Abstract
Staphylococcus caprae, a hemolytic coagulase-negative staphylococcus that is infrequently associated with humans, was initially detected in specimens from six infants in our neonatal intensive care unit due to phenotypic characteristics common to methicillin-resistant Staphylococcus aureus. These isolates were subsequently identified as S. caprae by the Automated RiboPrinter microbial characterization system. This prompted an 8-month retrospective investigation in our neonatal intensive care unit. S. caprae was the cause of 6 of 18 episodes of coagulase-negative staphylococcal bacteremia, was the most common coagulase-negative staphylococcus recovered from the nares of 6 of 32 infants surveyed in a methicillin-resistant S. aureus surveillance program, and was isolated from 1 of 37 health care providers' hands. Of 13 neonatal intensive care unit isolates tested, all were methicillin resistant and positive for the mecA gene. All 21 isolates were found to be a single strain by Automated RiboPrinter and pulsed-field gel electrophoresis with ApaI or SmaI digestion; ApaI was more discriminating in analyzing epidemiologically unrelated strains than Automated RiboPrinter or electrophoresis with SmaI. These findings extend the importance of S. caprae, emphasize its similarities to methicillin-resistant S. aureus, and demonstrate its ability to persist in an intensive care unit setting.
Coagulase-negative staphylococci are a major cause of nosocomially acquired bacteremia and other potential human infections in intensive care units, including neonatal intensive care units. Staphylococcus caprae, one of these species, is usually associated with animals, especially goats, and infrequently associated with humans. The infrequent human infections include bone and joint (3, 5, 13), sepsis/bacteremia (11, 14), recurring sepsis (16), urinary tract infections (18), endocarditis (18), and cases of acute otitis externa (12). Most infections have been hospital acquired, although a few community-acquired infections have been documented.
S. caprae was detected in specimens from six index infants in the neonatal intensive care unit, being recovered as part of an active methicillin-resistant Staphylococcus aureus nares surveillance program, as well as from a cerebrospinal fluid specimen and a vascular catheter of individual infants; the organisms detected were presumptively thought to be methicillin-resistant S. aureus due to shared phenotypic characteristics between methicillin-resistant S. aureus and S. caprae. Since the latter species is infrequently associated with humans, these findings prompted a retrospective epidemiological and microbiological study to determine the infection and colonization rates of S. caprae in our neonatal intensive care unit. Selected isolates were tested for the mecA gene to determine the mechanism of the methicillin resistance. In addition, we assessed whether the S. caprae isolates recovered were clonally related by genotyping with the RiboPrinter microbial characterization system (Qualicon, Inc.) and by a reference pulsed-field gel electrophoresis method.
MATERIALS AND METHODS
Organisms studied.
All coagulase-negative staphylococci recovered from blood over the 8-month period and blood isolates from 1 year before and 1 year after this period were studied. In addition, all hemolytic coagulase-negative staphylococci recovered from a one-time cycle of the monthly methicillin-resistant S. aureus nares surveillance cultures from infants in the neonatal intensive care unit and hand cultures from neonatal intensive care unit health care providers were also studied. Beta-hemolysis was read from the sheep blood agar plate for up to 48 h after the start of incubation.
A stock culture panel of 12 unrelated strains of S. caprae was also tested, including ATCC 51549, ATCC 33538, and 10 strains kindly provided by P. S. Roland at Alcon Laboratories, Fort Worth, Tex. These strains served as controls for the Automated RiboPrinter to ensure accurate identification; they also served as epidemiologically unrelated strains to ensure the discriminatory ability of the Automated RiboPrinter and pulsed-field gel electrophoresis genotyping assays.
Nares surveillance cultures.
The Johns Hopkins Hospital is a 1,026-bed tertiary-care institution. Our intensive care units house 208 beds, of which 36 are designated for neonatal care in our neonatal intensive care unit. All neonatal intensive care unit babies have their nares cultured monthly for methicillin-resistant S. aureus. This routine practice began in 1996. Nares cultures were performed by swabbing the anterior nares with one rayon swab (Becton Dickinson, Cockeysville, Md.) and plating them on 5% sheep blood agar plates (Becton Dickinson).
Hand surveillance cultures.
