Abstract
To investigate the extent to which drug resistance mutations are missed by standard genotyping methods, we analyzed the same plasma samples from 26 patients with suspected multidrug-resistant human immunodeficiency virus type 1 by using a newly developed single-genome sequencing technique and compared it to standard genotype analysis. Plasma samples were obtained from patients with prior exposure to at least two antiretroviral drug classes and who were on a failing antiretroviral regimen. Standard genotypes were obtained by reverse transcriptase (RT)-PCR and sequencing of the bulk PCR product. For single-genome sequencing, cDNA derived from plasma RNA was serially diluted to 1 copy per reaction, and a region encompassing p6, protease, and a portion of RT was amplified and sequenced. Sequences from 15 to 46 single viral genomes were obtained from each plasma sample. Drug resistance mutations identified by single-genome sequencing were not detected by standard genotype analysis in 24 of the 26 patients studied. Mutations present in less than 10% of single genomes were almost never detected in standard genotypes (1 of 86). Similarly, mutations present in 10 to 35% of single genomes were detected only 25% of the time in standard genotypes. For example, in one patient, 10 mutations identified by single-genome sequencing and conferring resistance to protease inhibitors (PIs), nucleoside analog reverse transcriptase inhibitors, and nonnucleoside reverse transcriptase inhibitors (NNRTIs) were not detected by standard genotyping methods. Each of these mutations was present in 5 to 20% of the 20 genomes analyzed; 15% of the genomes in this sample contained linked PI mutations, none of which were present in the standard genotype. In another patient sample, 33% of genomes contained five linked NNRTI resistance mutations, none of which were detected by standard genotype analysis. These findings illustrate the inadequacy of the standard genotype for detecting low-frequency drug resistance mutations. In addition to having greater sensitivity, single-genome sequencing identifies linked mutations that confer high-level drug resistance. Such linkage cannot be detected by standard genotype analysis.
The genetic diversity of human immunodeficiency virus type 1 (HIV-1) results from rapid, high-level virus turnover (approximately 1011 virions and 108 infected cells/day) and nucleotide misincorporation during replication of the HIV-1 genome by the error-prone reverse transcriptase (RT) (30, 32, 37, 39) and possibly by host cell RNA polymerase II. Many mutations do not have a large deleterious effect on viral fitness and thus accumulate during successive rounds of virus replication. The diversity of HIV-1 populations supports the hypothesis that important drug resistance mutations already exist in the virus population prior to the initiation of antiretroviral therapy, and mutations associated with HIV-1 drug resistance have been predicted to be present in drug-naïve patients at low frequencies (8). The clinical significance of preexisting, low-frequency mutations is not clearly defined, but preliminary data suggest that they may negatively affect response to initial and subsequent antiretroviral treatment regimens (20, 21, 34, 47).
Another important source of low-frequency drug resistance mutations is selection by antiretroviral therapy. Following removal of the selection pressure by either cessation of the drug or transmission of the virus to another untreated individual, mutations conferring resistance to the drug(s) often become undetectable in the virus population, albeit at variable rates (11, 13). Although the factors leading to loss of drug resistance mutations are not fully understood, such mutations rapidly reappear following reinitiation of the antiretroviral therapy and thus have clinical significance. Optimal management of treatment-experienced patients will therefore require the best possible understanding of the frequency and distribution of mutations in virus populations.
The most commonly employed methods of detection of drug-resistant variants in HIV-1 populations involve generating “bulk” RT-PCR product derived from multiple viral genomes extracted from plasma (18) followed by DNA sequencing (genotypic analysis) or measurement of the average effect on drug susceptibility after insertion of the RT-PCR product into a proviral HIV-1 clone (phenotypic analysis). Although these methods provide a composite of the sequences present, or their phenotypic properties, they are only able to detect mutants comprising a major fraction of the virus population (20) and cannot be used to determine linkage of mutations. To address these shortcomings, we developed a single-genome sequencing (SGS) technique, based on earlier limiting-dilution assays (4, 22, 44, 48), that allows more refined analyses of HIV-1 populations by obtaining DNA sequences derived from many single viral genomes in a plasma sample. DNA sequences derived from 20 to 40 single genomes are typically analyzed per sample, although the number of genomes obtained can be readily increased. In the present study, we compare the sensitivity of standard genotype analysis to SGS for detection of HIV-1 drug resistance mutations in plasma samples from patients with suspected multidrug-resistant HIV-1. Of 26 samples studied, 24 contained genomes with drug resistance mutations not detected by standard genotype analysis. In many cases, these mutations were linked on the same subset of genomes. Not only is SGS more sensitive for analyzing sequence diversity than standard genotype analysis, it can identify linkage of mutations on the same genome.
MATERIALS AND METHODS
Viruses.
Infectious molecular clones of HIV-1 89.6 and pNL4-3 were obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH).
Clinical samples.
Plasma samples were obtained from patients with suspected multidrug resistance who had prior exposure to at least two antiretroviral drug classes and who were on a failing antiretroviral regimen. Nineteen of the 26 plasma samples studied were obtained at study entry from patients enrolled in AIDS Clinical Trials Group (ACTG) protocol 398, which examined single versus dual protease inhibitor-containing regimens in 481 patients on a failing regimen containing a protease inhibitor (19). All patients had prior exposure to nucleoside analog reverse transcriptase inhibitors (NRTIs) and to one or more protease inhibitors (PIs); 44% also had prior exposure to nonnucleoside reverse transcriptase inhibitors (NNRTI). Of the 19 samples analyzed, 10 were from NNRTI-experienced patients and 9 were from NNRTI-naïve patients. ACTG protocol 398 was approved by the institutional review boards of the participating centers, and all patients provided written informed consent for participation in the study and testing of samples.
Seven plasma samples were collected from treatment-experienced patients on a failing regimen enrolled in studies at the NIAID Critical Care Medicine Department clinic at the Clinical Center of the NIH. All studies were approved by the institutional review board of NIAID, NIH, Bethesda, Md., and all individuals provided written informed consent.
Standard genotype.
Standard genotype analysis for samples from patients in ACTG 398 was obtained by using a ViroSeq version 2.0 kit (ABI, Foster City, Calif.) (patients 1 to 18 and 21). Standard genotypes for NIH samples were obtained by using Visible Genetics TruGene (Toronto, Ontario, Canada) (patients 19 and 23 to 26) or by testing at a reference laboratory (VircoLab Inc., Durham, N.C.) (patients 20 and 22). Mutations reported were those provided by standard analysis of the sequence output. Although further analysis of the raw sequence data might have identified mixtures or additional mutations, the goal was to compare the SGS method with standard sequence output and analysis.
Viral RNA extraction.
