Abstract
Single particle cryo-electron microscopy (cryoEM) is becoming widely adopted as a tool for structural characterization of biomolecules at near-atomic resolution. Vitrification of the sample to obtain a dense distribution of particles within a single field of view remains a major bottleneck for the success of such experiments. Here, we describe a simple and cost-effective method to increase the density of frozen-hydrated particles on grids with holey carbon support films. It relies on performing multiple rounds of sample application and blotting prior to plunge freezing in liquid ethane. We show that this approach is generally applicable and significantly increases particle density for a range of samples, such as small protein complexes, viruses and filamentous assemblies. The method is versatile, easy to implement, minimizes sample requirements and can enable characterization of samples that would otherwise resist structural studies using single particle cryoEM.
Keywords: Cryo-electron microscopy, Single particle, Vitrification
Single particle cryo-electron microscopy (cryoEM) is becoming an increasingly important tool in structural biology (Nogales and Scheres, 2015, Cheng et al., 2015). Due to recent advances in detector efficiency, microscope stability and image processing, the 3D structure of biomolecular complexes spanning a wide range of sizes, from haemoglobin to microtubules and intact viruses, can be reconstructed at (near-) atomic resolution (Bai et al., 2013, Brilot et al., 2012, Campbell et al., 2012, Campbell et al., 2015, Khoshouei et al., 2016, Liu et al., 2010, Nogales, 2015, Scheres, 2012, Walls et al., 2016a, Walls et al., 2016b).
Whereas sample requirements are less restrictive in cryoEM compared to X-ray crystallography or nuclear magnetic resonance, successful vitrification of protein complexes remains a major bottleneck (Grassucci et al., 2007). A solution of the particles under study is pipetted onto holey grids made of carbon-coated copper or gold (Quispe et al., 2007, Russo and Passmore, 2014). The holes in the grid support film become filled with particles suspended in buffer and the excess solution is blotted away using filter paper. The grid is then immediately plunged into a bath of liquid ethane, maintained at liquid nitrogen temperature (−180 °C) to vitrify the protein solution and preserve it in a near-native frozen-hydrated state (Dubochet, 1988). Numerous specimens, however, are reluctant to populate the holes and remain adsorbed on the carbon support film instead, even after significantly increasing the sample concentration. Furthermore, many protein complexes are precious, due to the difficulty to purify them in large quantities, such that using higher concentrations is not an option.
In an effort to improve the applicability of single particle cryoEM, many strategies have been developed for sample preparation. For instance, samples can be deposited on 2D crystals of streptavidin, captured on immuno-affinity supports or on a continuous carbon or graphene oxide film, prior to plunge-freezing in liquid ethane (Han et al., 2012, Kelly et al., 2008, Pantelic et al., 2010, Wagenknecht et al., 1988, Yu et al., 2016). However, those methods can be costly, technically challenging, reduce the contrast of the images (except for graphene oxide) and may introduce structural defects related to adsorption on a surface.
Here, we present a simple and cost-effective method that builds on the standard plunge freezing procedure to overcome the aforementioned shortcomings. Although previous reports mentioned the possibility of multiple sample applications to increase particle density in suspended vitreous ice (Cheng et al., 2015), this has never been described in detail to the best of our knowledge. We systematically tested this method on various samples and consistently observed that it greatly improved particle density in thin films of suspended vitreous ice, including cases in which ramping up the concentration failed to do so. We believe this procedure can benefit the exponentially growing numbers of single particle cryoEM practitioners who will inevitably run into samples that prove reluctant to evenly spread over holey cryoEM grids.
This method involves performing several successive rounds of sample application and blotting before plunge freezing. We tested up to four blotting rounds and observed a marked additive effect on particle density at each step. All sample applications and blotting steps can be carried out on the lab bench, similarly to what is done for negative staining sample preparation, until the final sample application, at which point the tweezers are mounted in a plunge-freezing apparatus to perform the final blotting step using standard procedures (Supplementary Movie S1). Alternatively, all rounds of sample application and blotting can be performed with the tweezers mounted in a plunge-freezing apparatus, if desired. We used Whatman No. 1 filter paper, which is the only extra requirement over our standard grid preparation procedure. Typically, the sample rests on the grid for a few seconds between successive rounds of sample application and blotting. During the final round, the sample is on the grid for 10–20 s, while running through the program of the Vitrobot. For all examples that follow, samples were vitrified using Protochips C-flat grids and imaged on an FEI T12 electron microscope equipped with a Gatan US4000 CCD camera, an FEI TF20 or an FEI Titan Krios, equipped with a Gatan K2 direct detector (Li et al., 2013). We used MotionCorr2 (Zheng et al., 2016) for movie frame alignment and binned all images by a factor of 4 for visualization.
