ABSTRACT
We recently demonstrated that the expression of the interferon regulatory factor 6 (IRF6) transcription factor in oral keratinocytes was stimulated by the periodontal pathogen Porphyromonas gingivalis. Here, we have established that IRF6 promotes the differentiation of oral keratinocytes in response to P. gingivalis. This was evidenced by the IRF6-dependent upregulation of specific markers of keratinocyte terminal differentiation (e.g., involucrin [IVL] and keratin 13 [KRT13]), together with additional transcriptional regulators of keratinocyte differentiation, including Grainyhead-like 3 (GRHL3) and Ovo-like zinc finger 1 (OVOL1). We have previously established that the transactivator function of IRF6 is activated by receptor-interacting protein kinase 4 (RIPK4). Consistently, the silencing of RIPK4 inhibited the stimulation of IVL, KRT13, GRHL3, and OVOL1 gene expression. IRF6 was shown to also regulate the stimulation of transglutaminase-1 (TGM1) gene expression by P. gingivalis, as well as that of small proline-rich proteins (e.g., SPRR1), which are covalently cross-linked by TGM1 to other proteins, including IVL, during cornification. The expression of the tight junction protein occludin (OCLN) was found to also be upregulated in an IRF6-dependent manner. IRF6 was demonstrated to be important for the barrier function of oral keratinocytes; specifically, silencing of IRF6 increased P. gingivalis-induced intercellular permeability and cell invasion. Taken together, our findings potentially position IRF6 as an important mediator of barrier defense against P. gingivalis.
KEYWORDS: differentiation, GRHL3, IRF6, keratinocyte, Porphyromonas gingivalis, RIPK4, barrier function
INTRODUCTION
The oral epithelium is an important physical and immunological barrier to pathogens (1). The integrity of the epithelium is maintained through continuous cycles of keratinocyte proliferation and differentiation, whereby basal keratinocytes intermittently exit the cell cycle and undergo terminal differentiation as they migrate toward the epithelium surface, concomitantly upregulating the expression of proteins that confer mechanical strength and elasticity to the epithelium (1–3). Depending on the anatomical location, terminally differentiated keratinocytes may also become cornified through the covalent cross-linking of IVL and other cornified envelope proteins by transglutaminases (e.g., TGM1) (1, 2).
IRF6 is a critical transcriptional regulator of keratinocyte differentiation (4–7). IRF6 promotes keratinocyte differentiation in part by inducing the expression of additional transcriptional regulators of differentiation, including GRHL3 and OVOL1 (5–7). GRHL3 promotes keratinocyte differentiation through its interaction with other transcription factors (e.g., LMO4) (8, 9) and by recruiting the Trithorax complex to the promoters of differentiation-associated genes (10). OVOL1 can promote keratinocyte differentiation by repressing the transcription of target genes, including c-Myc (11).
The transactivator function of IRF6 can be activated by RIPK4 (7, 12). Specifically, RIPK4 activates IRF6 by phosphorylating regulatory serine residues (Ser413 and Ser424) in its C-terminal domain, thereby enabling IRF6 and RIPK4 to function coordinately to promote the terminal differentiation of keratinocytes (7). Indeed, Irf6-deficient and Ripk4-deficient mice exhibit similar epidermal abnormalities, including the absence of the outermost cornified layers (4, 13).
We recently demonstrated that the expression of IRF6 in human oral keratinocytes was stimulated by Porphyromonas gingivalis (14). P. gingivalis is a key pathogen in chronic periodontitis, a highly prevalent chronic inflammatory disease in which the breakdown of periodontal tissues, including the resorption of alveolar bone, causes tooth loss (15, 16). P. gingivalis is found in the superficial layers of a polymicrobial subgingival biofilm against the epithelial lining of the periodontal pocket (17). P. gingivalis can impair epithelial barrier function by secreting proteinases (e.g., Kgp and RgpA/B) that degrade cell-cell and cell-extracellular matrix contacts (19–21); P. gingivalis can also invade epithelial cells (22, 23). The breakdown of epithelium tissues in periodontitis likely promotes episodes of bacteremia, which may exacerbate systemic inflammatory conditions (e.g., atherosclerosis and rheumatoid arthritis) (24). Consequently, the integrity of the oral epithelium is important for both oral and systemic health.