Between 250 and 1,800 μl of plasma (containing a minimum of 1,000 copies of HIV-1 RNA) was centrifuged at 16,000 × g for 1 h at 4°C. After centrifugation, the supernatant was removed, and the virion pellet was treated with 50 μl of 5 mM Tris-HCl (pH 8.0) containing 200 μg of proteinase K for 30 min at 55°C. The pellet was then treated with 200 μl of 5.8 M guanidinium isothiocyanate containing 200 μg of glycogen. After adding 270 μl of 100% isopropanol, the lysate was centrifuged at 21,000 × g for 15 min. The pellet was washed with 70% ethanol and resuspended in 40 μl of 5 mM Tris-HCl (pH 8.0) containing 1 μM dithiothreitol and 1,000 U of an RNase inhibitor (RNase Out; Invitrogen, Carlsbad, Calif.). The entire viral RNA extraction was used for cDNA synthesis.
Single-genome sequencing.
Five microliters of deoxynucleoside triphosphates (10 mM) and 5 μl of random hexamers (50 ng/μl; Promega, Madison, Wis.) were added to 40 μl of viral RNA, followed by a denaturation step at 65°C for 10 min. Fifty microliters of reverse transcription reaction mix was added to the denatured viral RNA mixture (final volume, 100 μl), giving the indicated final amounts or concentrations of the following components in sterile molecular-grade water: MgCl2 (5 mM), RNase Out (20 U), dithiothreitol (1 mM), 1× RT buffer (Invitrogen), and RT (100 U; Superscript II; Invitrogen). After 15 min at 25°C, the reaction mixture was incubated at 42°C for 40 min. Following completion of the reverse transcription step, the reaction mixture was heated to 85°C for 10 min, held at 25°C for 30 min, and then cooled to 4°C.
To obtain PCR products derived from single cDNA molecules, the resulting cDNA was serially diluted 1:3 in 2 mM Tris-HCl (pH 8.0) to a maximum dilution of 1:2,187. Ten separate real-time PCR amplifications were performed for each cDNA dilution as follows: 2 μl of the cDNA reaction product was added to 10 μl of PCR mix (final volume, 12 μl) containing the indicated final amounts or concentrations of the following components in sterile molecular-grade water: 1× quantitative PCR mix (Invitrogen), MgCl2 (increased to a final concentration of 6 mM), internal standard dye (ROX), and the primer-probe set (200 nm each) for HIV-1 quantification designed to bind to a conserved region of pol: 1849 forward (5′-GGGGGAATTGGAGGTTTTATCA-3′) and 3500 reverse (5′-CATTCCTGGCTTTAATTTTACTGGTACAGT-3′) primers and the detection probe 5′-FAM-TACAGTATTAGTAGGACCTACACCTGTCAACATAATTGGA-Q-3′, where FAM indicates a 6-carboxyfluorescein group and Q (quencher) indicates a 6-carboxytetramethylrhodamine group conjugated through a linker arm nucleotide as described previously (28). PCR product was detected by measuring the amount of fluorescence compared to that of a negative control. According to Poisson's distribution, the cDNA dilution yielding PCR product in 3 out of the 10 real-time PCRs contains 1 copy of cDNA per positive PCR about 80% of the time. Thus, 70 nested PCRs were then set up using the cDNA dilution yielding ∼30% positive reactions. The nested PCRs used High Fidelity Supermix (Invitrogen) with primers 1849 forward and 3500 reverse (described above), followed by a nested PCR with primers 1870 forward (5′-GAGTTTTGGCTGAAGCAATGAGCC-3′ ) and 3941 reverse (5′-TTAGTGGTACTACTTCTGTTAGTGTT3-′ ), producing a 1.33-kb amplicon containing the p6 region of gag, pro, and the first 900 nucleotides of pol. Positive nested PCRs were identified by agarose gel electrophoresis and sequenced by direct dideoxyterminator sequencing (Applied Biosystems) in both directions using overlapping internal primers. Sequences from each plasma sample were aligned and compared to the HIV-1 subtype B consensus sequence by using Clustal W software (7, 46). Sequencing reactions that identified more than one amplified genome were not included in analyses.
Fifteen to 23 sequences from each patient sample were initially analyzed. Further amplification and sequencing, to a maximum of 46 sequences, was performed only on those samples that were negative for drug resistance mutations in the initial analysis. Although this analytical strategy might lead to an underestimation of the total frequency of resistance mutations, it allowed us to maximize sensitivity of detection with a minimum amount of sequencing, appropriate for our goal of determining how many patient samples contained resistance mutations undetected in bulk sequences, not the absolute frequency of such mutations.
Analysis of assay-related error and recombination rates.
The HIV-1 gag-pro-pol region was amplified from two molecular clones (89.6 and pNL4-3) by using primers 1849 forward and 3500 reverse (described above). The resulting 1.6-kbp PCR product for each virus was cloned separately into the vector PCRscript II (Stratagene, La Jolla, Calif.). Plasmids were purified on columns with a QIAGEN kit and linearized by using NotI. The resulting DNA template was quantified and then transcribed with T7 RNA polymerase by using a Megascript kit (Ambion, Austin, Tex.). The RNA transcripts were diluted to 106 copies/μl and stored at −80°C.
One thousand copies of RNA transcript for both HIV-1 89.6 and pNL4-3 were mixed and used as a template in the single-genome sequencing procedure (described above). A total of 50 individual sequences (approximately 66,000 nucleotides) were analyzed for assay-related misincorporation and recombination.
RESULTS
Properties of the SGS assay.
To assess the extent of nucleotide misincorporation (polymerase errors) and intertemplate recombination that may occur during the assay, we mixed RNA transcripts from two different HIV-1 strains (89.6 and pNL4-3 [103 copies each]) and used the mixture as a template for the SGS assay (Table 1). Fifty individual sequences totaling 66,579 nucleotides were generated and compared to the published sequence of the reference strains (http://www.hiv.lanl.gov). This analysis showed five assay-related transitions (0.008%), two assay-related transversions (0.003%), and 15 frameshift mutations resulting from the addition of one or two A or T nucleotides to a preexisting run of A or T residues, yielding an overall assay error rate of 0.033%. Because analyses of patient samples never yielded frequencies of A or T additions higher than those of the controls, all sequences with such errors were corrected by their removal, leaving an assay error rate of 0.011%.
TABLE 1.
Properties of the SGS assaya
| Sequence | No. of genomes sequenced | No. of bases sequenced | No. of transitions (%) | No. of transversions (%) | No. of run extensions (%) | No. of recombinants (%) |
|---|---|---|---|---|---|---|
| 89.6 | 31 | 41,218 | 2 (0.0049) | 1 (0.0025) | 4 (0.0097) | 0 (<2) |
| PNL4-3 | 19 | 25,361 | 3 (0.012) | 1 (0.0039) | 11 (0.039) | 0 (<2) |
Approximately 1,000 copies of in vitro transcripts representing the gag-pro-pol region of each virus were mixed and subjected to SGS by the described protocol.