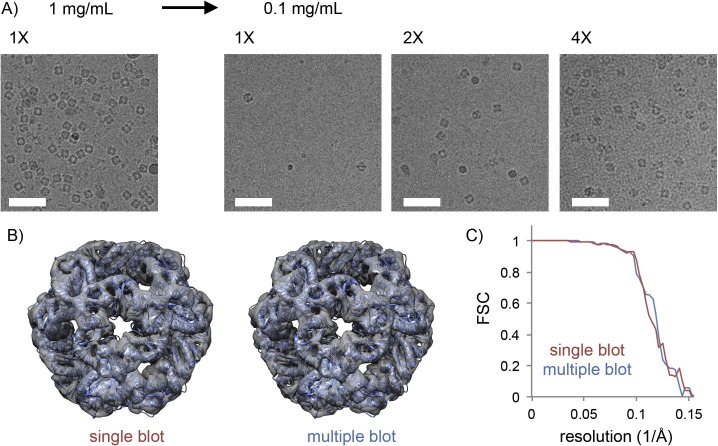
To demonstrate the efficacy of this method, we used a computationally designed octahedral protein cage (King et al., 2012), assembled from 24 identical subunits, as a test specimen (O3-33, Fig. 1 A). At 1 mg/mL, the O3-33 solution yielded an optimal particle density for single particle cryoEM imaging. Diluting the sample ten-fold significantly reduced the distribution of particles over suspended ice. This effect could be partially overcome after a second round of sample application and blotting. After the fourth blotting step, the particle density in suspended vitreous ice became comparable to that observed with grids prepared with a single application of a ten times higher protein concentration. We collected two O3-33 datasets of 100 micrographs each using grids prepared with a single or with multiple blotting steps. The 3D reconstructions of particles from either condition both reached a resolution of ∼7.5 Å, showing that the integrity of the particles is indistinguishable between the two procedures (Fig. 1B-C). These results illustrate that the method can greatly alleviate sample requirements for single particle cryoEM, both in terms of protein concentration and total amount required.
Fig. 1.
A) The effect of multiple rounds of sample application and blotting on particle density in thin films of suspended vitreous ice observed with samples of O3-33. Sample concentrations and number of application steps are indicated. Scale bars: 50 nm. B) 3D-reconstructions of O3-33 from grids prepared with a single or multiple rounds of blotting, with atomic coordinates from X-ray crystallography fitted (PDB ID 3VCD). C) Fourier Shell Correlation curves. Resolution of the maps (FSC = 0.143) using single and multiple blots is 7.8 and 7.3 Å, respectively.
To assess the versatility of this approach, we implemented this multiple blotting scheme for a range of protein complexes, several of which yielded insufficient particles in ice using standard vitrification procedures.
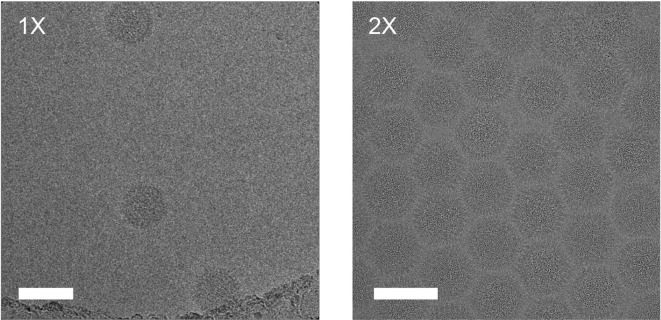
Adenoviruses are ∼1000 Å-wide non-enveloped icosahedral pathogens involved in respiratory, ocular and gastro-intestinal infections (Veesler et al., 2014). Samples of adenovirus serotype 26 virions showed a low propensity to evenly spread in thin films of suspended vitreous ice, which was problematic because a limited amount and concentration of sample was available for our cryoEM study. Using a double blotting strategy, we were able to overcome this issue, as attested by the densely packed virions in the micrograph presented in Fig. 2 , and to collect a large dataset which led to the determination of a structure at 3.7 Å resolution (manuscript in preparation).
Fig. 2.
Multiple rounds of sample application and blotting of human adenovirus (6 mg/mL). Scale bars: 100 nm.