In this study, we investigated the functional significance of the stimulation of IRF6 expression in oral keratinocytes by P. gingivalis. Our data indicate that IRF6 promotes the differentiation of oral keratinocytes in response to P. gingivalis, including upregulating the expression of additional transcriptional regulators of differentiation as well as proteins that directly mediate cornification and tight junction formation. Thus, the upregulation of IRF6 expression in response to P. gingivalis likely serves to promote barrier defense.
RESULTS
P. gingivalis stimulates the expression of differentiation-associated genes in oral keratinocytes in an IRF6-dependent manner.
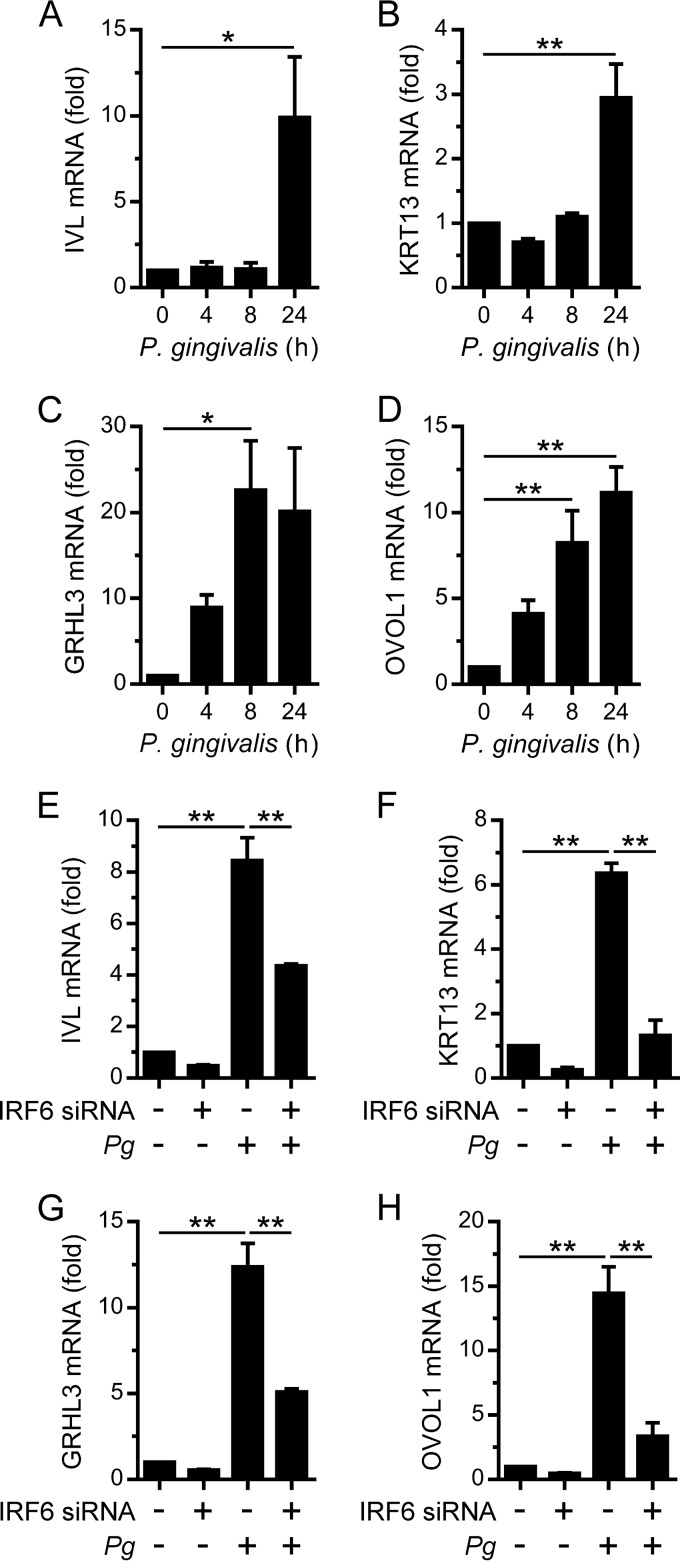
We recently demonstrated that the expression of the prodifferentiation transcription factor IRF6 in human oral keratinocytes (e.g., OKF6 cells) was stimulated by P. gingivalis (14). This prompted us to investigate whether oral keratinocytes may respond to P. gingivalis by further differentiating. Accordingly, OKF6 cells were cultured with P. gingivalis and changes in the gene expression levels of early (e.g., IVL) and late (e.g., KRT13) markers of keratinocyte terminal differentiation were measured. IVL gene expression and KRT13 gene expression were found to be stimulated by P. gingivalis (Fig. 1A and B). In addition to IRF6, GRHL3 and OVOL1 are also important transcriptional regulators of keratinocyte differentiation (8–11). Furthermore, IRF6 has been shown to directly regulate GRHL3 and OVOL1 gene expression (5, 6). Therefore, we also investigated the effects of P. gingivalis on GRHL3 and OVOL1 expression. GRHL3 gene expression and OVOL1 gene expression in OKF6 cells were found to be strongly stimulated by P. gingivalis (Fig. 1C and D). Given these results, we investigated whether IRF6 was important for the stimulation of IVL, KRT13, GRHL3, and OVOL1 gene expression. The transfection of OKF6 cells with an IRF6 siRNA, which reduced IRF6 mRNA levels by >80% (data not shown), inhibited the stimulation of IVL, KRT13, GRHL3, and OVOL1 gene expression by P. gingivalis (Fig. 1E to H). Taken together, these results suggest that IRF6 may, at least in part, promote the differentiation of oral keratinocytes in response to P. gingivalis by upregulating the expression of GRHL3 and OVOL1.
FIG 1.
IRF6-dependent stimulation of differentiation-associated gene expression in oral keratinocytes by P. gingivalis. (A to D) OKF6 cells were cultured with P. gingivalis (MOI of 100:1) for the times indicated, and IVL (A), KRT13 (B), GRHL3 (C), and OVOL1 (D) mRNA levels were then measured (n = 3). (E to H) OKF6 cells were transfected with an IRF6 (+) or control (–) siRNA. Thereafter, the cells were cultured with P. gingivalis (Pg) (MOI of 100:1) for 24 h, and IVL (E), KRT13 (F), GRHL3 (G), and OVOL1 (H) mRNA levels were then measured (n = 3). **, P < 0.01; *, P < 0.05.
Stimulation of differentiation-associated gene expression by P. gingivalis is TLR2 independent.
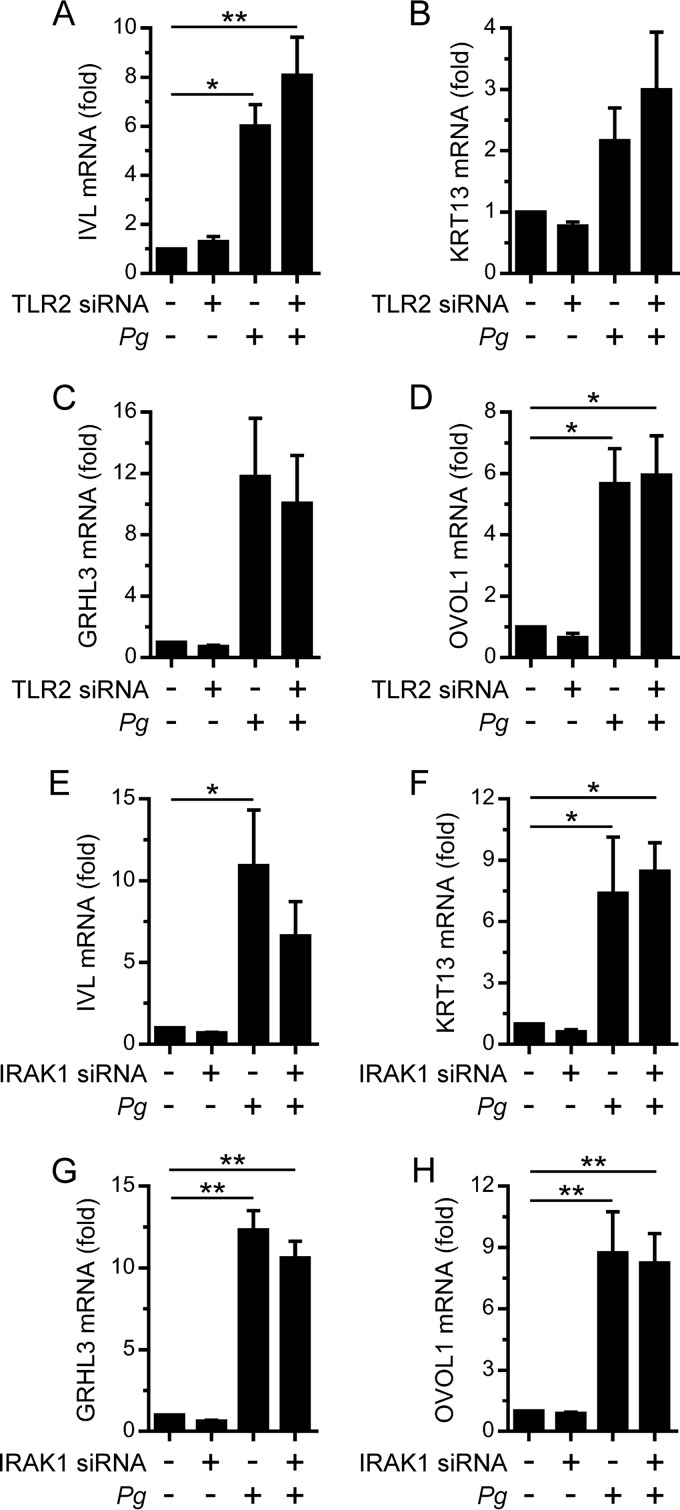
TLR2 is an important mediator of the innate immune response to P. gingivalis (25). Furthermore, the activation of IRF6 by TLR2 signaling in oral keratinocytes induces the expression of inflammatory cytokines (14, 26). Therefore, we investigated whether TLR2 signaling was important for the stimulation of IVL, KRT13, GRHL3, and OVOL1 gene expression by P. gingivalis. Significantly, the transfection of OKF6 cells with a TLR2 siRNA, which reduced TLR2 mRNA levels by >80% (data not shown), did not inhibit the stimulation of IVL, KRT13, GRHL3, or OVOL1 gene expression by P. gingivalis (Fig. 2A to D). The abrogation of TLR2-elicited responses was confirmed by demonstrating the ability of the siRNA to inhibit the stimulation of inflammatory cytokine expression (e.g., TNF and IL-6) by the TLR2 agonist FSL-1 (data not shown). IRAK1 is a key mediator of TLR2 signaling and can stimulate the transactivator function of IRF6 in oral keratinocytes (14, 26). We previously demonstrated that the silencing of IRAK1 in OKF6 cells inhibited the stimulation of inflammatory cytokine expression by FSL-1 (26). Therefore, we tested whether the silencing of IRAK1 affected the stimulation of IVL, KRT13, GRHL3, and OVOL1 gene expression by P. gingivalis. Consistent with the data presented above, the silencing of IRAK1 did not inhibit the upregulation of IVL, KRT13, GRHL3, and OVOL1 gene expression (Fig. 2E to H). These data indicate that IRF6 functions independently of TLR2 signaling to promote the differentiation of oral keratinocytes in response to P. gingivalis.