No recombination between the two reference strains was observed in the 50 sequences analyzed. The reference strains contain 38 nucleotide differences across the region analyzed, which equates to approximately 3% difference between the strains. Thus, assay-associated recombination with SGS was less than one crossover between two closely related templates in about 66,000 bp analyzed. Furthermore, gross rearrangements, such as deletion or reduplication of sequence, have never been observed in more than 3.5 million bases of the same region from more than 150 patient samples analyzed to date by SGS.
Comparison of SGS and standard genotypes.
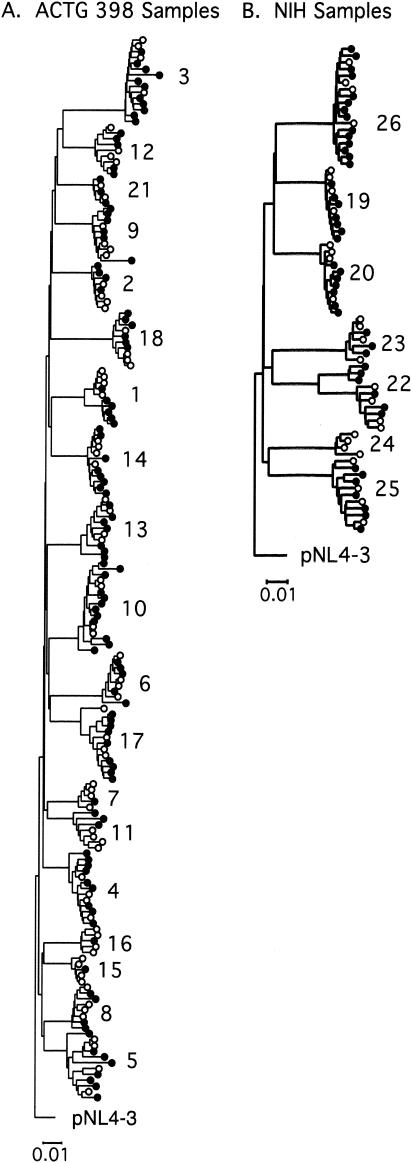
To compare the performance of the SGS assay to standard genotype analysis, plasma samples from 26 patients on failing antiretroviral regimens were analyzed by using both methods. All standard genotyping was done using methods routinely employed for clinical diagnostics. Between 15 and 46 single-genome sequences were analyzed for each patient sample. The absence of contamination or cross-contamination in these sets of sequences was verified by phylogenetic analysis (Fig. 1). In all cases, the sequences containing resistance mutations form a monophyletic group with wild-type sequences from the same patient, and sequences from each patient cluster separately from those from all other patients as well as a laboratory strain virus (pNL4-3). Bootstrap analysis of the phylogenetic trees gave values of ≥99% for each patient grouping (data not shown). With two exceptions, the branching pattern within any individual patient was not strongly supported by bootstrap analysis, consistent with a common origin and evolutionary history of all samples analyzed from each patient and excluding the possibility of any other origin of the sequences. In the two exceptional patients (1 and 22), the branches separating the two clades of virus were much shorter than those linking any patient groups to any another, again consistent with a common origin.
FIG. 1.
Phylogenetic analysis of patient HIV sequences. Phylogenetic trees of aligned sequences obtained by SGS from patients enrolled in the AIDS Clinical Trials Group study 398 (A) or the NIH clinic (B) were determined by using the neighbor-joining distance method, as implemented in the molecular evolutionary genetics analysis package (http://www.megasoftware.net). Patient identification is shown next to each group of sequences analyzed. All sequences containing resistance mutations (as defined in the Stanford database) that were identified by SGS but not found in the standard genotype were included in the analysis and are shown by filled circles. Four other sequences, which did not contain unique mutations, were chosen at random from each patient and are represented by open circles.
All of the mutations detected by standard genotype analysis were also detected by SGS (Table 2). By contrast, many mutations detected in single-genome sequences were not detected by standard genotypic analysis of the same samples. The primary drug resistance mutations not detected by standard genotypic analysis varied from patient to patient but included V82A, I84V, and L90M in protease and M41L, T69N, L74V, K103N, Y181C, and T215Y in reverse transcriptase. Several mutations (or polymorphisms) at sites associated with resistance, but not themselves known to cause resistance, were also observed. These mutations include M184K, V108G, and L210M in RT.
TABLE 2.
Comparison of standard genotype and single-genome sequencing
| Patientd | No. of resistance mutationsa
|
Resistance mutations found only by SGSb
|
||||
|---|---|---|---|---|---|---|
| PRe
|
RTc
|
|||||
| Bulk | SGS | Bulk | SGS | PR | RT | |
| 1N | 4 | 4 | 0, 0 | 7, 5 | A98S (33), K101E (33), V179A (7), Y181C (33), G190A (33), L210W (33), T215Y (33) | |
| 2N | 8 | 8 | 4, 0 | 6, 1 | A62V (12), K103N (3) | |
| 3N | 8 | 10 | 11, 0 | 17, 1 | D30G (3), I93V (3) | M41L (3), A62V (3), V75A (15), K103N (3), V106T (3), V108F (3), Y115D (3) |
| 4N | 2 | 5 | 5, 0 | 8, 0 | M36I (9), M46I (2), V82G (2) | D67N (2), M184K (2), T215D (2) |
| 5N | 6 | 10 | 1, 0 | 5, 0 | L10V (2), L24F (2), G73S (5), V82I (2) | A62T (2), K103E (2), F116C (2), V118I (2) |
| 6 | 4 | 5 | 2, 0 | 6, 0 | M46L (5) | E44K (2), A62T (2), D67N (2), K70R (2) |
| 7 | 5 | 5 | 1, 0 | 2, 0 | E44A (2) | |
| 8 | 1 | 3 | 2, 0 | 4, 0 | I47V (2), 184K (2) | V118D (2), L210M (2) |
| 9 | 8 | 9 | 5, 0 | 9, 1 | I93L (7) | F77L (2), K103N (2), V106I (2), V108G (2) |
| 10 | 3 | 3 | 1, 0 | 5, 0 | T69P (3), V106I (36), M184I (11), K219N (3) | |
| 11 | 5 | 6 | 3, 0 | 6, 0 | M36V (3) | M41L (3), V118I (3), T215F (5) |
| 12 | 8 | 11 | 6, 0 | 7, 0 | M46I (9), I54V (5), I84G (5) | V118I (5) |
| 13 | 2 | 4 | 1, 0 | 4, 1 | M36I (6), A71V (3) | E44K (3), L100I (3), P225L (3) |
| 14 | 2 | 7 | 5, 0 | 5, 0 | L10I (4), M46I (13), M46L (13), V82A (4), N88D (4) | |
| 15N | 1 | 2 | 0, 0 | 0, 0 | V77I (5) | |
| 16N | 1 | 2 | 0, 0 | 0, 0 | L10I (5) | |
| 17N | 6 | 8 | 6, 0 | 11, 1 | M36I (22), A71I (4) | T69N (4), K101E (30), K103E (4), K103R (4), F116S (4) |
| 18N | 8 | 11 | 6, 2 | 8, 3 | G73A (5), G73T (9), V82I (5) | L74V (5), Y181C (14) |
| 19N | 5 | 5 | 3, 1 | 4, 1 | D67N (30) | |
| 20 | 6 | 7 | 4, 1 | 7, 3 | M36I (10) | L74V (15), L100I (25), V108I (15) |
| 21N | 7 | 10 | 6, 1 | 6, 1 | M36I (10), G73S (5), L90M (5) | K101R (5) |
| 22N | 6 | 12 | 10, 2 | 14, 2 | L10V (20), M36V (5), M46I (15), I84V (15), L90M (15), 193L (15) | T69A (10), K101R (15), V179I (5), P225H (10) |
| 23N | 8 | 10 | 10, 3 | 10, 3 | K20T (7), L63S (7) | |
| 24N | 9 | 9 | 10, 2 | 10, 2 | ||
| 25N | 12 | 15 | 7, 2 | 13, 2 | K20I (13), K20T (7), I84L (7) | E44D (13), D67N (20), K70A (7), V75L (7), K103E (7), K103I (7) |
| 26N | 9 | 12 | 7, 2 | 12, 2 | L10I (6), L33I (3), L63T (3) | T69I (16), V75I (19), M184V (3), K219T (19), K219Q (3) |
Mutations in retroviral codons associated with drug resistance as designated by the Stanford database (24).