Hand cultures were performed by massaging both hands in 25 ml of brain heart infusion broth (Becton Dickinson) in a 1-gallon Ziploc bag for 1 min. A 100-μl aliquot was streaked on the surface of 5% sheep blood agar. Plates were incubated at 37°C and observed for hemolytic coagulase-negative staphylococci at 24 and 48 h.
Automated Riboprinter identification and typing.
All coagulase-negative staphylococci and hemolytic coagulase-negative staphylococci were identified to species level and genotyped by the automated Riboprinter microbial characterization system (Qualicon, Wilmington, Del.) according to the manufacturer's instructions. This system with the database available at the time of our study included 33 different species of coagulase-negative staphylococci. The S. caprae species was represented by only four different strains. Briefly, a single 24-h colony was tested. The instrument performs cell lysis, deproteinization, restriction digestion with EcoRI, gel electrophoresis, DNA transfer to a nylon membrane, hybridization with an extracellular matrix-labeled riboprobe, photography, scanning of the DNA patterns, and analysis by their software program. The Automated RiboPrinter is able to delineate strains based on ribogroups and also capable of identifying some bacterial species.
Biochemical (phenotypic) identification.
Since there were only four different strains of S. caprae in the Dupont identification database, a select panel of three isolates of S. caprae, including ATCC 51549 and the first and last clinical isolate recovered from the neonatal intensive care unit, were tested. Biochemical tests included slide coagulase, tube coagulase, nitrate, urea, trehalose, ornithine decarboxylase, polymyxin B resistance, pyrrolidonyl-α-naphthylamine hydrolysis novobiocin resistance, alkaline phosphatase, and nitrophenyl pyranogalactoside.
Pulsed-field gel electrophoresis.
The isolates were grown overnight in BHI broth, washed, suspended in a 0.8% InCert agarose, and pipetted into Bio-Rad molds. The cells in the agarose plugs were lysed with lysostaphin, proteinase K, and lauroylsarcosine. The plugs were washed three times with Tris-EDTA and then the DNA was digested with SmaI and ApaI (New England Biolabs, Beverly, Mass.). Electrophoresis was run with a Bio-Rad Genepath system (Bio-Rad). The parameters were 1% Bio-Rad pulsed-field gel electrophoresis agarose gel, 0.5× Tris-borate-EDTA running buffer at 14°C and 6 V with initial and final switch times of 5 to 50 s for 23.5 h. The gel was stained with ethidium bromide and visualized with UV light and scanned with Quantity One Image Capture software version 4.2.2 (Bio-Rad) and analyzed with the Molecular Analyst fingerprinting software version 1.61 (Bio-Rad). Any two isolates with identical pulsed-field gel electrophoresis patterns were interpreted as the same strain type; isolates that differed by more than three bands were considered epidemiologically unrelated (17).
Methicillin susceptibility testing.
Mean methicillin MICs were determined with oxacillin by the reference agar dilution method as per guidelines for S. aureus (10). Twofold dilutions ranging from 0.125 μg/ml up to 2.0 μg/ml were tested with Mueller Hinton agar supplemented with 2% NaCl. All organisms tested were also subcultured for purity, and the MICs were read after incubation at 30°C for 24 h. With the guidelines for S. aureus breakpoints, an organism was considered methicillin susceptible with an oxacillin MIC of ≤2 μg/ml or resistant with an MIC of >2 μg/ml.
mecA gene PCR.
The mecA gene was detected with a PCR assay developed by Murakami et al. (9). S. caprae DNA was extracted with the QIAmp DNA mini kit (Qiagen Inc., Valencia, Calif.). The mecA sequence-specific primers were mecA1 (5′-AAA ATC GAT GGT AAA GGT TGG C-3′) and mecA2 (5′-AGT TCT GCA GTA CCG GAT TTG C-3′). The Taq polymerase and all reagents for conventional PCR were purchased from Perkin Elmer. Thermocycling conditions were as follows: 94°C for 2 min, followed by 25 cycles of 94°C for 15 s, 55°C for 15 s, and 72°C for 15 s for each cycle. The mecA PCR product visualized by ethidium bromide staining was 533 bp in length.
RESULTS
Shared phenotypic and genotypic characteristics.