FIG 2.
TLR2-independent stimulation of differentiation-associated gene expression in oral keratinocytes by P. gingivalis. OKF6 cells were transfected with a TLR2 (+) or control (–) siRNA (A to D) or with an IRAK1 (+) or control (–) siRNA (E to H). Thereafter, the cells were cultured with P. gingivalis (MOI of 100:1) for 24 h, and IVL (A and E), KRT13 (B and F), GRHL3 (C and G), and OVOL1 (D and H) mRNA levels were then measured (n = 3). **, P < 0.01; *, P < 0.05.
RIPK4 is important for the stimulation of differentiation-associated gene expression by P. gingivalis.
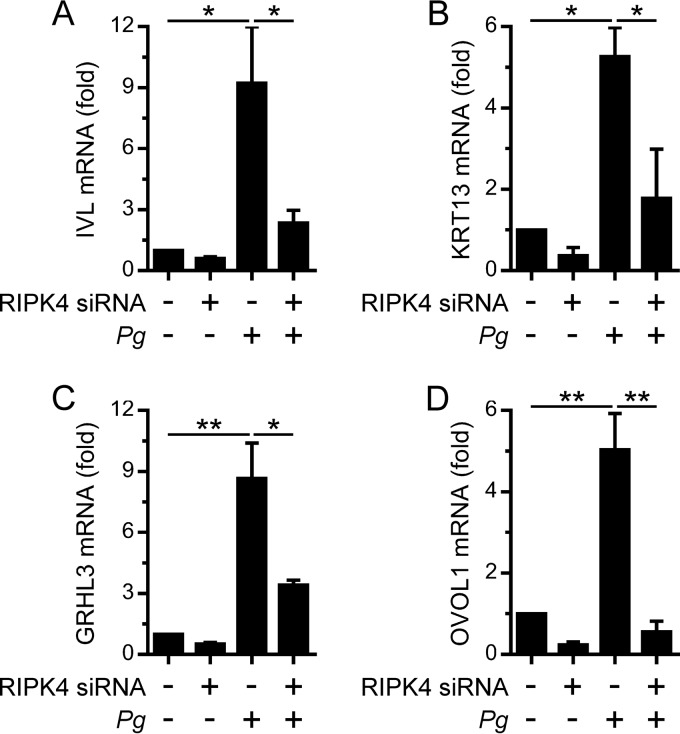
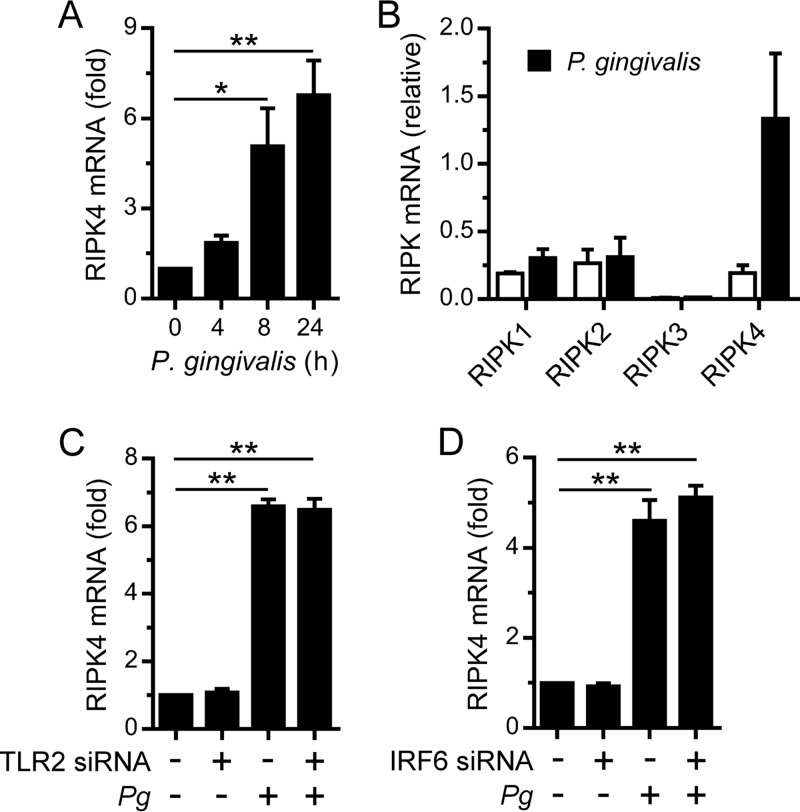
The transactivator function of IRF6 in oral keratinocytes can also be activated by RIPK4, for example, in response to PKC signaling (7, 12). Indeed, RIPK4 was important for the stimulation of IVL, KRT13, GRHL3, and OVOL1 gene expression by the PKC agonist PMA in OKF6 cells (7). Therefore, we sought to establish whether RIPK4 was important for the IRF6-mediated stimulation of differentiation-associated gene expression by P. gingivalis. The silencing of RIPK4 in OKF6 cells significantly inhibited the stimulation of IVL, KRT13, GRHL3, and OVOL1 gene expression (Fig. 3). Given these findings, and our recent demonstration that P. gingivalis stimulated IRF6 expression in OKF6 cells (14), we investigated whether RIPK4 expression might be similarly upregulated in response to P. gingivalis. As shown in Fig. 4A, RIPK4 gene expression in OKF6 cells was stimulated by P. gingivalis. In contrast, the gene expression levels of other RIPK family kinases, namely, RIPK1, RIPK2, and RIPK3, were unchanged (Fig. 4B), thereby indicating that the stimulation of RIPK4 gene expression by P. gingivalis was a specific response. Consistent with the data presented in Fig. 2, the stimulation of RIPK4 gene expression by P. gingivalis was TLR2 independent (Fig. 4C). IRF6 was shown by gene reporter assay to be capable of transactivating the RIPK4 gene promoter (27). Therefore, we tested whether IRF6 was important for the stimulation of RIPK4 gene expression by P. gingivalis. However, the stimulation of RIPK4 gene expression was found to be IRF6 independent (Fig. 4D). These findings indicate that oral keratinocytes may upregulate RIPK4 expression in response to P. gingivalis to enhance differentiation.
FIG 3.
RIPK4-dependent stimulation of differentiation-associated gene expression in oral keratinocytes by P. gingivalis. OKF6 cells were transfected with an RIPK4 (+) or control (–) siRNA. Thereafter, the cells were cultured with P. gingivalis (MOI of 100:1) for 24 h, and IVL (A), KRT13 (B), GRHL3 (C), and OVOL1 (D) mRNA levels were then measured (n = 3). **, P < 0.01; *, P < 0.05.