Mutations in boldface type are considered drug resistance mutations as designated by the Stanford database; parentheses show percentages of genomes containing mutations. NNRTI resistance mutations are underlined.
Numbers of NNRTI resistance mutations are underlined; numbers of NRTI mutations are not underlined.
All patients but one had failed a treatment regimen containing at least one PI and at least two NRTIs. Patients indicated by an N had also been treated with at least one NNRTI. Patient 20 was untreated but was infected with a multidrug-resistant strain.
PR, protease.
Percentage of sequences containing drug resistance mutations.
The detection of drug-resistant viral variants at a low frequency is important for monitoring the emergence of drug resistance mutations and for assessing the influence of prior therapy on the virus population. The sensitivity of the SGS method is limited only by the number of sequences obtained. In this case, we could detect mutations present in 2% of the genomes sequenced (Table 2). With a single exception, resistance mutations that were present in less than 10% of single genomes were not detected by standard genotype analysis. Mutations present in 10 to 35% of single genomes were detected only 25% of the time by standard genotyping, and the probability of their detection varied by mutation. For example, the sample from patient 19 contained two nucleoside RT inhibitor mutations: one mutation (D67N) present in 30% of genomes was not detected by standard genotype analysis, and the other mutation (V118I) present in 21% of genomes was detected by standard genotype analysis (Table 3). These results show that the SGS method is more sensitive than standard genotyping for detection of minor populations of drug-resistant variants in patient samples and suggest that the sensitivity of mutant detection by bulk analysis varies greatly from base to base. In addition, the analysis of multiple single genomes allows more refined comparisons of intra- and interpatient viral diversity (35).
TABLE 3.
Drug resistance mutations in the RT sequence of patient 19
| Genotype | Mutation at codona:
|
|||||
|---|---|---|---|---|---|---|
| 41 | 67 | 103 | 118 | 210 | 215 | |
| Wild type | M | D | K | V | L | T |
| Standard | L | N | I | W | Y | |
| SGS | ||||||
| 19-1 | L | N | W | Y | ||
| 19-2 | L | N | W | Y | ||
| 19-3 | L | N | W | Y | ||
| 19-4 | L | N | N | W | Y | |
| 19-5 | L | N | W | Y | ||
| 19-6 | L | N | I | W | Y | |
| 19-7 | L | N | W | Y | ||
| 19-8 | L | N | W | Y | ||
| 19-9 | L | N | W | Y | ||
| 19-10 | L | N | N | I | W | Y |
| 19-11 | L | N | W | Y | ||
| 19-12 | L | N | I | W | Y | |
| 19-13 | L | N | I | W | Y | |
| 19-14 | L | N | I | W | Y | |
| 19-15 | L | N | N | W | Y | |
| 19-16 | L | N | W | Y | ||
| 19-17 | L | N | N | W | Y | |
| 19-18 | L | N | W | Y | ||
| 19-19 | L | N | W | Y | ||
| 19-20 | L | N | N | W | Y | |
| 19-21 | L | N | W | Y | ||
| 19-22 | L | N | N | W | Y | |
| 19-23 | L | N | N | W | Y | |
NNRTI resistance mutations are underlined.
Linkage between specific mutations.
Since the frequency of assay-related recombination is very low, generation of multiple, single sequences from the same patient sample allows the analysis of genetic linkage of mutations, an analysis that cannot be performed by standard genotyping methods. Two patient samples showed clear linkage of resistance mutations (Tables 4 and 5). The sample from patient 22 had 10 resistance mutations conferring resistance to protease inhibitors and nucleoside and nonnucleoside RT inhibitors, none of which were detected by standard genotype analysis (Table 4). Each of these mutations was present in 5 to 20% of the 20 sequences analyzed. Fifteen percent of the sequences in this sample contained linked protease inhibitor resistance mutations (L10V, M46I, I84V, L90M, and I93L); that is, all five mutations were always present on the same genome. In the second example of linkage (patient 1), 33% of the genomes contained six linked NRTI and NNRTI resistance mutations (A98S, K101E, Y181C, G190A, L210W, T215Y), none of which were present in the standard genotype (Table 5). These examples demonstrate that the distribution of resistance mutations can be nonrandom in the virus population.
TABLE 4.