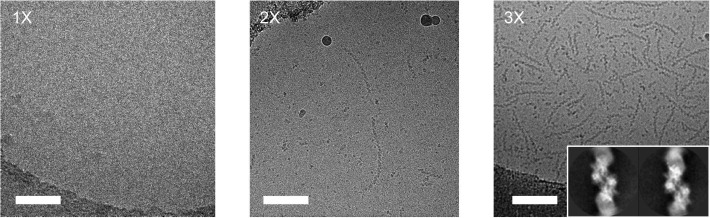
Yeast glucokinase 1 and human inosine-5′-monophosphate dehydrogenase are two key metabolic enzymes involved in carbohydrate and nucleotide metabolism, respectively. These two enzymes self-assemble as elongated homomeric filaments, which can only be structurally characterized by cryoEM due to their repetitive and flexible nature. Although sample availability was not a limiting factor for these two specimens, both proved reluctant to spread over the holes of cryoEM grids. We successfully enhanced particle distribution, following up to three consecutive applications of sample onto the grids, to achieve a particle density that was useful for high-resolution imaging (Fig. 3, Fig. 4 ). This is further illustrated by the details revealed in computed helical 2D averages of yeast glucokinase 1 filaments.
Fig. 3.
Multiple rounds of sample application and blotting of filaments of yeast glucokinase-1 (10 μM). Scale bars: 100 nm.
Fig. 4.
Multiple rounds of sample application and blotting of filaments of inosine-5′-monophosphate dehydrogenase. Scale bars: 100 nm.
Detergents or amphipols are oftentimes used at low concentrations to supplement a protein solution prior to grid preparation with the aim of reducing aggregation and/or promoting more isotropic particle orientations (Lyumkis et al., 2013, Chowdhury et al., 2015). However, this strategy requires much higher concentrations of protein than one would need in the absence of such additives, to ensure the sample will uniformly spread over holey carbon grids. Similar problems are encountered in cryoEM studies of detergent or amphipol solubilized membrane proteins.
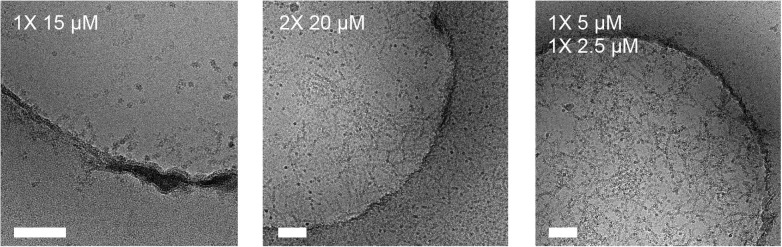
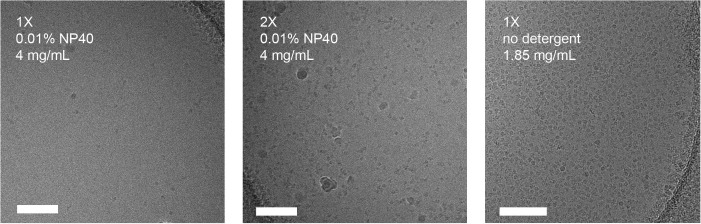
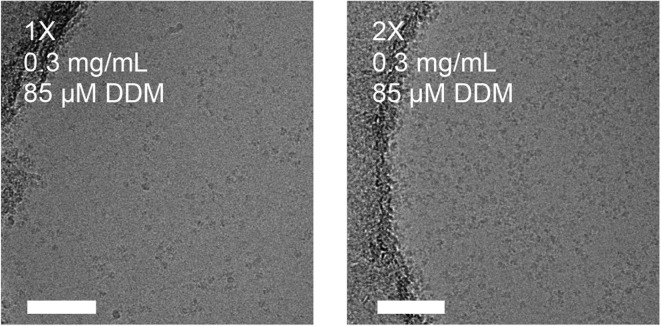
We used the mouse hepatitis virus (MHV) spike (S) glycoprotein ectodomain (Walls et al., 2016a, Walls et al., 2016c) to illustrate the usefulness of our approach when vitrifying samples in presence of detergents (see Fig. 5 ). A single round of sample application and blotting of the MHV S ectodomain in the presence of 0.01% NP40 yielded few particles in suspended vitreous ice, even at concentrations up to 4 mg/mL. After the second round of blotting, many more particles could be observed in the ice. We would like to emphasize that in the absence of detergent, a single sample application resulted in a very dense distribution of particles, even at less than half the concentration. We obtained similar results with the HIV envelope glycoprotein ectodomain in the presence of 85 µM of dodecyl-β-D-maltoside (a sample known to be prone to aggregation upon vitrification, Lyumkis et al., 2013). Although few particles were detected in vitreous ice after a single round of blotting, good particle density was achieved after the second sample application, thereby alleviating the need for large amounts of protein (Fig. 6 ).
