NIH-NIAID.
Footnotes
ASSOCIATED CONTENT
Supporting Information.
The Supporting Information is available free of charge on the ACS Publications website at at DOI: Additional figures and data.
The authors declare no competing financial interest.
REFERENCES
- 1.Fauci AS. 25 years of HIV. Nature. 2008;453:289–290. doi: 10.1038/453289a. [DOI] [PubMed] [Google Scholar]
- 2.Koff WC, Johnson PR, Watkins DI, Burton DR, Lifson JD, Hasenkrug KJ, McDermott AB, Schultz A, Zamb TJ, Boyle R, Desrosiers RC. HIV vaccine design: insights from live attenuated SIV vaccines. Nat. Immunol. 2006;7:19–23. doi: 10.1038/ni1296. [DOI] [PubMed] [Google Scholar]
- 3.Daniel MD, Kirchhoff F, Czajak SC, Sehgal PK, Desrosiers RC. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science. 1992;258:1938–1941. doi: 10.1126/science.1470917. [DOI] [PubMed] [Google Scholar]
- 4.Connor RI, Montefiori DC, Binley JM, Moore JP, Bonhoeffer S, Gettie A, Fenamore EA, Sheridan KE, Ho DD, Dailey PJ, Marx PA. Temporal analyses of virus replication, immune responses, and efficacy in rhesus macaques immunized with a live, attenuated simian immunodeficiency virus vaccine. J. Virol. 1998;72:7501–7509. doi: 10.1128/jvi.72.9.7501-7509.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tenner-Racz K, Hennig CS, Ueberla K, Stoiber H, Ignatius R, Heeney J, Steinman RM, Racz P. Early protection against pathogenic virus infection at a mucosal challenge site after vaccination with attenuated simian immunodeficiency virus. Proc. Natl. Acad. Sci. U. S. A. 2004;101:3017–3022. doi: 10.1073/pnas.0308677101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reynolds MR, Weiler AM, Weisgrau KL, Piaskowski SM, Furlott JR, Weinfurter JT, Kaizu M, Soma T, Leon EJ, MacNair C, Leaman DP, Zwick MB, Gostick E, Musani SK, Price DA, Friedrich TC, Rakasz EG, Wilson NA, McDermott AB, Boyle R, Allison DB, Burton DR, Koff WC, Watkins DI. Macaques vaccinated with live-attenuated SIV control replication of heterologous virus. J. Exp. Med. 2008;205:2537–2550. doi: 10.1084/jem.20081524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reynolds MR, Weiler AM, Piaskowski SM, Kolar HL, Hessell AJ, Weiker M, Weisgrau KL, Leon EJ, Rogers WE, Makowsky R, McDermott AB, Boyle R, Wilson NA, Allison DB, Burton DR, Koff WC, Watkins DI. Macaques vaccinated with simian immunodeficiency virus SIVmac239Δnef delay acquisition and control replication after repeated low-dose heterologous SIV challenge. J. Virol. 2010;84:9190–9199. doi: 10.1128/JVI.00041-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baba TW, Jeong YS, Penninck D, Bronson R, Greene MF, Ruprecht RM. Pathogenicity of live, attenuated SIV after mucosal infection of neonatal macaques. Science. 1995;267:1820–1825. doi: 10.1126/science.7892606. [DOI] [PubMed] [Google Scholar]
- 9.Hofmann-Lehmann R, Vlasak J, Williams AL, Chenine A-L, McClure HM, Anderson DC, O'Neil S, Ruprecht RM. Live attenuated, nef-deleted SIV is pathogenic in most adult macaques after prolonged observation. AIDS. 2003;17:157–166. doi: 10.1097/00002030-200301240-00004. [DOI] [PubMed] [Google Scholar]
- 10.Anderson JC, Clarke EJ, Arkin AP, Voigt CA. Environmentally controlled invasion of cancer cells by engineered bacteria. J. Mol. Biol. 2006;355:619–627. doi: 10.1016/j.jmb.2005.10.076. [DOI] [PubMed] [Google Scholar]
- 11.Liu CC, Qi L, Yanofsky C, Arkin AP. Regulation of transcription by unnatural amino acids. Nat. Biotechnol. 2011;29:164–168. doi: 10.1038/nbt.1741. [DOI] [PubMed] [Google Scholar]