Linkage of specific protease codons in patient 22
| Genotype | Mutation at codon:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 20 | 36 | 46 | 48 | 54 | 63 | 71 | 82 | 84 | 90 | 93 | |
| Wild type | L | K | M | M | G | I | L | A | V | I | L | I |
| Standard | I | M | I | V | T | P | V | A | ||||
| SGS | ||||||||||||
| 22-1 | I | M | I | V | T | P | V | A | ||||
| 22-2 | I | M | I | V | T | P | V | A | ||||
| 22-3 | V | M | I | P | V | M | L | |||||
| 22-4 | I | M | I | V | T | P | V | A | ||||
| 22-5 | I | M | I | V | T | P | V | A | ||||
| 22-6 | I | M | I | V | T | P | V | A | ||||
| 22-7 | I | M | I | V | T | P | V | A | ||||
| 22-8 | I | M | I | V | T | P | V | A | ||||
| 22-9 | V | M | I | P | V | M | L | |||||
| 22-10 | I | M | I | V | T | P | V | A | ||||
| 22-11 | I | M | I | V | T | P | V | A | ||||
| 22-12 | V | M | I | P | V | M | L | |||||
| 22-13 | V | M | I | V | T | P | V | A | ||||
| 22-14 | I | M | I | V | T | P | V | A | ||||
| 22-15 | I | M | I | V | T | P | V | A | ||||
| 22-16 | I | M | I | V | T | P | V | A | ||||
| 22-17 | I | M | I | V | T | P | V | A | ||||
| 22-18 | I | M | V | T | P | V | A | |||||
| 22-19 | I | M | I | V | T | P | V | A | ||||
| 22-20 | I | M | I | V | T | P | V | A | ||||
TABLE 5.
Linkage of specific RT codons in patient 1
| Genotype | Mutation at codona:
|
||||||
|---|---|---|---|---|---|---|---|
| 98 | 101 | 179 | 181 | 190 | 210 | 215 | |
| Wild type | A | K | V | Y | G | L | T |
| Standard | |||||||
| SGS | |||||||
| 1-1 | S | E | C | A | Y | ||
| 1-2 | |||||||
| 1-3 | |||||||
| 1-4 | |||||||
| 1-5 | A | ||||||
| 1-6 | S | E | C | A | Y | ||
| 1-7 | |||||||
| 1-8 | S | E | C | A | W | Y | |
| 1-9 | |||||||
| 1-10 | S | E | C | A | W | Y | |
| 1-11 | |||||||
| 1-12 | |||||||
| 1-13 | |||||||
| 1-14 | |||||||
| 1-15 | S | E | C | A | Y | ||
NNRTI resistance mutations are underlined.
DISCUSSION
Antiretroviral therapy that does not suppress HIV-1 replication to <50 RNA copies/ml of plasma usually results in selection of HIV-1 variants encoding drug resistance mutations (5, 41), leading to treatment failure. The detection of these drug-resistant variants is important for the selection of subsequent treatment regimens that are most likely to be successful (12). Consequently, standard HIV genotyping has emerged as an important technique in treatment monitoring (3, 14, 15, 17, 36). The most commonly employed method of HIV-1 genotyping involves DNA sequencing of bulk RT-PCR product amplified from many viral genomes extracted from plasma, which generates a composite sequence of the virus population. Although mixtures at specific nucleotide positions can be detected in composite sequences, detection is unreliable for variants that constitute <30% of the virus population (18). More sensitive methods are thus needed to characterize the diversity of drug-resistant HIV-1, particularly in treatment-experienced patients. We therefore developed the SGS method to refine the analysis of HIV-1 diversity and to assess the linkage of specific drug resistance mutations or polymorphisms.
The SGS approach relies on sequencing of PCR product obtained from amplification of single DNA molecules obtained by dilution of cDNA derived from plasma viral RNA. This approach, a variant of a method reported previously (4, 22, 44, 48), was expected to have several important advantages over the more commonly used method of sequencing DNA clones derived from bulk PCR product (2, 9, 10, 23, 26, 38, 42, 43). First, the large majority of errors introduced during PCR should be undetectable, and thus the assay-based error rate should approximate that of the RT step. With DNA cloning of PCR product, PCR errors that accumulate during amplification are copied in the cloned DNA. Second, PCR-based recombination should not occur with a single cDNA as a template for amplifications but is more likely to occur by using multiple cDNA molecules as a template. Third, resampling of multiple clones derived from the same initial molecule should not be possible, whereas this problem can arise with amplification and cloning from PCR products, unless each clone is obtained from a separate PCR (27). The latter approach, used by a number of investigators (9, 10), is cumbersome and does not resolve other problems such as accumulation of mutations during PCR and PCR-based recombination.
Control studies using in vitro transcripts derived from cloned reference viruses as well as analysis of patient samples confirmed these expected advantages of SGS. The SGS method has a background substitution error rate of 0.011%, or 1.1 assay-related errors/10,000 nucleotides amplified, in addition to a 0.022% rate of elongation of runs of A or T residues. Because of the design of the assay, we attributed this residual error rate to the reverse transcription step, consistent with extensions of runs of adenines being the most common errors observed (45). Although errors also introduced by T7 RNA polymerase during synthesis of the control templates could not be excluded, the actual substitution rate of the assay can only be less than or equal to that observed (0.011%).
To assess recombination during the SGS assay, we performed control experiments in which transcripts from two different HIV-1 genotypes were mixed in equal portions and used as a template for the SGS method. These control studies revealed no crossovers in 50 genomes sequenced, consistent with an assay recombination rate of <1/66,000 nucleotides analyzed. These control data indicate that the genetic diversity observed among sequences derived from patient samples is not due to assay-related errors or recombination. This conclusion is strongly supported by the presence of linked drug resistance mutations in sequences from patients 1 and 22 (Tables 4 and 5) and several others that we have analyzed (data not shown). We also observed no cases of intragenic recombination in the form of deletion or duplication of sequence.
The absence of bias due to resampling is supported by the observation that in over 2,700 sequences obtained from patients with long-term, untreated infection by this method, including in some cases over 60 sequences from the same plasma sample, no two identical sequences have been observed.
Based on the results of others (29), we believe that the rates of assay-based mutation and recombination are significantly lower in SGS than in methods based on cloning PCR products. To our knowledge, however, comparable control studies at single-copy sensitivity have not been reported for these other approaches. Furthermore, the SGS assay is reproducible, and separate analyses of the same sample reveals similar minority variants. For example, in the case of patient 26, the original dilution used for SGS analysis was too high and only 11 single genomes were amplified from this dilution, of which six low-frequency mutations in retroviral codons associated with drug resistance where identified. SGS analysis of this sample at a lower dilution produced 21 additional single genomes, and four of the six low-frequency mutations found in the earlier SGS analysis were also found in low frequencies in this second sampling (data not shown).
The amplicon selected for the SGS analysis was an approximately 1,300-bp sequence, flanked by highly conserved primer sites, extending from the p6 region of gag through pro and 300 codons of pol. This region includes the sites of all mutations giving rise to PI resistance as well as most RT inhibitor resistance mutations. It is also much better conserved among isolates than sequences in env, making evolutionary analysis more straightforward.
