Abstract
A fluorogenic-probe hydrolysis (TaqMan)-reverse transcriptase (RT) PCR for classical swine fever virus (CSFV) was evaluated for diagnostic sensitivity and specificity by using clinical samples obtained from the Dominican Republic, where the disease is enzootic. The sensitivity of this test, using nasal swab samples taken from both symptomatic and asymptomatic animals, exceeded the diagnostic sensitivity of virus isolation (100% versus 72.4%, respectively) with little loss of specificity (98.9% versus 100%, respectively). At the herd level, three of four infected farms were identified by virus isolation, while the CSFV real-time RT-PCR assay identified all four infected premises. This simple and accurate test permits rapid detection of CSFV in affected herds.
Classical swine fever (CSF) is a highly contagious and often fatal disease of swine. Classical Swine Fever Virus (CSFV), the causative agent of CSF, is a member of the genus Pestivirus in the family Flaviviridae (22). CSFV is an enveloped virus with a 12.5-kb single-stranded RNA genome of positive polarity (9). The genome encodes a 4,000-amino-acid polyprotein that is co- and posttranslationally processed by viral and cellular proteases into 12 polypeptides (15). Both ends of the genome contain untranslated regions (UTR), which are highly conserved among virus isolates (3, 13, 14, 20, 24).
Although not present in the United States, CSF is distributed worldwide. In the Western hemisphere the disease occurs in Central and South America, southern Mexico, and the Caribbean basin. Sporadic outbreaks of CSF are frequently recorded in the European Union. Countries affected by the disease experience an international ban on trade of live animals and pork products. As a result, the European Union has had a stamping-out/nonvaccination policy since 1990 (23). The CSF outbreak in The Netherlands in 1997 to 1998 resulted in 11 million pigs being destroyed at a cost of over 2 billon US dollars (17). In large part, the disease status of these animals was unknown (5).
Rapid and precise detection of CSFV is critical for disease containment. Current diagnostic methods, including detection of viral antigens in tonsils by using fluorescent antibody (18), antigen capture enzyme-linked immunosorbent assay (4, 21), and detection of genomic RNA by reverse transcription-PCR (3, 8, 11, 12, 24), are relatively rapid diagnostic tests; however, these techniques require centralized laboratory facilities and clinical specimen submissions, which delay disease diagnosis, thus affecting the efficiency of emergency disease management measures.
A rapid, presumptive diagnosis at the site of a suspected disease outbreak would be extremely useful for controlling CSF. To address this need, a fluorogenic-probe hydrolysis (TaqMan)-reverse transcriptase (RT) PCR assay for CSFV was evaluated with samples from experimentally infected swine (19). The assay's sensitivity equalled or exceeded the sensitivity of virus isolation (VI). Viral RNA was detected in nasal and tonsil scraping samples 2 to 4 days prior to the onset of clinical disease. In addition, the assay can be performed in 2 h or less, thus providing a rapid method for the diagnosis of CSF.
Here we evaluated the diagnostic sensitivity and specificity of this test using clinical samples collected from swine holdings in the Dominican Republic (DR). In the DR, CSFV is enzootic in the domestic pig population, although actual disease incidence is unknown. A large part of the national herd is vaccinated regularly with live attenuated and, more recently, CSFV E2 subunit vaccines (2).
A total of 449 nasal swabs were collected from nine farms in the central region of the DR (Table 1). Samples were collected with sterile cotton swabs (Fisherbrand, Fisher Scientific, Pittsburgh, Pa.) and placed into 1.5-ml sterile tubes containing 1 ml of Dulbecco's minimal essential medium (Gibco, Grand Island, N.Y.) plus antibiotics and antimycotic (Gibco). Tubes were placed in dry ice, transported to the laboratory, and kept at −70°C until processed. The CSFV real-time RT-PCR assay was performed as described previously (19). SK6 swine kidney cell monolayers were used for VI. Monolayers were inoculated with 250 μl of filtered (0.22 μm) nasal swab transport media and then incubated at 37°C in 5% CO2. Inoculated cell cultures were passed twice more at 4-day intervals by transferring freeze-thawed cultures onto fresh SK6 cell monolayers before being considered negative for virus. CSFV was detected in cultures by peroxidase staining (1), using the CSFV monoclonal antibody WH303 (7) and a Vectastain ABC kit (Vector Laboratories, Burlingame, Calif.).
TABLE 1.