- 12.Rovner AJ, Haimovich AD, Katz SR, Li Z, Grome MW, Amiram M, Patel JR, Gallagher RR, Isaacs FJ, Gassaway BM, Rinehart J. Recoded organisms engineered to depend on synthetic amino acids. Nature. 2015;518:89–93. doi: 10.1038/nature14095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mandell DJ, Kuznetsov G, Norville JE, Gregg CJ, Lajoie MJ, Mee MT, Takeuchi R, Stoddard BL, Church GM. Biocontainment of genetically modified organisms by synthetic protein design. Nature. 2015;518:55–60. doi: 10.1038/nature14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang N, Li Y, Niu W, Sun M, Cerny R, Li Q, Guo J. Construction of a live-attenuated HIV-1 vaccine through genetic code expansion. Angew. Chem., Int. Ed. 2014;53:4867–4871. doi: 10.1002/anie.201402092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verhoef K, Marzio G, Hillen W, Bujard H, Berkhout B. Strict control of human immunodeficiency virus type 1 replication by a genetic switch: Tet for Tat. J. Virol. 2001;75:979–987. doi: 10.1128/JVI.75.2.979-987.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith SM, Khoroshev M, Marx PA, Orenstein J, Jeang K-T. Constitutively dead, conditionally live HIV-1 genomes: ex vivo implications for a live virus vaccine. J. Biol. Chem. 2001;276:32184–32190. doi: 10.1074/jbc.M101604200. [DOI] [PubMed] [Google Scholar]
- 17.Berkhout B, Marzio G, Verhoef K. Control over HIV-1 replication by an antibiotic; a novel vaccination strategy with a drug-dependent virus. Virus Res. 2002;82:103–108. doi: 10.1016/s0168-1702(01)00399-9. [DOI] [PubMed] [Google Scholar]
- 18.Das AT, Zhou X, Vink M, Klaver B, Berkhout B. Conditional live virus as a novel approach towards a safe live attenuated HIV vaccine. Expert Rev. Vaccines. 2002;1:293–301. doi: 10.1586/14760584.1.3.293. [DOI] [PubMed] [Google Scholar]
- 19.Das AT, Verhoef K, Berkhout B. A conditionally replicating virus as a novel approach toward an HIV vaccine. Methods Enzymol. 2004;388:359–379. doi: 10.1016/S0076-6879(04)88028-5. [DOI] [PubMed] [Google Scholar]
- 20.Marzio G, Verhoef K, Vink M, Berkhout B. In vitro evolution of a highly replicating, doxycycline-dependent HIV for applications in vaccine studies. Proc. Natl. Acad. Sci. U. S. A. 2001;98:6342–6347. doi: 10.1073/pnas.111031498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou X, Vink M, Klaver B, Verhoef K, Marzio G, Das AT, Berkhout B. The Genetic Stability of a Conditional Live HIV-1 Variant Can Be Improved by Mutations in the Tet-On Regulatory System That Restrain Evolution. J. Biol. Chem. 2006;281:17084–17091. doi: 10.1074/jbc.M513400200. [DOI] [PubMed] [Google Scholar]
- 22.Zhou X, Vink M, Berkhout B, Das AT. Modification of the Tet-On regulatory system prevents the conditional-live HIV-1 variant from losing doxycycline-control. Retrovirology. 2006;3:82. doi: 10.1186/1742-4690-3-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das AT, Zhou X, Vink M, Klaver B, Verhoef K, Marzio G, Berkhout B. Viral Evolution as a Tool to Improve the Tetracycline-regulated Gene Expression System. J. Biol. Chem. 2004;279:18776–18782. doi: 10.1074/jbc.M313895200. [DOI] [PubMed] [Google Scholar]
- 24.Si L, Xu H, Zhou X, Zhang Z, Tian Z, Wang Y, Wu Y, Zhang B, Niu Z, Zhang C, Fu G, Xiao S, Xia Q, Zhang L, Zhou D. Generation of influenza A viruses as live but replication-incompetent virus vaccines. Science. 2016;354:1170–1173. doi: 10.1126/science.aah5869. [DOI] [PubMed] [Google Scholar]