As we anticipated, SGS was more sensitive than standard genotype analysis for detection of a broad range of clinically important drug resistance mutations in pro and pol. In our analysis of 26 antiretroviral drug-experienced patients, 24 of the plasma samples analyzed contained between 1 and 10 drug resistance mutations identified by SGS that were not detected by standard genotype. Many mutations detected only by SGS were primary mutations that appear early in the evolution of resistance to a particular inhibitor. Drug resistance mutations present in less than 10% of single genomes were almost never detected by standard genotype analysis, and mutations present in 10 to 35% of single genomes were more often missed (75% of the time) than detected (25% of the time). These findings agree with the results of a previous study of standard genotypes that showed highly variable detection of mutants in plasma samples containing 25% mutant virus (40). Our results also support the finding that there is a profound underestimation of HIV-1 resistance mutations when standard genotype analysis is used (29). While it is possible that modifications to the standard genotyping protocols used here (such as use of random hexamer primers) could provide greater sensitivity, our goal was to compare SGS to commonly used analytical methods, not to improve on them.
Complex patterns of primary and secondary drug resistance mutations are often detected in plasma samples from treatment-experienced patients by standard genotype analysis. However, the genetic linkage of resistance mutations within and between drug classes is poorly defined. The analysis of multiple single genomes from an individual patient can reveal patterns of linkage that cannot be discerned in standard genotype analysis. Two of the 26 patient samples analyzed by SGS showed distinct linkage of multiple mutations. The first sample contained five linked protease inhibitor mutations in 3 of 20 genomes sequenced. Two of the linked mutations were primary protease mutations (I84V and L90M), and the other three mutations were secondary mutations (L10V, M46I, and I93L). This finding confirms recent predictions based upon analysis of standard genotypes from many protease inhibitor-experienced patients that specific protease mutations can be linked (16, 20).
The second patient sample showed five linked RT inhibitor mutations in 33% of the genomes sequenced. Four of the five linked mutations are associated with resistance to nonnucleoside reverse transcriptase inhibitors (A98S, K101E, Y181C, and G190A); the remaining linked mutation (T215Y) is associated with resistance to zidovudine, a nucleoside reverse transcriptase inhibitor. This result indicates that linkage of resistance mutations is not limited to only one class of drugs. Importantly, composite sequencing was unable to identify these specific mutations or their linkages in either patient.
Our analysis of this highly drug-experienced patient group reveals that the large majority has mutations conferring resistance to important groups of antiviral drugs at levels substantially less than 100%. Like many patients failing antiviral therapy, this group has a long and complex treatment history with many changes in the drugs used. It is well known that resistance to specific antiviral drugs declines (albeit at highly variable rates) with time after their discontinuation (1, 11, 25). It is therefore likely that low levels of resistance mutations observed are due to drugs that have failed in the past and have been discontinued.
The clinical significance of low levels of mutations conferring resistance to antiviral drugs remains to be fully assessed. Preliminary evidence suggests that minor drug-resistant variants which may be missed by standard genotyping can lead to the failure of subsequent treatment regimens (6, 31, 33). Clearly, there must be a threshold level below which the risk of failure declines to that of drug-naïve patients. However, the levels of such variants detected in many patients in this study are well above those seen in untreated patients (S. Palmer, V. Boltz, F. Maldarelli, J. Mellors, and J. M. Coffin, unpublished results). Furthermore, the risk of failure is almost certainly much greater if mutations conferring resistance to multiple drugs are linked on the same genomes, as is the case with some of the patients analyzed here. Further studies designed to relate the presence, frequency, and linkage of mutations detected by SGS to virologic response are under way.
In conclusion, we have developed a single-genome sequencing method that is more sensitive than standard genotyping methods for the identification of minor populations of drug-resistant mutations in patient samples. In addition to its greater sensitivity, SGS permits detection of linked mutations that confer high-level drug resistance. Analysis of multiple single genomes provides a new level of sensitivity for understanding the evolution of viral populations and response to antiretroviral therapy.
Acknowledgments
We thank Valerie Boltz and Ann Wiegand for helpful discussion.
This work was supported by grants to J.W.M. from the National Institute of Allergy and Infectious Diseases (U01AI38858) and from the National Cancer Institute (SAIC contract 20XS190A).
REFERENCES
- 1.Albrecht, D., B. Zollner, H. H. Feucht, T. Lorenzen, R. Laufs, A. Stoehr, and A. Plettenberg. 2002. Reappearance of HIV multidrug-resistance in plasma and circulating lymphocytes after reintroduction of antiretroviral therapy. J. Clin. Virol. 24:93-98. [DOI] [PubMed] [Google Scholar]
- 2.Bacheler, L. T., E. D. Anton, P. Kudish, D. Baker, J. Bunville, K. Krakowski, L. Bolling, M. Aujay, X. V. Wang, D. Ellis, M. F. Becker, A. L. Lasut, H. J. George, D. R. Spalding, G. Hollis, and K. Abremski. 2000. Human immunodeficiency virus type 1 mutations selected in patients failing efavirenz combination therapy. Antimicrob. Agents Chemother. 44:2475-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baxter, J. D., D. L. Mayers, D. N. Wentworth, J. D. Neaton, M. L. Hoover, M. A. Winters, S. B. Mannheimer, M. A. Thompson, D. I. Abrams, B. J. Brizz, J. P. Ioannidis, T. C. Merigan, et al. 2000. A randomized study of antiretroviral management based on plasma genotypic antiretroviral resistance testing in patients failing therapy. AIDS 14:F83-F93. [DOI] [PubMed] [Google Scholar]