All 21 isolates of S. caprae recovered from the neonatal intensive care unit recovered over the 8-month period had characteristics similar to methicillin-resistant S. aureus. They were all beta-hemolytic and had slide coagulase test results that were difficult to interpret, appearing positive at times. The antibiogram of the neonatal intensive care unit S. caprae isolates also resembled the patterns for methicillin-resistant S. aureus, including resistance to clindamycin, erythromycin, penicillin, and oxacillin by agar dilution. Additionally, a subgroup of 13 of the 21 neonatal intensive care unit-related S. caprae isolates were tested for mecA to determine the mechanism of methicillin resistance. All 13 of the neonatal intensive care unit-related isolates tested were mecA gene positive. In contrast, all 12 epidemiologically unrelated isolates were methicillin susceptible by agar dilution and mecA gene negative.
Infections.
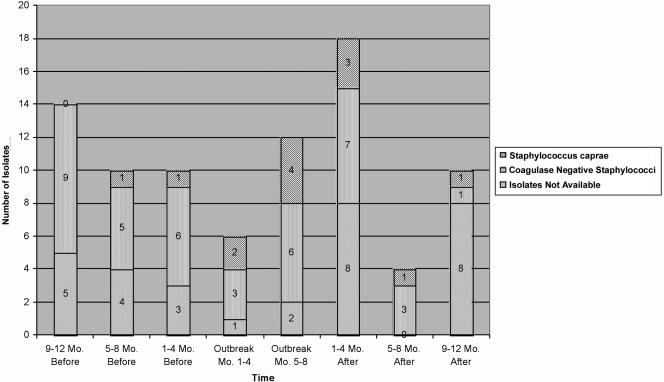
Infections were based solely on positive culture results. S. caprae infections were identified by identifying the species of all isolates of coagulase-negative staphylococci recovered from blood specimens from infants in the neonatal intensive care unit over the 8-month study period (Table 1). S. haemolyticus, S. hominis, S. epidermidis, and S. caprae were recovered. S. caprae represented 6 of 18 (33%) of the cases of coagulase-negative staphylococcal bacteremia. These six cases plus the original index cases from the vascular catheter and cerebrospinal fluid shunt totaled eight possible infections over the 8-month study period. To assess whether there was an increase of coagulase-negative staphylococci due to S. caprae, available isolates from the year prior and after the 8-month study period were identified to species level (Fig. 1). The data imply that the organism was introduced in the neonatal intensive care unit 5 to 8 months before the study period and has decreased to one isolate 5 to 8 months after the study period. Fortunately, none of the 13 bacteremic patients died due to S. caprae.
TABLE 1.
Coagulase-negative staphylococcal species recovered from blood cultures from neonates over the 8-month study period
| Species | No. of neonates positivea |
|---|---|
| S. caprae | 6 |
| S. epidermidis | 7 |
| S. haemolyticus | 2 |
| S. hominis | 3 |
Two patients had two different species recovered from blood.
Sixteen neonates were tested.
FIG. 1.
Isolates from neonatal intensive care unit patients.
Colonization.
S. caprae colonization was defined and determined by a positive nares culture. All hemolytic coagulase-negative staphylococci recovered from methicillin-resistant S. aureus nares surveillance cultures from neonatal intensive care unit infants performed during one of the 8 months were identified to species level (Table 2). A total of 6 of 32 (19%) neonatal intensive care unit infants were colonized with S. caprae. These six plus the four index children with positive nares cultures yielded a total of at least 10 infants colonized.
TABLE 2.
Staphylococcus caprae colonization: hemolytic coagulase-negative staphylococci recovered from neonate nares surveillance cultures and health care provider hand cultures
| Species | No. of positive individuals
|
|
|---|---|---|
| Neonates (n = 32) | Health care providers (n = 37) | |
| S. caprae | 6 | 1 |
| S. capitis | 0 | 6 |
| S. epidermidis | 1 | 2 |
| S. haemolyticus | 1 | 1 |
| S. hominis | 2 | 1 |
| S. lugdunensis | 0 | 1 |
| S. warneri | 4 | 12 |
| Total | 14 | 24 |
Thirty-seven neonatal intensive care unit health care providers (approximately 33%) volunteered to have hand cultures performed to assess S. caprae colonization (Table 2). Seven species of hemolytic coagulase-negative staphylococci were recovered from 24 of the 37 (65%) health care providers, but only one (2.7%) was positive for S. caprae.