Fig. 5.
Multiple rounds of sample application and blotting of coronavirus MHV spike glycoprotein ectodomain in the presence of 0.01% NP40. Scale bars: 100 nm.
Fig. 6.
Multiple rounds of sample application and blotting of HIV envelope glycoprotein ectodomain (0.3 mg/mL) in the presence of 85 μM dodecyl-maltoside. Scale bars: 100 nm.
Our observations indicate that the blotting procedure increases the concentration of particles inside the grid holes compared to the free solution of particles. For instance, at the concentration of O3-33 used in the upper-left panel of Fig. 1a (2 μM), one should only expect up to two particles in the projected volume of ice if it corresponded directly to the sample concentration in the test-tube (assuming an ice thickness on the order of 40 nm). Instead, dozens of particles can be observed, suggesting that blotting actively forces the particles into the holes. We put forward that successive rounds of sample application and blotting are additive in this regard, which would explain how the multiple blotting procedure produces ice with a higher concentration of particles.
In conclusion, using a simple and cost-effective trick, one can increase the applicability of single particle cryoEM to protein complexes that cannot be purified in large quantities and/or that behave poorly upon vitrification. Multiple steps of sample application can force particles into the vitreous iced-filled holes of a cryoEM grid to minimize sample requirements for structural studies.
Data deposition
The two O3-33 reconstructions have been deposited to the EMDB with accession numbers EMD-8613 (single blot) and EMD-8614 (multiple blots).
Acknowledgements
Research reported in this publication was supported by the National Institute of General Medical Sciences (NIGMS) under award number 1R01GM120553-01 (to D.V.), 1R01GM118396-01 (to J.M.K.), R01AI070771 to VSR, T32GM008268 (to A.J.B. and A.C.W.). J.S. acknowledges support from the Netherlands Organization for Scientific Research (NWO, Rubicon 019.2015.2.310.006) and the European Molecular Biology Organisation (EMBO, ALTF 933-2015). We are grateful to Neil King (University of Washington) for providing the O3-33 construct.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jsb.2017.02.008.
Appendix A. Supplementary data
References
- Bai X., Fernandez I.S., McMullan G., Scheres S.H.W. Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. eLife. 2013;2:e00461. doi: 10.7554/eLife.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brilot A.F., Chen J.Z., Cheng A., Pan J., Harrison S.C., Potter C.S., Carragher B., Henderson R., Grigorieff N. Beam-induced motion of vitrified specimen on holey carbon film. J. Struct. Biol. 2012;177:630–637. doi: 10.1016/j.jsb.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell M.G., Cheng A., Brilot A.F., Moeller A., Lyumkis D., Veesler D., Pan J., Harrison S.C., Potter C.S., Carragher B., Grigorieff N. Movies of ice-embedded particles enhance resolution in electron cryo-microscopy. Structure. 2012;20:1823–1828. doi: 10.1016/j.str.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell M.G., Veesler D., Cheng A., Potter C.S., Carragher B. 2.8 Å resolution reconstruction of the Thermoplasma acidophilum 20 s proteasome using cryo-electron microscopy. eLife. 2015;4:e06380. doi: 10.7554/eLife.06380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Grigorieff N., Penczek P.A., Walz T. A primer to single-particle cryo-electron microscopy. Cell. 2015;161:439–449. doi: 10.1016/j.cell.2015.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S., Ketcham S.A., Schroer T.A., Lander G.C. Structural organization of the dynein-dynactin complex bound to microtubules. Nat. Struct. Mol. Biol. 2015;22:345–347. doi: 10.1038/nsmb.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubochet J. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- Grassucci R.A., Taylor D.J., Frank J. Preparation of macromolecular complexes for cryo-electron microscopy. Nat. Protoc. 2007;2:3239–3246. doi: 10.1038/nprot.2007.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B., Walton R.W., Song A., Hwu P., Stubbs M.T., Yannone S.M., Arbeláez P., Dong M., Glaeser R.M. Electron microscopy of biotinylated protein complexes bound to streptavidin monolayer crystals. J. Struct. Biol. 2012;180:249–253. doi: 10.1016/j.jsb.2012.04.025. [DOI] [PubMed] [Google Scholar]