- 25.Wyand MS, Manson KH, Garcia-Moll M, Montefiori D, Desrosiers RC. Vaccine protection by a triple deletion mutant of simian immunodeficiency virus. J Virol. 1996;70:3724–3733. doi: 10.1128/jvi.70.6.3724-3733.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hryckiewicz K, Bura M, Kowala-Piaskowska A, Bolewska B, Mozer-Lisewska I. HIV RNA splicing. HIV & AIDS Review. 2011;10:61–64. [Google Scholar]
- 27.Hubner W, Chen P, Del PA, Liu Y, Gordon RE, Chen BK. Sequence of human immunodeficiency virus type 1 (HIV-1) Gag localization and oligomerization monitored with live confocal imaging of a replication-competent, fluorescently tagged HIV-1. J Virol. 2007;81:12596–12607. doi: 10.1128/JVI.01088-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hubner W, McNerney GP, Chen P, Dale BM, Gordon RE, Chuang FYS, Li X-D, Asmuth DM, Huser T, Chen BK. Quantitative 3D video microscopy of HIV transfer across T cell virological synapses. Science. 2009;323:1743–1747. doi: 10.1126/science.1167525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen GB, Gandhi RT, Davis DM, Mandelboim O, Chen BK, Strominger JL, Baltimore D. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity. 1999;10:661–671. doi: 10.1016/s1074-7613(00)80065-5. [DOI] [PubMed] [Google Scholar]
- 30.Collins KL, Chen BK, Kalams SA, Walker BD, Baltimore D. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature. 1998;391:397–401. doi: 10.1038/34929. [DOI] [PubMed] [Google Scholar]
- 31.Chatterjee A, Guo J, Lee HS, Schultz PG. A genetically encoded fluorescent probe in mammalian cells. J. Am. Chem. Soc. 2013;135:12540–12543. doi: 10.1021/ja4059553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Evans DT, Bricker JE, Desrosiers RC. A novel approach for producing lentiviruses that are limited to a single cycle of infection. J. Virol. 2004;78:11715–11725. doi: 10.1128/JVI.78.21.11715-11725.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Evans DT, Bricker JE, Sanford HB, Lang S, Carville A, Richardson BA, Piatak M, Jr, Lifson JD, Mansfield KG, Desrosiers RC. Immunization of macaques with single-cycle simian immunodeficiency virus (SIV) stimulates diverse virus-specific immune responses and reduces viral loads after challenge with SIVmac239. J. Virol. 2005;79:7707–7720. doi: 10.1128/JVI.79.12.7707-7720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Falkensammer B, Rubner B, Hiltgartner A, Wilflingseder D, Stahl HC, Kuate S, Ueberla K, Norley S, Strasak A, Racz P, Stoiber H. Role of complement and antibodies in controlling infection with pathogenic simian immunodeficiency virus (SIV) in macaques vaccinated with replication-deficient viral vectors. Retrovirology. 2009;6:60. doi: 10.1186/1742-4690-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuate S, Stahl-Hennig C, ten HP, Heeney J, Uberla K. Single-cycle immunodeficiency viruses provide strategies for uncoupling in vivo expression levels from viral replicative capacity and for mimicking live-attenuated SIV vaccines. Virology. 2003;313:653–662. doi: 10.1016/s0042-6822(03)00388-x. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook JF, Russell DW, editors. Molecular cloning: A laboratory manual, third edition. Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- 37.Wang F, Flanagan J, Su N, Wang L-C, Bui S, Nielson A, Wu X, Vo H-T, Ma X-J, Luo Y. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan Z, Kang G, Ma F, Lu W, Fan W, Li Q, Fennessey CM, Keele BF. Recapitulating Cross-Species Transmission of Simian Immunodeficiency Virus SIVcpz to Humans by Using Humanized BLT Mice. J Virol. 2016;90:7728–7739. doi: 10.1128/JVI.00860-16. [DOI] [PMC free article] [PubMed] [Google Scholar]