FIG 4.
Stimulation of RIPK4 expression in oral keratinocytes by P. gingivalis. (A) OKF6 cells were cultured with P. gingivalis (MOI of 100:1) for the times indicated, and RIPK4 mRNA levels were then measured (n = 3). (B) OKF6 cells were cultured with P. gingivalis (MOI of 100:1) for 24 h, and RIPK1, RIPK2, RIPK3, and RIPK4 mRNA levels were then measured (n = 3). (C and D) OKF6 cells were transfected with a TLR2 siRNA (C) or an IRF6 siRNA (D). Thereafter, the cells were cultured with P. gingivalis (MOI of 100:1) for 24 h, and RIPK4 mRNA levels were then measured (n = 3). **, P < 0.01; *, P < 0.05.
IRF6 is important for the stimulation of barrier function gene expression by P. gingivalis.
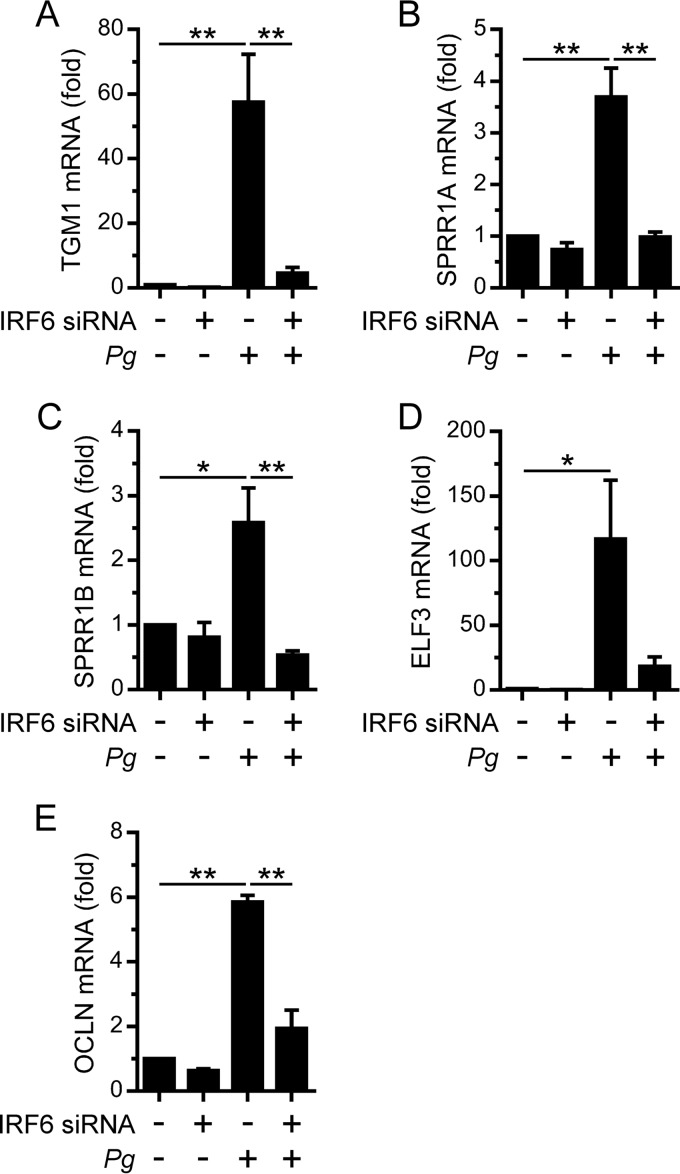
The covalent cross-linking of IVL and small proline-rich proteins (SPRR) by TGM1 is important for the cornification of terminally differentiated keratinocytes (1, 2). For example, Tgm1-deficient mice have impaired epidermal barrier function due to defective keratinocyte cornification (28). We recently established that IRF6 forms a hierarchal regulatory network with the GRHL3 and ELF3 transcription factors to regulate TGM1 and SPRR1 gene expression downstream of RIPK4 signaling (29). Therefore, we investigated the effects of P. gingivalis on TGM1 and SPRR1 gene expression. P. gingivalis stimulated the upregulation of TGM1 gene expression in OKF6 cells; furthermore, the upregulation of TGM1 expression was IRF6 dependent (Fig. 5A). The gene expression levels of SPRR1A and SPRR1B were similarly stimulated in an IRF6-dependent manner (Fig. 5B and C). P. gingivalis also stimulated the IRF6-dependent upregulation of ELF3 gene expression (Fig. 5D), consistent with IRF6 mediating the stimulation of SPRR1A and SPRR1B gene expression by P. gingivalis via its regulation of ELF3 expression. In addition to cornification, the formation of specialized intercellular junctions, such as tight junctions, can also enhance the barrier function of epithelia (1). Therefore, we investigated the effects of P. gingivalis on the expression levels of the tight junction-associated proteins OCLN, CLDN1, and TJP1. OCLN gene expression in OKF6 cells was stimulated in an IRF6-dependent manner by P. gingivalis (Fig. 5E). In contrast, the gene expression levels of CLDN1 and TJP1 were not stimulated (data not shown). These findings suggest that IRF6 may promote barrier function in response to P. gingivalis by regulating the expression of proteins that mediate cornification and tight junction formation.
FIG 5.
IRF6-dependent stimulation of barrier function gene expression in oral keratinocytes by P. gingivalis. OKF6 cells were transfected with an IRF6 (+) or control (–) siRNA. Thereafter, the cells were cultured with P. gingivalis (MOI of 100:1) for 24 h, and TGM1 (A), SPRR1A (B), SPRR1B (C), ELF3 (D), and OCLN (E) mRNA levels were then measured (n = 3). **, P < 0.01; *, P < 0.05.
IRF6 enhances the barrier function of oral keratinocytes.