Detection of CSFV in clinical samples by CSFV real-time RT-PCR assay, VI, and nested PCR
| Farm | Location | CSF clinical signs | No. of samples | Vaccinea | No. of samples
|
CSFV RNA detected by nested PCRb | ||
|---|---|---|---|---|---|---|---|---|
| VI positive | Real-time PCR positive | VI negative PCR positive | ||||||
| 1 | Licey | No | 22 | Marker/subunit | 0 | 2 | 2 | ND (0/2) |
| 2 | Licey | No | 19 | LAV | 0 | 0 | ||
| 3 | Licey | Yes | 57 | None | 3 | 5 | 2 | DR02.3 (2/2) |
| 4 | Licey | No | 18 | LAV | 0 | 0 | ||
| 5 | Licey | Yes | 112 | LAV | 45 | 52 | 7 | DR02.6 (7/7) |
| 6 | Licey | No | 10 | LAV | 0 | 1 | 1 | ND (0/1) |
| 7 | La Vega | Yes | 102 | Marker/subunit | 0 | 1 | 1 | DR02.1 (0/1) |
| 8 | La Paloma | No | 65 | Marker/subunit | 0 | 1 | 1 | ND (1/1) |
| 9 | La Paloma | Yes | 44 | LAV | 2 | 7 | 5 | DR02.6 (1/5); DR02.4 (4/5) |
| Total | 449 | 50 | 69 | 19 | 15/19 | |||
“Marker/subunit” indicates that at the time of sample collection a E2 CSFV vaccine was in use, before that live attenuated vaccine had been used. LAV, live attenuated vaccine.
Values are number of samples with indicated RNA/number of VI-negative PCR-positive samples. ND, not detected.
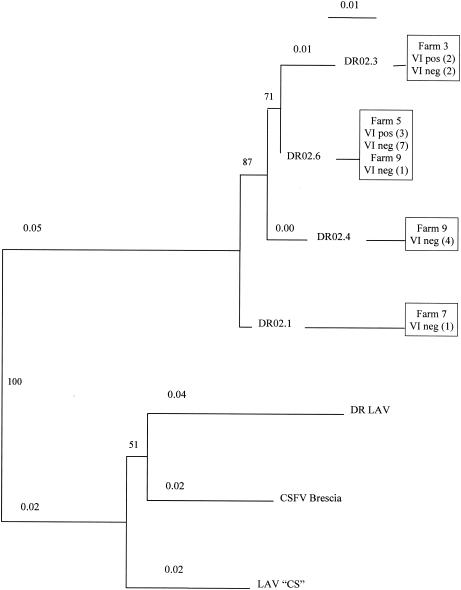
Of 449 samples, 69 were positive by the CSFV real-time RT-PCR assay (Table 1); of these, 50 were VI positive. The 19 samples that tested positive by real-time PCR and negative by VI were evaluated further for the presence of CSFV RNA by a nested PCR. The first PCR consisted of the amplification of a 777-bp segment encompassing the 5′ UTR and regions of Npro, using the primer pair 5′GTCGTCAGTAGTTCGACG 3′ and 5′ATGCTCTTTTGGGGCTAT 3′. Nested PCR was performed with the primers 5′TCTCTGCTGTACATGGCA 3′ and 5′TATCCTTTTGGTCACCTC 3′, which generated a 355-bp amplicon. PCR products obtained were sequenced on a PRISM 3700 automated DNA sequencer (PE Biosystems, Foster City, Calif.). Sequence data were assembled with the Phrap software program, with confirmatory assemblies performed by using CAP3 (10). Of the 19 VI-negative samples examined, CSFV RNA was detected by nested PCR in 15 (Fig. 1; Table 1). Four samples that were negative by nested PCR were assumed to be CSFV real-time RT-PCR assay false positives (Tables 1 and 2). Notably, these samples were obtained from farms (no. 1, 6, and 8) that lacked evidence of clinical disease and from which virus was not isolated (Table 1; Fig. 1). The real-time PCR and nested PCR assays do not target the same genomic region (5′UTR and Npro, respectively). Thus, there is a possibility that the discrepancy between tests results obtained might be due to differences between the sensitivity of real-time PCR and that of the first round of amplification in the nested PCR assay. The DR isolates clustered together and differed from the live attenuated vaccine currently used in the DR (Pestiffa, Merial). Isolates were unique to a particular farm, with the exception of farm 9, where two different viruses were detected (Table 1). Additional samples that were CSFV real-time RT-PCR assay positive and VI positive were also sequenced (n = 5). Two VI-positive samples from farm 3 and three VI-positive samples from farm 5 were similar to nested-PCR-positive VI-negative samples obtained from those farms (Fig. 1; Table 1).