- Kelly D.F., Abeyrathne P.D., Dukovski D., Walz T. The affinity grid: a pre-fabricated EM grid for monolayer purification. J. Mol. Biol. 2008;382:423–433. doi: 10.1016/j.jmb.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshouei, M., Radjainia, M., Baumeister, W., Danev, R., 2016. Cryo-EM structure of haemoglobin at 3.2 A determined with the Volta phase plate. bioRxiv 087841. [DOI] [PMC free article] [PubMed]
- King N.P., Sheffler W., Sawaya M.R., Vollmar B.S., Sumida J.P., André I., Gonen T., Yeates T.O., Baker D. Computational design of self-assembling protein nanomaterials with atomic level accuracy. Science. 2012;336:1171–1174. doi: 10.1126/science.1219364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Mooney P., Zheng S., Booth C.R., Braunfield M.B., Gubbens S., Agard D.A., Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods. 2013;10(6):584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Jin L., Koh S.B.S., Atanasov I., Schein S., Wu L., Zhou Z.H. Atomic structure of human adenovirus by Cryo-EM reveals interactions among protein networks. Science. 2010;329:1038–1043. doi: 10.1126/science.1187433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyumkis D., Julien J., De Val N., Cupo A., Potter C.S., Klasse P., Burton D.R., Sanders R.W., Moore J.P., Carragher B., Wilson I.A., Ward A.B. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E. An electron microscopy journey in the study of microtubule structure and dynamics. Protein Sci. 2015;24:1912–1919. doi: 10.1002/pro.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E., Scheres S.H.W. Cryo-EM: a unique tool for the visualization of macromolecular complexity. Mol. Cell. 2015;58:677–689. doi: 10.1016/j.molcel.2015.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantelic R.S., Meyer J.C., Kaiser U., Baumeister W., Plitzko J.M. Graphene oxide: a substrate for optimizing preparations of frozen-hydrated samples. J. Struct. Biol. 2010;170:152–156. doi: 10.1016/j.jsb.2009.12.020. [DOI] [PubMed] [Google Scholar]
- Quispe J., Damiano J., Mick S.E., Nackashi D.P., Fellmann D., Ajero T.G., Carragher B., Potter C.S. An improved holey carbon film for cryo-electron microscopy. Microsc. Microanal. 2007;13(5):365–371. doi: 10.1017/S1431927607070791. [DOI] [PubMed] [Google Scholar]
- Russo C., Passmore L. Electron microscopy: Ultrastable gold substrates for electron cryomicroscopy. Science. 2014;346(6215):1377–1380. doi: 10.1126/science.1259530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres S.H.W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veesler D., Cupelli K., Burger M., Gräber P., Stehle T., Johnson J.E. Single-particle EM reveals plasticity of interactions between the adenovirus penton base and integrin αVβ3. Proc. Natl. Acad. Sci. U.S.A. 2014;111(24):8815–8819. doi: 10.1073/pnas.1404575111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenknecht T., Grassucci R., Frank J. Electron microscopy and computer image averaging of ice-embedded large ribosomal subunits from Escherichia coli. J. Mol. Biol. 1988;199:137–147. doi: 10.1016/0022-2836(88)90384-1. [DOI] [PubMed] [Google Scholar]
- Walls A.C., Tortorici M.A., Bosch B.J., Frenz B., Rottier P.J.M., DiMaio F., Rey F.A., Veesler D. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016;531:114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Tortorici M.A., Frenz B., Snijder J., Rey F.A., DiMaio F., Bosch B.J., Veesler D. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat. Struct. Mol. Biol. 2016;23(10):899–905. doi: 10.1038/nsmb.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Tortorici M.A., Bosch B.J., Frenz B., Rottier P.J.M., DiMaio F., Rey F.A., Veesler D. Crucial steps in the structure determination of a coronavirus spike glycoprotein using cryo-electron microscopy. Protein Sci. 2016 doi: 10.1002/pro.3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Li K., Huang P., Jiang X., Jiang W. Antibody-based affinity cryoelectron microscopy at 2.6-Å resolution. Structure. 2016;24:1984–1990. doi: 10.1016/j.str.2016.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, S., Palovcak, E., Armache, J., Cheng, Y., Agard, D., 2016. Anisotropic Correction of Beam-induced Motion for Improved Single-particle Electron Cryo-microscopy. bioRxiv 061960. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.