S. caprae identification and typing.
Twenty-eight S. caprae isolates from the neonatal intensive care unit and the 12 epidemiologically unrelated isolates were identified and typed by the Automated RiboPrinter and also typed by pulsed-field gel electrophoresis; 32 (26 of 28 isolates from the neonatal intensive care unit and 6 of 12 epidemiologically unrelated isolates) of the 40 (80%) were identified as S. caprae by the Automated RiboPrinter. The remaining eight were not given an identification by the Automated RiboPrinter, but when these isolates were compared to the library of Dupont identification patterns, the best match for each of these eight was S. caprae. Three isolates, including the American Type Culture Collection strain, were identified as S. caprae by phenotypic assays. Of the biochemical tests, the nitrate, urea, trehalose (delayed), pyrrolidonyl-α-naphthylamine hydrolysis (11 to 89%), and alkaline phosphatase (delayed) tests were positive. Negative results were observed with the slide coagulase, tube coagulase, ornithine decarboxylase, polymyxin B resistance, and novobiocin resistance tests.
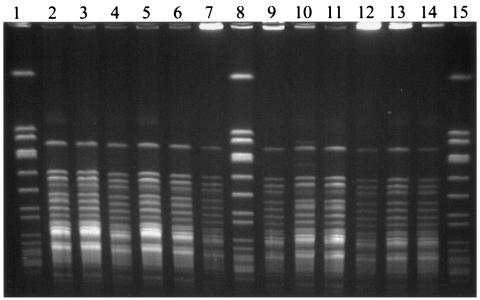
Typing of the 28 neonatal intensive care unit-related isolates by the Automated RiboPrinter system placed all in a single ribogroup. With SmaI, one cluster was also delineated by pulsed-field gel electrophoresis: one strain type with 26 members and two epidemiologically related types with one member each. These three pulsed-field gel electrophoresis types were all epidemiologically related and were within three bands. However, when ApaI was used, all 28 isolates were identical (Fig. 2).
FIG. 2.
Pulsed-field gel electrophoresis. Lanes: 1, 8, and 15, size markers; 10 and 12, isolates from nares surveillance cultures; 13, vascular catheter isolate; 2, 3, 5, 6, 7, and 11, nares surveillance cultures; 4 and 9, blood culture isolates; 14, hand culture isolate from a health care worker.
To assess the discriminatory ability of the typing assays, the panel of 12 epidemiologically unrelated isolates was tested. Eleven ribogroups were delineated by the Automated RiboPrinter, one ribogroup with two members and 10 ribogroups with a single member each. One of the unrelated strains was placed in the same ribogroup as the neonatal intensive care unit outbreak strain. The pulsed-field gel electrophoresis assay with SmaI delineated 9 of the panel of 12 unique types; but with ApaI, all 12 were found to be unique strain types. None of the 12 epidemiologically unrelated pulsed-field gel electrophoresis types matched the neonatal intensive care unit outbreak strain.
DISCUSSION
Coagulase-negative staphylococci are a major cause of bloodstream infections in hospitalized patients, especially in intensive care settings; the neonatal intensive care unit is no exception (11, 14). In one neonatal intensive care unit study, S. epidermidis, S. haemolyticus, and S. warneri but no S. caprae isolates were recovered from cultured blood (11). In another report, there was a single case report of S. caprae sepsis that occurred in a neonate (14); therefore, our data documenting the extremely high percentage (33%) of S. caprae as a cause of neonatal coagulase-negative staphylococcal bacteremia are an important new finding. The analysis of coagulase-negative staphylococci before and after the 8-month study period did not reveal an increase in S. caprae. Interestingly the S. caprae appear to have been introduced earlier in the study period and decreased 5 to 8 months after the 8-month study period. Fortunately, there was not any evidence of mortality due to S. caprae. In addition, our data extend the anatomic sites of infection to include a case of cerebrospinal fluid shunt infection in a neonate.
Since many laboratories (including ours) do not routinely identify coagulase-negative staphylococci to species level, the true incidence and types of infections associated with S. caprae as well as other Staphylococcus species are relatively unknown. In other studies where staphylococci from all patient populations were identified to species level, S. caprae was present in most, but certainly not as a predominant species. In one study, only 1 of 617 staphylococci recovered from clinical specimens was S. caprae (4), and in another study, 10.7% of 1,230 staphylococci were S. caprae (7). Additionally, only 1 of 107 coagulase-negative staphylococci recovered from clinical specimens and 2.6% of 2,887 bacteria recovered from patients with acute otitis externa were S. caprae (12).