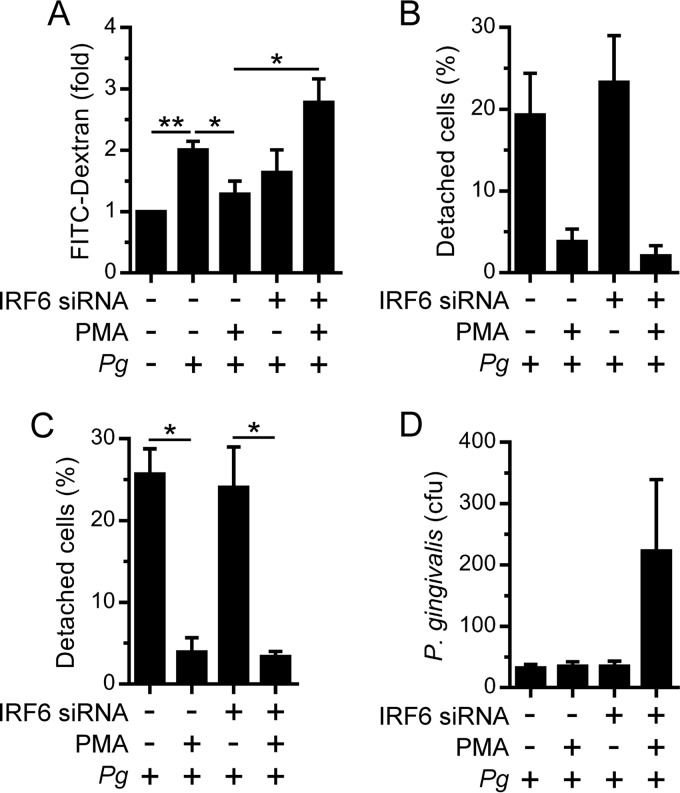
P. gingivalis can impair epithelial barrier function by secreting proteinases (e.g., Kgp and RgpA/B) which degrade proteins that regulate cell-cell and cell-extracellular matrix interactions (19–21). Therefore, we investigated the importance of IRF6-mediated differentiation for the barrier function of OKF6 cells. FITC-dextran was used to assess the effect of P. gingivalis on the intercellular permeability of OKF6 cells. P. gingivalis was found to increase the intercellular permeability of OKF6 cells (Fig. 6A). We have previously demonstrated that PMA upregulates IRF6 expression in OKF6 cells and also stimulates their differentiation via the RIPK4-mediated activation of IRF6 (7). Therefore, to determine whether the upregulation of IRF6 expression in OKF6 cells promotes barrier function, we tested the effect of PMA on P. gingivalis-induced intercellular permeability. Treating OKF6 cells with PMA significantly inhibited the stimulation of intercellular permeability by P. gingivalis (Fig. 6A). Furthermore, the inhibition of P. gingivalis-induced intercellular permeability by PMA was IRF6 dependent (Fig. 6A). PMA also reduced the detachment of OKF6 cells by P. gingivalis (Fig. 6B); the proportion of cells that were weakly adherent was also reduced (Fig. 6C). The inhibitory effect of PMA on P. gingivalis-induced cell detachment/reduced adherence was IRF6 independent (Fig. 6B and C). P. gingivalis can also invade epithelial cells (22, 23). Therefore, we investigated whether the silencing of IRF6 affected the invasion of OKF6 cells by P. gingivalis. As shown in Fig. 6D, the silencing of IRF6 increased the invasion of PMA-treated OKF6 cells by P. gingivalis. Taken together, these results indicate that IRF6 promotes the barrier function of oral keratinocytes.
FIG 6.
Regulation of oral keratinocyte barrier function by IRF6. OKF6 cells were transfected with an IRF6 (+) or control (–) siRNA. Thereafter, the cells were treated with 100 ng/ml PMA (+) or 0.1% DMSO (–) for 24 h. (A) Cells growing on a Transwell insert were incubated with P. gingivalis (MOI of 100:1) and 1 mg/ml FITC-dextran for 60 min, and FITC-dextran levels in the lower chamber were then measured (n = 4). (B and C) Cells growing in a 12-well plate were cultured with P. gingivalis (MOI of 100:1) for 24 h. (B) Detached cells were collected and enumerated (n = 2). (C) The remaining adherent cells in the experiment performed as described for panel B were incubated in PBS-EDTA for 60 min, and the numbers of detached cells were then enumerated (n = 2). In panels B and C, the numbers of detached cells are presented as a percentage of the total (detached plus attached) cell number. (D) Cells growing in a 12-well plate were cultured with P. gingivalis (MOI of 100:1) for 2 h, and bacterial invasion was then measured by antibiotic protection assay (n = 4). **, P < 0.01; *, P < 0.05.
DISCUSSION
In this study, we have established that IRF6 promotes the differentiation of oral keratinocytes in response to the periodontal pathogen P. gingivalis. Our data suggest that IRF6 confers increased structural integrity to oral keratinocytes by promoting the expression of proteins that mediate cornification (e.g., IVL, SPRR1, and TGM1) and tight junction formation (e.g., OCLN). IRF6 was demonstrated to be important for the barrier function of oral keratinocytes; specifically, silencing of IRF6 resulted in increased P. gingivalis-induced intercellular permeability and cell invasion. Accordingly, our findings potentially posit IRF6 as an important regulator of oral barrier defense.
We recently demonstrated that the expression of IRF6 in oral keratinocytes (e.g., OKF6 cells) was stimulated by P. gingivalis (14). We have shown here that IRF6 is important for the stimulation of IVL and KRT13 gene expression in OKF6 cells by P. gingivalis. In addition to these markers of keratinocyte terminal differentiation, IRF6 was also shown to be important for the upregulation of other transcriptional regulators of differentiation, including GRHL3. We have previously established that IRF6 is activated by RIPK4 signaling in differentiating keratinocytes (7, 12); specifically, RIPK4 stimulates the transactivator function of IRF6 by phosphorylating regulatory serine residues in its C-terminal domain (7). Significantly, the silencing of RIPK4 in OKF6 cells inhibited the stimulation of IVL, KRT13, and GRHL3 gene expression in response to P. gingivalis. Although further studies will be required to determine how RIPK4 is activated by P. gingivalis, RIPK4 likely promotes subsequent keratinocyte terminal differentiation by activating IRF6.
IVL is a key component of the cornified envelope of terminally differentiated keratinocytes, including those of the gingival epithelium, and functions as a scaffold to which other structural proteins (e.g., envoplakin) as well as lipids are covalently attached by transglutaminases (e.g., TGM1) (1, 2). The cross-linking of small proline-rich proteins (SPRRs) further strengthens the cornified envelope (1, 2). The gene expression levels of TGM1 and SPRR1 in OKF6 cells were stimulated in an IRF6-dependent manner by P. gingivalis. We recently established in PMA-stimulated OKF6 cells that IRF6 forms a hierarchal regulatory network with the GRHL3 and ELF3 transcription factors to promote IVL, TGM1, and SPRR1 gene expression downstream of RIPK4 signaling (7, 29). Significantly, a role for GRHL3 in mediating the repair of epidermal lesions arising from immune-mediated barrier dysregulation has been demonstrated (30). Thus, the IRF6-dependent stimulation of GRHL3 gene expression in response to P. gingivalis may serve to promote the integrity of oral epithelia.
The assembling of keratin proteins into filaments also enhances the integrity of epithelia; furthermore, the specific keratins expressed can change as keratinocytes differentiate (1, 2). Accordingly, IRF6 may promote barrier function by also regulating the expression of specific keratins, for example, KRT13. The forkhead box O (FOXO) transcription factors FOXO1 and FOXO3 (FOXO1/3) were recently shown to regulate the stimulation of KRT1 and KRT10 expression by P. gingivalis (31). A functional relationship between IRF6 and FOXO1/3 has not been reported. However, FOXO1 can interact with NOTCH and CSL in differentiating myoblasts (32), and IRF6 expression in differentiating keratinocytes is regulated by NOTCH and CSL (33). Accordingly, it will be important to establish whether IRF6 functions downstream of FOXO1/3 to regulate the expression of specific keratins.
Epithelial barrier function is also enhanced by the formation of tight junctions. P. gingivalis has been shown to cause the proteolytic degradation of the tight junction protein OCLN in epithelial cells (19). Therefore, our demonstration of the IRF6-dependent upregulation of OCLN gene expression in response to P. gingivalis may be a host compensatory mechanism to promote the maintenance of tight junctions. Interestingly, the upregulation of OCLN gene expression was a specific response, as the gene expression of other tight junction proteins, such as CLDN1 and TJP1, was not stimulated.