FIG. 1.
Comparative sequence analysis of CSFV isolates from the DR. 5′ UTR/Npro region sequences (see the text) were aligned by using ClustalW. Unrooted trees were generated by using the neighbor-joining algorithm with a Poisson correction for multiple substitutions. DR LAV, CSFV live attenuated vaccine currently used in the DR; LAV “CS”, live CSFV attenuated vaccine from Russia; DR02.1, DR02.3, DR02.4, and DR02.6, DR field isolates; pos, positive; neg, negative. The number of isolates sequenced is shown in parentheses.
TABLE 2.
Performance of the CSFV real-time RT-PCR assay after evaluation of VI-negative CSFV real-time RT-PCR-positive samples by RT-nested PCR
| Test indexa | % | 95% Confidence interval |
|---|---|---|
| Sensitivity | 100 (65/65) | 100-100 |
| Specificity | 98.9 (380/384) | 97.9-99.9 |
| Positive predictive value | 94.3 (65/69) | 88.6-99.7 |
| Negative predictive value | 100 (380/380) | 100-100 |
| False-negative rate | 0.00 (0/65) | 0.0-0.0 |
| False-positive rate | 5.7 (4/69) | 1.8-14.9 |
| Overall accuracy | 99.1 (445/449) | 98.9-99.9 |
Sensitivity, number of positive samples over number of true-positive samples; specificity, number of negative samples over number of true-negative samples; positive predictive value, number of samples that were positive and had CSFV; negative predictive value, number of samples that were negative and did not have CSFV; false-negative rate, number of negative samples confirmed positive over true-positive samples; false-positive rate, number of positive samples confirmed negative over true-positive samples; overall accuracy, total of true-positive and true-negative samples over the total number of samples.
Analysis of the cycle threshold (Ct) values of the CSFV real-time RT-PCR-positive samples indicated that all samples with Ct values below 38 (46 of 69) were CSFV positive by either VI or nested PCR. The four false-positive samples were identified beyond cycle 38, one sample was identified between cycles 38 and 40, and three samples were identified between cycles 40 and 45 (Fig. 2). All samples with Ct values above 45 were CSFV negative by the three tests used here.
FIG. 2.
Relationship between (Ct) values of real-time PCR-positive samples and CSFV status of the samples. Filled diamonds represent samples that were positive by both CSFV RT-PCR assay and VI (n = 50). Empty squares represent samples that were CSFV RT-PCR assay positive, VI negative, and nested PCR positive (n = 15). Filled triangles represent false-positive samples. These samples were CSFV RT-PCR assay positive but negative by both VI and nested PCR (n = 4).
Between cycles 40 and 45, a total of eight samples were positive (Fig. 2). Three of these samples were false positives (75% of false-positive results), one sample was VI positive, and four samples were confirmed by nested PCR and sequencing as CSFV positive. Thus, samples testing positive between cycles 40 and 45 require the use of a second confirmatory test, in this case nested PCR, to determine true sample status.
In this diagnostic evaluation, CSFV real-time RT-PCR assay outperformed VI, which is considered the diagnostic standard for CSF (5, 16, 18). The performances of VI and CSFV real-time RT-PCR were estimated as previously described (University of Oklahoma Health Sciences Center [www.fammed.ouhsc.edu/robhamm/cdmcalc.htm]). The diagnostic sensitivity of the CSFV real-time RT-PCR assay (100%) was higher than that of VI (72.4%) with little loss of specificity (Tables 2 and 3). The false-negative rate of VI in this evaluation was 27.54%, compared with no false negatives observed with the CSFV real-time RT-PCR assay (Tables 2 and 3). Importantly, the overall accuracy of the CSFV real-time RT-PCR assay (99.14%) was higher than that of VI (95.77%) (Tables 2 and 3).
TABLE 3.