Our cluster of S. caprae in neonates would not have been discovered except for the initial isolates from neonatal intensive care unit methicillin-resistant S. aureus nares surveillance that were considered presumptive methicillin-resistant S. aureus due to shared characteristics. They were all beta-hemolytic and gave equivocal coagulase results. They were methicillin resistant with an antibiotic profile similar to that of methicillin-resistant S. aureus. The identification of S. caprae was made by Automated RiboPrinter and confirmed by phenotypic (biochemical) assays.
The mechanism of methicillin resistance of some strains of S. caprae is known to be associated with the presence of the mecA gene (6, 9, 15). Interestingly, a subset of our neonatal intensive care unit strains were tested and were all mecA positive, whereas the 12 epidemiologically unrelated strains were methicillin susceptible by agar dilution and negative for mecA. S. caprae methicillin resistance varies widely. When data from the literature were totaled, 13% were resistant to methicillin (6, 9, 12, 18).
An important finding was that the S. caprae detected from infants in our neonatal intensive care unit over the 8-month study period were members of a single clone. Pulsed-field gel electrophoresis with SmaI, pulsed-field gel electrophoresis with ApaI, and the Automated RiboPrinter all supported this conclusion; all 28 were members of the same cluster by each assay. The analysis of the panel of 12 epidemiologically unrelated strains used to demonstrate discrimination revealed that pulsed-field gel electrophoresis with SmaI and the Automated RiboPrinter method were less discriminating than pulsed-field gel electrophoresis with ApaI. SmaI-pulsed-field gel electrophoresis delineated 9 of 12 as unique with three band differences for interpretation. The Automated RiboPrinter delineated 11 of the 12 as unique. Pulsed-field gel electrophoresis with ApaI was highly discriminating, delineating all 12 isolates as unique. Therefore, for this species, we recommend pulsed-field gel electrophoresis with ApaI digestion or screening by Automated RiboPrinter followed by testing with a second more sensitive assay for isolates considered identical.
These molecular epidemiologic data support the idea that transmission or persistence of this organism occurred over the 8-month study period, although the species did not cause an increase in the number of coagulase-negative staphylococcal bloodstream infections. Since the mode of transmission of coagulase-negative staphylococci is unknown, environmental cultures were not performed. In addition, the health care provider hand cultures did not contribute to our understanding of the transmission of this organism; perhaps nares cultures would have been helpful.
Clonality of coagulase-negative staphylococci has been reported in the literature; a study of two neonatal intensive care unit units has shown that 33 and 45% of the coagulase-negative staphylococci were clonal (19). This is not surprising with the challenges in patient care in a neonatal intensive care unit. One could hypothesize that the persistence of this clone over the study period would be related to its ability to adhere to human cells, extracellular matrices, and/or hardware in the hospital environment. S. caprae recently has been shown to have genes similar to other staphylococci that contribute to its ability to colonize humans and to produce biofilms. These include an atlC gene, which encodes an autolysin responsible for mediating adhesion of the organism to fibronectin, and an operon of four ica genes that is associated with this organism's ability to produce a biofilm (1, 2).
In conclusion, data from this study extend our knowledge of this interesting species that is known to cause infections in animals. In our neonatal intensive care unit, S. caprae persisted for at least 28 months. Fortunately, the number decreased over the 12 months poststudy. These data also emphasize the value of molecular epidemiology studies in delineating clonality for use in outbreak investigations.
Acknowledgments
We thank P. S. Roland from the University of Texas Southwestern Medical Center at Dallas for kindly providing 10 strains of Staphylococcus caprae. We thank the neonatal intensive care unit staff for being so cooperative during this study. We also thank Karen Carroll for providing a critical review of the manuscript.