The differentiation of OKF6 cells with PMA increased their resistance to P. gingivalis-induced intercellular permeability. Significantly, the silencing of IRF6 expression prevented the increase in resistance, consistent with IRF6 promoting the barrier function of oral keratinocytes. PMA-differentiated OKF6 cells were also more resistant to P. gingivalis-induced cell detachment; however, this occurred independently of IRF6, thereby indicating that PKC signaling also regulates keratinocyte adhesion via additional pathways. Interestingly, PMA-induced differentiation did not affect the invasion of OKF6 cells by P. gingivalis, except when IRF6 expression was silenced. While not addressed in this study, it is possible that as keratinocytes differentiate in response to PMA, IRF6 may not only promote the expression of proteins that enhance barrier function (e.g., IVL and TGM1) but also inhibit the expression of proteins which might be utilized by P. gingivalis to facilitate invasion. For instance, integrin proteins have been shown to play a role in cell invasion by P. gingivalis (34), and IRF6 was recently reported to inhibit the expression of alpha-3 integrin in differentiating keratinocytes (5). Finally, although OKF6 cells retain the ability to differentiate like primary cells (35) and have been used in similar studies exploring the interaction of P. gingivalis with oral keratinocytes (36–38), the possibility that subtle differences in the differentiation program between OKF6 cells and primary oral keratinocytes exist needs to be considered.
Collectively, our findings indicate that IRF6 promotes the differentiation of oral keratinocytes in response to P. gingivalis. Given that IRF6 also regulates the expression of inflammatory cytokines by oral keratinocytes (14, 26, 39), IRF6 may be a key mediator of host defense against P. gingivalis and possibly other pathogens. Indeed, genetic deletion of IRF6 in the oral epithelium increased the susceptibility of mice to oral colonization by pathogens (40). IRF6 also regulates the differentiation and expression of inflammatory cytokines by epidermal keratinocytes (5, 7, 41). Thus, roles for IRF6 in host defense likely extend beyond oral epithelia.
MATERIALS AND METHODS
Abbreviations.
ANOVA, analysis of variance; BHI, brain heart infusion; CLDN1, claudin 1; CT, threshold cycle; DMSO, dimethyl sulfoxide; EGF, epidermal growth factor; ELF3, E74-like factor-3; FITC, fluorescein isothiocyanate; FOXO1/3, Forkhead box O proteins 1 and 3; GRHL3, Grainyhead-like 3; IL-6, interleukin-6; HPRT, hypoxanthine guanine phosphoribosyl transferase; IRAK1, IL-1 receptor-associated kinase-1; IRF6, interferon regulatory factor 6; IVL, involucrin; Kgp, lysine-specific gingipain; KRT, keratin; MOI, multiplicity of infection; OCLN, occludin; OVOL1, Ovo-like zinc finger 1; PBS, phosphate-buffered saline; PMA, phorbol 12-myristate 13-acetate; qPCR, quantitative real-time PCR; Rgp, arginine-specific gingipain; RIPK4, receptor-interacting protein kinase 4; SEM, standard errors of the means; siRNA, small interfering RNA; SPRR, small proline-rich protein; TGM1, transglutaminase-1; TJP1, tight junction protein 1; TNF, tumor necrosis factor; TLR2, Toll-like receptor 2.
Reagents.
Keratinocyte serum-free medium and supplements (human EGF, bovine pituitary extract, and GlutaMax-1), Opti-MEM I reduced-serum medium, Lipofectamine RNAiMAX transfection reagent, and Silencer Select RIPK4 siRNA were from Life Technologies. The ON-TARGETplus IRAK1, IRF6, and TLR2 siRNAs were from GE Healthcare. BHI medium was from BD Biosciences, and defibrinated horse blood was from Equicell. Hemin, menadione, metronidazole, FITC-dextran (10 kDa), and PMA were from Sigma-Aldrich.
P. gingivalis and culture conditions.
Freeze-dried cultures of P. gingivalis (ATCC 33277) were obtained from the culture collection of the Melbourne Dental School (The University of Melbourne). The bacterium was maintained on horse blood agar plates at 37°C in an anaerobic atmosphere of 5% H2, 80% N2, and 15% CO2. Bacterial colonies were used to inoculate BHI medium supplemented with 0.5 mg/ml cysteine, 5 μg/ml hemin, and 5 μg/ml menadione.
Oral keratinocyte culture conditions.
Human OKF6/TERT-2 oral keratinocytes (35) (here referred to as OKF6 cells) were cultured in keratinocyte serum-free medium supplemented with 0.4 ng/ml human EGF, 25 μg/ml bovine pituitary extract, 0.4 mM CaCl2, and 2 mM GlutaMax-1. The cells were maintained at 37°C in a humidified atmosphere of 5% CO2.
Bacterial challenge of oral keratinocytes.
P. gingivalis cell concentrations were determined spectrophotometrically and confirmed retrospectively by counting viable cell colonies on horse blood agar plates. The bacteria were harvested by centrifugation at 7,000 × g for 20 min at 4°C and resuspended in keratinocyte growth medium. OKF6 cells were cultured with P. gingivalis at an MOI of 100:1.
RNA purification and reverse transcription.
Total RNA was purified using a ReliaPrep RNA Cell miniprep system (Promega), which included an on-column DNase treatment step. RNA was reverse transcribed using random primers and GoScript reverse transcriptase (Promega) according to the manufacturer's instructions. Briefly, 500 ng RNA was incubated in 20 μl of reaction buffer supplemented with MgCl2, PCR nucleotide mix, RNasin RNase inhibitor, 500 ng random primers, and 1 μl GoScript reverse transcriptase for 5 min at 25°C, 60 min at 42°C, and 15 min at 75°C. The cDNA was typically diluted 1:4 with water prior to analysis by quantitative real-time PCR.
Quantitative real-time PCR.
qPCR was performed in duplicate or triplicate using GoTaq qPCR master mix (Promega) and predeveloped TaqMan assays (Life Technologies) for the following proteins (genes): ELF3 (Hs00963881_m1), GRHL3 (Hs00297962_m1), IRAK1 (Hs01018347_m1), IRF6 (Hs00196213_m1), IVL (Hs00902520_m1), KRT13 (Hs00999762_m1), OCLN (Hs00170162_m1), OVOL1 (Hs00190060_m1), RIPK1 (Hs00169407_m1), RIPK2 (Hs01572686_m1), RIPK3 (Hs00179132_m1), RIPK4 (Hs01062501_m1), SPRR1A (Hs00954595_s1), SPRR1B (Hs00824893_m1), and TGM1 (Hs00165929_m1). PCR was performed on a QuantStudio 7 Flex real-time PCR system (Life Technologies). The data were normalized against the HPRT gene, and changes in gene expression were calculated using the ΔCT method.
RNA interference-mediated gene silencing.
A reverse transfection protocol was used for siRNA transfection of OKF6 cells (7, 26). Briefly, the siRNAs were diluted to 120 nM with 100 μl Opti-MEM I reduced-serum medium, mixed with 100 μl Opti-MEM containing 1 μl Lipofectamine RNAiMAX transfection reagent, and incubated at room temperature for 15 to 20 min. OKF6 cells (2 × 105 cells in 1 ml keratinocyte serum-free medium supplemented with 0.4 ng/ml human EGF, 25 μg/ml bovine pituitary extract, 0.4 mM CaCl2, and 2 mM GlutaMax-1) were seeded in 12-well plates and cultured with the transfection cocktail overnight. Thereafter, the medium was replaced and the cells cultured with P. gingivalis 24 h later (i.e., 48 h posttransfection).
Intercellular permeability assay.