Comparison of performance of VI with CSFV real-time RT-PCR assay
| Test indexa | % | 95% confidence interval |
|---|---|---|
| Sensitivity | 72.4 (50/69) | 61.9-83.0 |
| Specificity | 100 (380/380) | 100-100 |
| Positive predictive value | 100 (50/50) | 100-100 |
| Negative predictive value | 95.2 (380/399) | 93.1-97.3 |
| False negative rate | 27.5 (19/69) | 17.0-31.9 |
| False positive rate | 0.0 (0/50) | 0.0-0.0 |
| Overall accuracy | 95.7 (430/449) | 93.9-97.6 |
Sensitivity, number of positive samples over number of true-positive samples; specificity, number of negative samples over number of true-negative samples; positive predictive value, number of samples that were positive and had CSFV; negative predictive value, number of samples that were negative and did not have CSFV; false-negative rate, number of negative samples confirmed positive over true-positive samples; false-positive rate, number of positive samples confirmed negative over true-positive samples; overall accuracy, total of true-positive and true-negative samples over the total number of samples.
Analysis of these data at the herd level indicates that CSFV was detected by CSFV real-time RT-PCR assay on four farms (farms 3, 5, 7, and 9) (Table 1), while only three farms were positive by VI (farms 3, 5, and 9) (Table 1). CSFV real-time RT-PCR detected CSFV on farms 1, 3, 5, 6, 7, 8, and 9. Farms 1, 6, and 8 were confirmed as CSFV negative by the confirmatory nested PCR assay. Thus, the CSFV real-time RT-PCR assay allowed the identification of a fourth CSFV-infected farm, farm 7, which was not identified by VI. A low predictive value for VI has been previously noted during a CSF outbreak in The Netherlands where only 4.5% of infected herds were detected by VI (6, 17). Similarly, a lower sensitivity for VI was observed by us previously, using experimentally infected animals (19).
In summary, the CSFV real-time RT-PCR assay had a higher predictive value than the current diagnostic standard, VI. Notably, the high sensitivity of the test is accompanied by little loss of specificity. The assay allowed rapid identification of CSF-affected farms by using a nasal swab sample. This diagnostic tool should prove useful during a CSF outbreak, when decisions regarding infection status need to be made rapidly to curtail disease transmission.
Acknowledgments
We thank Felix Del Orbe (“LAVECEN”, Santo Domingo, Dominican Republic) and Luis Ramos (Dirección General de Ganadería, Secretaría de Estado de Agricultura, Dominican Republic) for providing all field support. We thank Pedro Hansen and Diego López from Instituciones Pecuarias Dominicanas S.A. for their assistance. We thank Richard Pacer, Veterinary Attaché, USDA, APHIS, IS, Dominican Republic, for coordinating field activities.
REFERENCES
- 1.Afshar, A., G. C. Dulac, and A. Bouffard. 1989. Application of peroxidase labelled antibody assays for detection of porcine IgG antibodies to hog cholera and bovine viral diarrhea viruses. J. Virol. Methods 23:253-261. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous. 2002. Control y erradicación del Cólera Porcino. Secretaría de Ganadería, Ministerio de Agricultura, República Dominicana.
- 3.Boye, M., S. Kamstrup, and K. Dalsgaard. 1991. Specific sequence amplification of bovine virus diarrhea virus (BVDV) and hog cholera virus and sequencing of BVDV nucleic acid. Vet. Microbiol. 29:1-13. [DOI] [PubMed] [Google Scholar]
- 4.Clavijo, A., E. M. Zhou, S. Vydelingum, and R. Heckert. 1998. Development and evaluation of a novel antigen capture assay for the detection of classical swine fever virus antigens. Vet. Microbiol. 60:155-168. [DOI] [PubMed] [Google Scholar]
- 5.de Smit, A. J. 2000. Classical swine fever. efficacy of marker vaccines and laboratory diagnosis. Ph.D. thesis. University of Utrecht, Utrecht, The Netherlands.