REFERENCES
- 1.Allignet, J., J. Galdbart, A. Morvan, K. G. H. Dyke, P. Vaudaux, S. Aubert, N. Desplaces, and N. El Solh. 1999. Tracking adhesion factors in Staphylococcus caprae strains responsible for human bone infections following implantation of orthopedic material. Microbiology 145:2033-2042. [DOI] [PubMed] [Google Scholar]
- 2.Allignet, J., S. Aubert, K. G. H. Dyke, and N. El Solh. 2001. Staphylococcus caprae strains carry determinants known to be involved in pathogenicity: a gene encoding an autolysin-binding fibronectin and the ica operon involved in biofilm formation. Infect. Immun. 69:712-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanc, V., J. Picaud, E. Legros, M. Bes, J. Etienne, D. Moatti, and M. F. Raynaud. 1999. Infection after total hip replacement by Staphylococcus caprae: case report and review of the literature. Pathol. Biol. 47:409-413. [PubMed] [Google Scholar]
- 4.Couto, I., S. Pereira, M. Miragaia, I. S. Sanches, and H. de Lencastre. 2001. Identification of clinical staphylococcal isolates from humans by internal transcribed spacer PCR. J. Clin. Microbiol. 39:3099-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eisner, H. A., G. P. Dahmen, R. Laufs, and D. Mack. 1998. Intra-articular empyema due to Staphylococcus caprae following arthroscopic cruciate ligament repair. J. Infect. 37:66-67. [DOI] [PubMed] [Google Scholar]
- 6.Kanda, K., E. Suzuki, K. Hiramatsu, T. Oguri, H. Miura, T. Ezaki, and T. Yokota. 1991. Identification of a methicillin-resistant strain of Staphylococcus caprae from a human clinical specimen. Antimicrob. Agents Chemother. 35:174-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawamura, Y., X. Hou, F. Sultana, K. Hirose, M. Miyake, S. Shu, and T. Ezaki. 1998. Distribution of Staphylococcus species among human clinical specimens and emended description of Staphylococcus caprae. J. Clin. Microbiol. 36:2038-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lang, S., M. A. Livesley, P. A. Lambert, J. Elliott, and T. S. J. Elliott. 1999. The genomic diversity of coagulase-negative staphylococci associated with nosocomial infections. J. Hosp. Infect. 43:187-193. [DOI] [PubMed] [Google Scholar]
- 9.Murakami, K., W. Minamide, K. Wada, E. Nakamura, and H. Teraoka. 1991. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J. Clin. Microbial. 29:2240-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.National Committee for Clinical Laboratory Standards. 2004. Performance standards for antimicrobial susceptibility testing; fourteenth informational supplement. NCCLS document M100-S14 (ISBN 1-56238-516-X). National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 11.Raimundo, O., H. Heussler, J. B. Bruhn, S. Suntrarachun, N. Kelly, M. A. Deighto, and S. M. Garland. 2002. Molecular epidemiology of coagulase-negative staphylococcal bacteremia in a newborn intensive care unit. J. Hosp. Infect. 51:33-42. [DOI] [PubMed] [Google Scholar]
- 12.Roland, P. S., and D. W. Stroman. 2002. Microbiology of acute otitis externa. Laryngoscope 112:1166-1177. [DOI] [PubMed] [Google Scholar]
- 13.Shuttleworth, R., R. J. Behme, A. McNabb, and W. D. Colby. 1997. Human isolates of Staphylococcus caprae: association with bone and joint infections. J. Clin. Microbiol. 35:2537-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spellerberg, B., K. Steidel, R. Lutticken, and G. Haase. 1998. Isolation of Staphylococcus caprae from blood cultures of a neonate with congenital heart disease. Eur. J. Clin. Microbiol. Infect. Dis. 17:61-62. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki, E., K. Hiramatsu, and T. Yokota. 1992. Survey of methicillin-resistant clinical strains of coagulase-negative staphylococci for mecA gene distribution. Antimicrob. Agents Chemother. 36:429-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takemura, K., S. Takagi, T. Baba, Y. Goto, and H. Nonogi. 2000. A 72-year-old man with recurrent sepsis due to Staphylococcus caprae. J. Cardiol. 36:269-271. [PubMed] [Google Scholar]
- 17.Tenover, F. C., R. Arbeit, R. Goering, P. A. Mickelsen, B. E. Murray, D. H. Pershing, and B. Swaminaghan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vandenesch, F., S. J. Eykyn, M. Bes, H. Meugnier, J. Fleurette, and J. Etienne. 1995. Identification and ribotypes of Staphylococcus caprae isolates isolated as human pathogens and from goat milk. J. Clin. Microbiol. 33:888-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vermont, C. L., N. G. Hartwig, A. Fleer, P. de Man, H. Verbrugh, J. van den Anker, R. de Groot, and A. van Belkum. 1998. Persistence of clones of coagulase-negative staphylococci among premature neonates in neonatal intensive care units: two-center study of bacterial genotyping and patient risk factors. J. Clin. Microbiol. 36:2485-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]