OKF6 cells were grown to confluence on 0.4-μm-diameter insertions in a 12-mm-diameter Transwell plate (Sigma-Aldrich). P. gingivalis (MOI of 100:1) and 1 mg/ml FITC-dextran (10 kDa) were added to the upper chamber of the insertions, and the plate was incubated at 37°C for 60 min. FITC-dextran in the lower chamber was measured on a plate reader (Victor3; PerkinElmer) with a 485-nm/535-nm excitation-emission filter set.
Cell detachment assay.
OKF6 cells were cultured with P. gingivalis (MOI of 100:1) in a 12-well plate for 24 h at 37°C. The growth medium was collected, and the well was gently washed with room-temperature PBS. The growth medium and PBS were combined, and the cells were collected by centrifugation. Thereafter, the cells were resuspended in fresh growth medium and viable cells enumerated using trypan blue. The cells that remained attached following the PBS wash were incubated in PBS supplemented with 10 mM EDTA for 60 min on a rocking platform. Detached cells were collected and enumerated as described above.
Cell invasion assay.
OKF6 cells were cultured with P. gingivalis (MOI of 100:1) in a 12-well plate for 2 h at 37°C. Extracellular bacteria were killed by incubating the cells in the presence of 25 μg/ml metronidazole for 60 min. Thereafter, the cells were washed with PBS and lysed with Milli-Q water. Duplicate aliquots of the cell lysates were spread on horse blood agar plates and incubated at 37°C under anaerobic conditions. Bacterial colonies were enumerated after 7 days.
Statistical analysis.
Data combined from three or more independent biological replicate experiments are presented as means ± SEM. Statistical analyses were performed using GraphPad Prism 7. Differences between two groups were evaluated using the Student t test. For multiple comparisons, statistical analysis was performed by ANOVA with Dunnett's (time course experiments) or Sidak's (gene silencing experiments) post hoc test. A P value of <0.05 was considered to be statistically significant.
ACKNOWLEDGMENTS
We thank J. Rheinwald (Harvard Medical School, Cambridge, MA) for generously providing the OKF6/TERT-2 cell line used in this study.
This research was supported by the Australian Government, Department of Industry, Innovation and Science, and National Health and Medical Research Council (project grant 628769).
REFERENCES
- 1.Presland RB, Dale BA. 2000. Epithelial structural proteins of the skin and oral cavity: function in health and disease. Crit Rev Oral Biol Med 11:383–408. doi: 10.1177/10454411000110040101. [DOI] [PubMed] [Google Scholar]
- 2.Candi E, Schmidt R, Melino G. 2005. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol 6:328–340. doi: 10.1038/nrm1619. [DOI] [PubMed] [Google Scholar]
- 3.Polimeni G, Xiropaidis AV, Wikesjo UM. 2006. Biology and principles of periodontal wound healing/regeneration. Periodontol 2000 41:30–47. doi: 10.1111/j.1600-0757.2006.00157.x. [DOI] [PubMed] [Google Scholar]
- 4.Ingraham CR, Kinoshita A, Kondo S, Yang B, Sajan S, Trout KJ, Malik MI, Dunnwald M, Goudy SL, Lovett M, Murray JC, Schutte BC. 2006. Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6). Nat Genet 38:1335–1340. doi: 10.1038/ng1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Botti E, Spallone G, Moretti F, Marinari B, Pinetti V, Galanti S, De Meo PD, De Nicola F, Ganci F, Castrignano T, Pesole G, Chimenti S, Guerrini L, Fanciulli M, Blandino G, Karin M, Costanzo A. 2011. Developmental factor IRF6 exhibits tumor suppressor activity in squamous cell carcinomas. Proc Natl Acad Sci U S A 108:13710–13715. doi: 10.1073/pnas.1110931108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de la Garza G, Schleiffarth JR, Dunnwald M, Mankad A, Weirather JL, Bonde G, Butcher S, Mansour TA, Kousa YA, Fukazawa CF, Houston DW, Manak JR, Schutte BC, Wagner DS, Cornell RA. 2013. Interferon regulatory factor 6 promotes differentiation of the periderm by activating expression of Grainyhead-like 3. J Investig Dermatol 133:68–77. doi: 10.1038/jid.2012.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwa MQ, Huynh J, Aw J, Zhang L, Nguyen T, Reynolds EC, Sweet MJ, Hamilton JA, Scholz GM. 2014. Receptor-interacting protein kinase 4 and interferon regulatory factor 6 function as a signaling axis to regulate keratinocyte differentiation. J Biol Chem 289:31077–31087. doi: 10.1074/jbc.M114.589382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ting SB, Caddy J, Hislop N, Wilanowski T, Auden A, Zhao LL, Ellis S, Kaur P, Uchida Y, Holleran WM, Elias PM, Cunningham JM, Jane SM. 2005. A homolog of Drosophila grainy head is essential for epidermal integrity in mice. Science 308:411–413. doi: 10.1126/science.1107511. [DOI] [PubMed] [Google Scholar]
- 9.Yu Z, Lin KK, Bhandari A, Spencer JA, Xu X, Wang N, Lu Z, Gill GN, Roop DR, Wertz P, Andersen B. 2006. The Grainyhead-like epithelial transactivator Get-1/Grhl3 regulates epidermal terminal differentiation and interacts functionally with LMO4. Dev Biol 299:122–136. doi: 10.1016/j.ydbio.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 10.Hopkin AS, Gordon W, Klein RH, Espitia F, Daily K, Zeller M, Baldi P, Andersen B. 2012. GRHL3/GET1 and trithorax group members collaborate to activate the epidermal progenitor differentiation program. PLoS Genet 8:e1002829. doi: 10.1371/journal.pgen.1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nair M, Teng A, Bilanchone V, Agrawal A, Li B, Dai X. 2006. Ovol1 regulates the growth arrest of embryonic epidermal progenitor cells and represses c-myc transcription. J Cell Biol 173:253–264. doi: 10.1083/jcb.200508196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwa MQ, Huynh J, Reynolds EC, Hamilton JA, Scholz GM. 2015. Disease-associated mutations in IRF6 and RIPK4 dysregulate their signalling functions. Cell Signal 27:1509–1516. doi: 10.1016/j.cellsig.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 13.Holland P, Willis C, Kanaly S, Glaccum M, Warren A, Charrier K, Murison J, Derry J, Virca G, Bird T, Peschon J. 2002. RIP4 is an ankyrin repeat-containing kinase essential for keratinocyte differentiation. Curr Biol 12:1424–1428. doi: 10.1016/S0960-9822(02)01075-8. [DOI] [PubMed] [Google Scholar]
- 14.Huynh J, Scholz GM, Aw J, Kwa MQ, Achuthan A, Hamilton JA, Reynolds EC. 2016. IRF6 regulates the expression of IL-36gamma by human oral epithelial cells in response to Porphyromonas gingivalis. J Immunol 196:2230–2238. doi: 10.4049/jimmunol.1501263. [DOI] [PubMed] [Google Scholar]
- 15.Darveau RP. 2010. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol 8:481–490. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- 16.Hajishengallis G, Darveau RP, Curtis MA. 2012. The keystone-pathogen hypothesis. Nat Rev Microbiol 10:717–725. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zijnge V, van Leeuwen MBM, Degener JE, Abbas F, Thurnheer T, Gmur R, Harmsen HJM. 2010. Oral biofilm architecture on natural teeth. PLoS One 5:e9321. doi: 10.1371/journal.pone.0009321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reference deleted.