- 6.de Smit, A. J., P. L. Eble, E. P. de Kluijver, M. Bloemraad, and A. Bouma. 1999. Laboratory decision-making during the classical swine fever epidemic of 1997-1998 in The Netherlands. Prev. Vet. Med. 42:185-199. [DOI] [PubMed] [Google Scholar]
- 7.Edwards, S., V. Moennig, and G. Wensvoort. 1991. The development of an international reference panel of monoclonal antibodies for the differentiation of hog cholera virus from other pestiviruses. Vet. Microbiol. 29:101-108. [DOI] [PubMed] [Google Scholar]
- 8.Harding, M., C. Lutze-Wallace, I. Prud'Homme, X. Zhong, and J. Rola. 1994. Reverse transcriptase-PCR assay for detection of hog cholera virus. J. Clin. Microbiol. 32:2600-2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horzinek, M. C. 1991. Pestiviruses—taxonomic perspectives. Arch. Virol. Suppl. 3:1-5. [PubMed] [Google Scholar]
- 10.Huang, X., and A. Madan. 1999. CAP3: a DNA sequence assembly program. Genome Res. 9:868-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katz, J. B., J. F. Ridpath, and S. R. Bolin. 1993. Presumptive diagnostic differentiation of hog cholera virus from bovine viral diarrhea and border disease viruses by using a cDNA nested-amplification approach. J. Clin. Microbiol. 31:565-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu, S. T., S. N. Li, D. C. Wang, S. F. Chang, S. C. Chiang, W. C. Ho, Y. S. Chang, and S. S. Lai. 1991. Rapid detection of hog cholera virus in tissues by the polymerase chain reaction. J. Virol. Methods 35:227-236. [DOI] [PubMed] [Google Scholar]
- 13.Lowings, P., G. Ibata, J. Needham, and D. Paton. 1996. Classical swine fever virus diversity and evolution. J. Gen. Virol. 77(Pt. 6):1311-1321. [DOI] [PubMed] [Google Scholar]
- 14.McGoldrick, A., J. P. Lowings, G. Ibata, J. J. Sands, S. Belak, and D. J. Paton. 1998. A novel approach to the detection of classical swine fever virus by RT-PCR with a fluorogenic probe (TaqMan). J. Virol. Methods 72:125-135. [DOI] [PubMed] [Google Scholar]
- 15.Meyers, G., H. J. Thiel, and T. Rumenapf. 1996. Classical swine fever virus: recovery of infectious viruses from cDNA constructs and generation of recombinant cytopathogenic defective interfering particles. J. Virol. 70:1588-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pearson, J. E. 1992. Hog cholera diagnostic techniques. Comp. Immunol. Microbiol. Infect. Dis. 15:213-219. [DOI] [PubMed] [Google Scholar]
- 17.Pluimers, F. H., P. W. de Leeuw, J. A. Smak, A. R. Elbers, and J. A. Stegeman. 1999. Classical swine fever in The Netherlands 1997-1998: a description of organisation and measures to eradicate the disease. Prev. Vet. Med. 42:139-155. [DOI] [PubMed] [Google Scholar]
- 18.Ressang, A. A., and G. F. de Boer. 1968. A comparison between the cell culture, frozen tissue section, impression and mucosal smear techniques for fluorescent antibody in the diagnosis of hog cholera. Netherlands J. Vet. Sci. 1:72. [Google Scholar]
- 19.Risatti, G. R., J. D. Callahan, W. M. Nelson, and M. V. Borca. 2003. Rapid detection of classical swine fever virus by a portable real-time reverse transcriptase PCR assay. J. Clin. Microbiol. 41:500-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schroeder, B. A., and T. C. Balassu-Chan. 1990. Specific sequence amplification of bovine viral diarrhoea virus nucleic acid. Arch. Virol. 111:239-246. [DOI] [PubMed] [Google Scholar]
- 21.Shannon, A. D., C. Morrissy, S. G. Mackintosh, and H. A. Westbury. 1993. Detection of hog cholera virus antigens in experimentally-infected pigs using an antigen-capture ELISA. Vet. Microbiol. 34:233-248. [DOI] [PubMed] [Google Scholar]
- 22.Wengler, G., D. W. Bradley, M. S. Colett, F. X. Heinz, R. W. Schlesinger, and J. H. Strauss. 1995. Flaviviridae, p. 415-427. In F. A. Murphy, C. M. Fauquet, D. H. L. Bishop, S. A. Ghabrial, A. W. Jarbis, G. P. Martelli, M. A. Mayo, and M. D. Summers (ed.), Virus taxonomy. Sixth Report of the International Committee on Taxonomy of Viruses. Springer-Verlag, New York, N.Y.
- 23.Westergaard, J. M. 1994. Attitude of the European Community to vaccines, p. 13-20. In P. R. Wood, P. Willadsen, J. E. Vercoe, R. M. Hoskinson, and D. Demayer (ed.), Vaccines in agriculture: immunological applications to animal health and production. CSIRO, East Melbourne, Australia.
- 24.Wirtz, B., J. D. Tratchin, H. K. Muller, and D. B. Mitchell. 1993. Detection of hog cholera virus and differentiation from another pestiviruses by polymerase chain reaction. J. Clin. Microbiol. 31:1148-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]