- 4.Brown, A. J., and A. Cleland. 1996. Independent evolution of the env and pol genes of HIV-1 during zidovudine therapy. AIDS 10:1067-1073. [PubMed] [Google Scholar]
- 5.Carpenter, C. C., M. A. Fischl, S. M. Hammer, M. S. Hirsch, D. M. Jacobsen, D. A. Katzenstein, J. S. Montaner, D. D. Richman, M. S. Saag, R. T. Schooley, M. A. Thompson, S. Vella, P. G. Yeni, and P. A. Volberding. 1997. Antiretroviral therapy for HIV infection in 1997. Updated recommendations of the International AIDS Society-USA panel. JAMA 277:1962-1969. [PubMed] [Google Scholar]
- 6.Charpentier, C., D. E. Dwyer, F. Mammano, D. Lecossier, F. Clavel, and A. J. Hance. 2004. Role of minority populations of human immunodeficiency virus type 1 in the evolution of viral resistance to protease inhibitors. J. Virol. 78:4234-4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coffin, J. M. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483-489. [DOI] [PubMed] [Google Scholar]
- 9.Condra, J. H., D. J. Holder, W. A. Schleif, O. M. Blahy, R. M. Danovich, L. J. Gabryelski, D. J. Graham, D. Laird, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, T. Yang, J. A. Chodakewitz, P. J. Deutsch, R. Y. Leavitt, F. E. Massari, J. W. Mellors, K. E. Squires, R. T. Steigbigel, H. Teppler, and E. A. Emini. 1996. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J. Virol. 70:8270-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Condra, J. H., W. A. Schleif, O. M. Blahy, L. J. Gabryelski, D. J. Graham, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, D. Titus, T. Yang, H. Teppler, K. E. Squires, P. J. Deutsch, and E. A. Emini. 1995. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature (London) 374:569-571. [DOI] [PubMed] [Google Scholar]
- 11.Deeks, S. G., T. Wrin, T. Liegler, R. Hoh, M. Hayden, J. D. Barbour, N. S. Hellmann, C. J. Petropoulos, J. M. McCune, M. K. Hellerstein, and R. M. Grant. 2001. Virologic and immunologic consequences of discontinuing combination antiretroviral-drug therapy in HIV-infected patients with detectable viremia. N. Engl. J. Med. 344:472-480. [DOI] [PubMed] [Google Scholar]
- 12.DeGruttola, V., L. Dix, R. D'Aquila, D. Holder, A. Phillips, M. Ait-Khaled, J. Baxter, P. Clevenbergh, S. Hammer, R. Harrigan, D. Katzenstein, R. Lanier, M. Miller, M. Para, S. Yerly, A. Zolopa, J. Murray, A. Patick, V. Miller, S. Castillo, L. Pedneault, and J. Mellors. 2000. The relation between baseline HIV drug resistance and response to antiretroviral therapy: re-analysis of retrospective and prospective studies using a standardized data analysis plan. Antivir. Ther. 5:41-48. [DOI] [PubMed] [Google Scholar]
- 13.Delaugerre, C., M. A. Valantin, M. Mouroux, M. Bonmarchand, G. Carcelain, C. Duvivier, R. Tubiana, A. Simon, F. Bricaire, H. Agut, B. Autran, C. Katlama, and V. Calvez. 2001. Re-occurrence of HIV-1 drug mutations after treatment re-initiation following interruption in patients with multiple treatment failure. AIDS 15:2189-2191. [DOI] [PubMed] [Google Scholar]
- 14.Demeter, L. M., R. D'Aquila, O. Weislow, E. Lorenzo, A. Erice, J. Fitzgibbon, R. Shafer, D. Richman, T. M. Howard, Y. Zhao, E. Fisher, D. Huang, D. Mayers, S. Sylvester, M. Arens, K. Sannerud, S. Rasheed, V. Johnson, D. Kuritzkes, P. Reichelderfer, A. Japour, et al. 1998. Interlaboratory concordance of DNA sequence analysis to detect reverse transcriptase mutations in HIV-1 proviral DNA. Virol. Methods 75:93-104. [DOI] [PubMed] [Google Scholar]
- 15.Durant, J., P. Clevenbergh, P. Halfon, P. Delgiudice, S. Porsin, P. Simonet, N. Montagne, C. A. Boucher, J. M. Schapiro, and P. Dellamonica. 1999. Drug-resistance genotyping in HIV-1 therapy: the VIRADAPT randomised controlled trial. Lancet 353:2195-2199. [DOI] [PubMed] [Google Scholar]
- 16.Dykes, C., J. Najjar, R. J. Bosch, M. Wantman, M. Furtado, S. Hart, S. M. Hammer, and L. M. Demeter. 2004. Detection of drug-resistant minority variants of HIV-1 during virologic failure of indinavir, lamivudine, and zidovudine. J. Infect. Dis. 189:1091-1096. [DOI] [PubMed] [Google Scholar]
- 17.EuroGuidelines Group for HIV Resistance. 2001. Clinical and laboratory guidelines for the use of HIV-1 drug resistance testing as part of treatment management: recommendations for the European setting. AIDS 15:309-320. [DOI] [PubMed] [Google Scholar]
- 18.Gunthard, H. F., J. K. Wong, C. C. Ignacio, D. V. Havlir, and D. D. Richman. 1998. Comparative performance of high-density oligonucleotide sequencing and dideoxynucleotide sequencing of HIV type 1 pol from clinical samples. AIDS Res. Hum. Retrovir. 14:869-876. [DOI] [PubMed] [Google Scholar]
- 19.Hammer, S. M., F. Vaida, K. K. Bennett, M. K. Holohan, L. Sheiner, J. J. Eron, L. J. Wheat, R. T. Mitsuyasu, R. M. Gulick, F. T. Valentine, J. A. Aberg, M. D. Rogers, C. N. Karol, A. J. Saah, R. H. Lewis, L. J. Bessen, C. Brosgart, V. DeGruttola, and J. W. Mellors. 2002. Dual vs single protease inhibitor therapy following antiretroviral treatment failure: a randomized trial. JAMA 288:169-180. [DOI] [PubMed] [Google Scholar]
- 20.Hance, A. J., V. Lemiale, J. Izopet, D. Lecossier, V. Joly, P. Massip, F. Mammano, D. Descamps, F. Brun-Vezinet, and F. Clavel. 2001. Changes in human immunodeficiency virus type 1 populations after treatment interruption in patients failing antiretroviral therapy. J. Virol. 75:6410-6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Havlir, D. V., S. Eastman, A. Gamst, and D. D. Richman. 1996. Nevirapine-resistant human immunodeficiency virus: kinetics of replication and estimated prevalence in untreated patients. J. Virol. 70:7894-7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmes, E. C., L. Q. Zhang, P. Simmonds, C. A. Ludlam, and A. J. Brown. 1992. Convergent and divergent sequence evolution in the surface envelope glycoprotein of human immunodeficiency virus type 1 within a single infected patient. Proc. Natl. Acad. Sci. USA 89:4835-4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holterman, L., R. Dubbes, J. Mullins, J. Haaijman, and J. Heeney. 2000. A strategy for cloning infectious molecular clones of retroviruses from serum or plasma. J. Virol. Methods 84:37-48. [DOI] [PubMed] [Google Scholar]
- 24.Kuiken, C., B. Korber, and R. W. Shafer. 2003. HIV sequence databases. AIDS Rev. 5:52-61. [PMC free article] [PubMed] [Google Scholar]