- 19.Katz J, Sambandam V, Wu JH, Michalek SM, Balkovetz DF. 2000. Characterization of Porphyromonas gingivalis-induced degradation of epithelial cell junctional complexes. Infect Immun 68:1441–1449. doi: 10.1128/IAI.68.3.1441-1449.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katz J, Yang QB, Zhang P, Potempa J, Travis J, Michalek SM, Balkovetz DF. 2002. Hydrolysis of epithelial junctional proteins by Porphyromonas gingivalis gingipains. Infect Immun 70:2512–2518. doi: 10.1128/IAI.70.5.2512-2518.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hintermann E, Haake SK, Christen U, Sharabi A, Quaranta V. 2002. Discrete proteolysis of focal contact and adherens junction components in Porphyromonas gingivalis-infected oral keratinocytes: a strategy for cell adhesion and migration disabling. Infect Immun 70:5846–5856. doi: 10.1128/IAI.70.10.5846-5856.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamont RJ, Chan A, Belton CM, Izutsu KT, Vasel D, Weinberg A. 1995. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect Immun 63:3878–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakagawa I, Inaba H, Yamamura T, Kato T, Kawai S, Ooshima T, Amano A. 2006. Invasion of epithelial cells and proteolysis of cellular focal adhesion components by distinct types of Porphyromonas gingivalis fimbriae. Infect Immun 74:3773–3782. doi: 10.1128/IAI.01902-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hajishengallis G. 2015. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol 15:30–44. doi: 10.1038/nri3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burns E, Bachrach G, Shapira L, Nussbaum G. 2006. Cutting edge: TLR2 is required for the innate response to Porphyromonas gingivalis: activation leads to bacterial persistence and TLR2 deficiency attenuates induced alveolar bone resorption. J Immunol 177:8296–8300. doi: 10.4049/jimmunol.177.12.8296. [DOI] [PubMed] [Google Scholar]
- 26.Kwa MQ, Nguyen T, Huynh J, Ramnath D, De Nardo D, Lam PY, Reynolds EC, Hamilton JA, Sweet MJ, Scholz GM. 2014. Interferon regulatory factor 6 differentially regulates Toll-like receptor 2-dependent chemokine gene expression in epithelial cells. J Biol Chem 289:19758–19768. doi: 10.1074/jbc.M114.584540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Groote P, Tran HT, Fransen M, Tanghe G, Urwyler C, De Craene B, Leurs K, Gilbert B, Van Imschoot G, De Rycke R, Guerin CJ, Holland P, Berx G, Vandenabeele P, Lippens S, Vleminckx K, Declercq W. 2015. A novel RIPK4-IRF6 connection is required to prevent epithelial fusions characteristic for popliteal pterygium syndromes. Cell Death Differ 22:1012–1024. doi: 10.1038/cdd.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuki M, Yamashita F, Ishida-Yamamoto A, Yamada K, Kinoshita C, Fushiki S, Ueda E, Morishima Y, Tabata K, Yasuno H, Hashida M, Iizuka H, Ikawa M, Okabe M, Kondoh G, Kinoshita T, Takeda J, Yamanishi K. 1998. Defective stratum corneum and early neonatal death in mice lacking the gene for transglutaminase 1 (keratinocyte transglutaminase). Proc Natl Acad Sci U S A 95:1044–1049. doi: 10.1073/pnas.95.3.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scholz GM, Sulaiman NS, Al Baiiaty S, Kwa MQ, Reynolds EC. 2016. A novel regulatory relationship between RIPK4 and ELF3 in keratinocytes. Cell Signal 28:1916–1922. doi: 10.1016/j.cellsig.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 30.Gordon WM, Zeller MD, Klein RH, Swindell WR, Ho H, Espetia F, Gudjonsson JE, Baldi PF, Andersen B. 2014. A GRHL3-regulated repair pathway suppresses immune-mediated epidermal hyperplasia. J Clin Invest 124:5205–5218. doi: 10.1172/JCI77138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li S, Dong G, Moschidis A, Ortiz J, Benakanakere MR, Kinane DF, Graves DT. 2013. P. gingivalis modulates keratinocytes through FOXO transcription factors. PLoS One 8:e78541. doi: 10.1371/journal.pone.0078541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitamura T, Kitamura YI, Funahashi Y, Shawber CJ, Castrillon DH, Kollipara R, DePinho RA, Kitajewski J, Accili D. 2007. A Foxo/Notch pathway controls myogenic differentiation and fiber type specification. J Clin Invest 117:2477–2485. doi: 10.1172/JCI32054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Restivo G, Nguyen BC, Dziunycz P, Ristorcelli E, Ryan RJ, Ozuysal OY, Di Piazza M, Radtke F, Dixon MJ, Hofbauer GF, Lefort K, Dotto GP. 2011. IRF6 is a mediator of Notch pro-differentiation and tumour suppressive function in keratinocytes. EMBO J 30:4571–4585. doi: 10.1038/emboj.2011.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yilmaz O, Watanabe K, Lamont RJ. 2002. Involvement of integrins in fimbriae-mediated binding and invasion by Porphyromonas gingivalis. Cell Microbiol 4:305–314. doi: 10.1046/j.1462-5822.2002.00192.x. [DOI] [PubMed] [Google Scholar]
- 35.Dickson MA, Hahn WC, Ino Y, Ronfard V, Wu JY, Weinberg RA, Louis DN, Li FP, Rheinwald JG. 2000. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol 20:1436–1447. doi: 10.1128/MCB.20.4.1436-1447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giacaman RA, Nobbs AH, Ross KF, Herzberg MC. 2007. Porphyromonas gingivalis selectively up-regulates the HIV-1 coreceptor CCR5 in oral keratinocytes. J Immunol 179:2542–2550. doi: 10.4049/jimmunol.179.4.2542. [DOI] [PubMed] [Google Scholar]
- 37.Dommisch H, Chung WO, Jepsen S, Hacker BM, Dale BA. 2010. Phospholipase C, p38/MAPK, and NF-kappaB-mediated induction of MIP-3alpha/CCL20 by Porphyromonas gingivalis. Innate Immun 16:226–234. doi: 10.1177/1753425909339237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramage G, Lappin DF, Millhouse E, Malcolm J, Jose A, Yang J, Bradshaw DJ, Pratten JR, Culshaw S. 22 June 2016. The epithelial cell response to health and disease associated oral biofilm models. J Periodont Res doi: 10.1111/jre.12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwa MQ, Scholz GM, Reynolds EC. 2016. RIPK4 activates an IRF6-mediated proinflammatory cytokine response in keratinocytes. Cytokine 83:19–26. doi: 10.1016/j.cyto.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 40.Tamasas B, Cox TC. 2017. Massively increased caries susceptibility in an Irf6 cleft lip/palate model. J Dent Res 96:315–322. doi: 10.1177/0022034516679376. [DOI] [PubMed] [Google Scholar]
- 41.Ramnath D, Tunny K, Hohenhaus DM, Pitts CM, Bergot AS, Hogarth PM, Hamilton JA, Kapetanovic R, Sturm RA, Scholz GM, Sweet MJ. 2015. TLR3 drives IRF6-dependent IL-23p19 expression and p19/EBI3 heterodimer formation in keratinocytes. Immunol Cell Biol 93:771–779. doi: 10.1038/icb.2015.77. [DOI] [PubMed] [Google Scholar]