- 25.Lawrence, J., D. L. Mayers, K. H. Hullsiek, G. Collins, D. I. Abrams, R. B. Reisler, L. R. Crane, B. S. Schmetter, T. J. Dionne, J. M. Saldanha, M. C. Jones, and J. D. Baxter. 2003. Structured treatment interruption in patients with multidrug-resistant human immunodeficiency virus. N. Engl. J. Med. 349:837-846. [DOI] [PubMed] [Google Scholar]
- 26.Learn, G. H., D. Muthui, S. J. Brodie, T. Zhu, K. Diem, J. I. Mullins, and L. Corey. 2002. Virus population homogenization following acute human immunodeficiency virus type 1 infection. J. Virol. 76:11953-11959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, S. L., A. G. Rodrigo, R. Shankarappa, G. H. Learn, L. Hsu, O. Davidov, L. P. Zhao, and J. I. Mullins. 1996. HIV quasispecies and resampling. Science 273:415-416. [DOI] [PubMed] [Google Scholar]
- 28.Livak, K. J., S. J. Flood, J. Marmaro, W. Giusti, and K. Deetz. 1995. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 4:357-362. [DOI] [PubMed] [Google Scholar]
- 29.Malet, I., M. Belnard, H. Agut, and A. Cahour. 2003. From RNA to quasispecies: a DNA polymerase with proofreading activity is highly recommended for accurate assessment of viral diversity. J. Virol. Methods 109:161-170. [DOI] [PubMed] [Google Scholar]
- 30.Mansky, L. M., and H. M. Temin. 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 69:5087-5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mellors, J., S. Palmer, D. Nissley, M. Kearney, E. Halvas, C. Bixby, L. Demeter, S. Eshleman, K. Bennett, F. Vaida, J. Coffin, and S. Hammer. 2003. Low frequency nonnucleoside reverse transcriptase inhibitor (NNRTI)-resistant variants contribute to failure of efavirenz-containing regimens in NNRTI-experienced patients with negative standard genotypes for NNRTI genotypes. Antivir. Ther. 8:S150. [Google Scholar]
- 32.Menendez-Arias, L. 2002. Molecular basis of fidelity of DNA synthesis and nucleotide specificity of retroviral reverse transcriptases. Prog. Nucleic Acid Res. Mol. Biol. 71:91-147. [DOI] [PubMed] [Google Scholar]
- 33.Metzner, K. J., S. Bonhoeffer, M. Fischer, R. Karanicolas, K. Allers, B. Joos, R. Weber, B. Hirschel, L. G. Kostrikis, H. F. Günthard, et al. 2003. Emergence of minor populations of human immunodeficiency virus type 1 carrying the M184V and L90M mutations in subjects undergoing structured treatment interruptions. J. Infect. Dis. 188:1433-1443. [DOI] [PubMed] [Google Scholar]
- 34.Nájera, I., A. Holguín, M. E. Quiñones-Mateu, M. A. Muñoz-Fernández, R. Nájera, C. López-Galíndez, and E. Domingo. 1995. pol gene quasispecies of human immunodeficiency virus: mutations associated with drug resistance in virus from patients undergoing no drug therapy. J. Virol. 69:23-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palmer, S., M. Kearney, F. Maldarelli, S. Kottilil, D. Lucey, J. Metcalf, D. Rock, M. VanHoutte, L. Michels, K. Hertogs, J. Mellors, and J. Coffin. 2002. Genetics of HIV-1 populations in acute and chronic infection. Antivir. Ther. 7:S57. [Google Scholar]
- 36.Paul, S. M., and V. S. Jorden. 2003. HIV resistance testing. A clinical tool. N. J. Med. 100:44-49. (Quiz, 100: 75-76). [PubMed] [Google Scholar]
- 37.Preston, B. D., B. J. Poiesz, and L. A. Loeb. 1988. Fidelity of HIV-1 reverse transcriptase. Science 242:1168-1171. [DOI] [PubMed] [Google Scholar]
- 38.Richman, D. D., D. Havlir, J. Corbeil, D. Looney, C. Ignacio, S. A. Spector, J. Sullivan, S. Cheeseman, K. Barringer, D. Pauletti, C.-K. Shih, M. Myers, and J. Griffin. 1994. Nevirapine resistance mutations of human immunodeficiency virus type 1 selected during therapy. J. Virol. 68:1660-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts, J. D., K. Bebenek, and T. A. Kunkel. 1988. The accuracy of reverse transcriptase from HIV-1. Science 242:1171-1173. [DOI] [PubMed] [Google Scholar]
- 40.Schuurman, R., D. Brambilla, T. de Groot, D. Huang, S. Land, J. Bremer, I. Benders, and C. A. Boucher. 2002. Underestimation of HIV type 1 drug resistance mutations: results from the ENVA-2 genotyping proficiency program. AIDS Res. Hum. Retrovir. 18:243-248. [DOI] [PubMed] [Google Scholar]
- 41.Shafer, R. W., M. A. Winters, S. Palmer, and T. C. Merigan. 1998. Multiple concurrent reverse transcriptase and protease mutations and multidrug resistance of HIV-1 isolates from heavily treated HIV-1 infected patients. Ann. Intern. Med. 128:906-911. [DOI] [PubMed] [Google Scholar]
- 42.Shankarappa, R., P. Gupta, G. H. Learn, Jr., A. G. Rodrigo, C. R. Rinaldo, Jr., M. C. Gorry, J. I. Mullins, P. L. Nara, and G. D. Ehrlich. 1998. Evolution of human immunodeficiency virus type 1 envelope sequences in infected individuals with differing disease progression profiles. Virology 241:251-259. [DOI] [PubMed] [Google Scholar]
- 43.Shankarappa, R., J. B. Margolick, S. J. Gange, A. G. Rodrigo, D. Upchurch, H. Farzadegan, P. Gupta, C. R. Rinaldo, G. H. Learn, X. He, X. L. Huang, and J. I. Mullins. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489-10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simmonds, P., P. Balfe, C. A. Ludlam, J. O. Bishop, and A. J. Brown. 1990. Analysis of sequence diversity in hypervariable regions of the external glycoprotein of human immunodeficiency virus type 1. J. Virol. 64:5840-5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Svarovskaia, E. S., S. R. Cheslock, W. H. Zhang, W. S. Hu, and V. K. Pathak. 2003. Retroviral mutation rates and reverse transcriptase fidelity. Front. Biosci. 8:d117-d134. [DOI] [PubMed] [Google Scholar]
- 46.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTALW: improving the sensitivity of progressive multiple-sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winters, M. A., J. M. Schapiro, J. Lawrence, and T. C. Merigan. 1998. Human immunodeficiency virus type 1 protease genotypes and in vitro protease inhibitor susceptibilities of isolates from individuals who were switched to other protease inhibitors after long-term saquinavir treatment. J. Virol. 72:5303-5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, L. Q., P. Simmonds, C. A. Ludlam, and A. J. Brown. 1991. Detection, quantification and sequencing of HIV-1 from the plasma of seropositive individuals and from factor VIII concentrates. AIDS 5:675-681. [DOI] [PubMed] [Google Scholar]