Abstract
We used broad-range PCR amplification and sequencing to detect and identify bacterial DNA in 156 valves of patients treated for infective endocarditis (IE). Bacterial DNA was found more frequently in patients who underwent valve replacement while on antibiotic treatment for IE (60%) than in patients who had completed antibiotic treatment for IE (37%; P = 0.02). We found specific bacterial DNA in valves removed from 11 of 30 patients who had completed antibiotic treatment for IE. Six had no histological evidence of IE. The presence of DNA was significantly correlated with the presence of histologic lesions (P = 0.001) and with the presence of bacteria detected by Gram staining (P < 0.001). Bartonella and streptococci were detected for much longer after antibiotic treatment by PCR than other species (P = 0.047 and 0.04, respectively), and coagulase-negative staphylococci were detected for much shorter periods (P = 0.02). The finding that bacterial DNA was more likely to be detected in valves of patients with active IE than in patients who had completed antibiotic treatment for IE shows that bacterial DNA is cleared slowly. There was no significant correlation between the duration of antibiotic therapy and the presence of bacterial DNA in valves. Since the persistence of bacterial DNA in valves does not necessarily indicate the persistence of viable bacteria, the detection of bacterial DNA in valves from IE patients should be interpreted with caution, in particular in those patients with a past history of treated IE.
Direct amplification of bacterial DNA from normally sterile sites by using broad-range primers provides an alternative approach to the detection of pathogens. Amplification and sequencing of the 16S rRNA gene in DNA extracted from cardiac-valve tissue has been used to provide etiological diagnoses in patients with culture-negative endocarditis (8, 11, 13, 15, 30, 35, 47). Consequently, Bosshard et al. and Millar et al. proposed that the Duke criteria should be modified to include a molecularly based diagnosis of noncultured causal agents as an additional major criterion (4, 28).
One problem is that bacterial DNA can still be detected after bacterial cultures become sterile. In patients with septic arthritis, bacterial DNA can be found in synovial tissue for long periods after cultures become negative (44). The persistence of bacterial DNA despite effective antibiotic treatment has also been demonstrated in patients with pulmonary tuberculosis (14, 49) and leptospirosis (3, 27). DNA is sufficiently stable that it can be amplified by PCR for long periods after bacteria are no longer viable (7, 25). As examples, PCR can be used to detect DNA in cells from mummies (31), Yersinia pestis DNA was found in 500-year-old human dental pulp (37), and mycobacterial DNA was detected in ancient human corpses that had macroscopic signs of tuberculosis (43) and leprosy (36). In a recent study, the presence of histological signs of infective endocarditis (IE) correlated with the presence of bacterial DNA in heart valve tissue, and when patients had no histological evidence of IE, PCR amplification was always negative (10). A recent study has shown that the presence of Gram-staining organisms in heart valves of patients cured of IE does not predict the presence of viable organisms (29). However, it has not been determined whether DNA amplification from valve tissue is predictive of an active infectious process. If this were the case, it might represent a tool to assess the efficiency of antibiotic therapy.
Since 1994, we have routinely used broad-range PCR amplifying the 16S rRNA-encoding gene and sequencing to detect and identify bacterial DNA in heart valve samples. In this report, our goal was to evaluate the role of antibiotic treatment duration in the persistence of bacterial DNA during endocarditis. We describe the experiments we performed to determine whether amplification of bacterial DNA in valve tissue was correlated with the duration of antibiotic treatment in patients with IE.
MATERIALS AND METHODS
Patients.
In a prospective study, we enrolled patients with endocarditis diagnosed according to the modified Duke's criteria described by Li et al. (23). There was a known etiology for their IE (by culture or serology), and the patients had been or were being treated with antibiotics when they had valve replacement surgery between 1 April 1994 and 31 April 2003. Infections with Bartonella spp. were diagnosed by culture or amplification of Bartonella DNA from valve tissue or whole blood or by serology with immunoglobulin G titers of ≥1:800 by microimmunofluorescence (38). For IE due to viridans streptococci or enterococci, effective treatment was defined using the recommendations of Wilson et al. (48), and for IE due to staphylococci, effective treatment was defined using the recommendations of Rubinstein and Carbon (40). For Bartonella spp., the recommendations of Raoult et al. were used to define effective treatment (38). For other organisms, antibiotic treatment was regarded as effective if antibiotics were administered intravenously according to the results of antimicrobial sensitivity testing. The days of prescription of parenteral antimicrobial treatment with more than one active antibiotics were counted. Oral agents taken after completion of effective treatment were included in the duration of treatment. Patients who received antibiotics only orally and those receiving insufficient doses of antibiotics were considered to have had insufficient treatment. Patients with a history of IE but for whom the type and duration of antibiotic therapy were not known were considered to have been treated effectively if they had no signs of active endocarditis at the time of surgery. Patients with IE due to multiple bacteria or with fungal endocarditis were excluded from the study. Patients with Coxiella burnetii endocarditis were excluded because it is a chronic condition that responds poorly to therapy (22) and would have biased the study. Patients were also excluded if the sequencing results indicated an etiological agent different from that obtained by culture or serology. Our selection criteria enabled us to group patients into those who, at the time of valve replacement surgery, had active IE and were receiving antibiotic treatment and those who had completed antibiotic treatment at the time of surgery.
Microbiology.
Three aerobic (BACTEC PLUS Aerobic/F) and anaerobic (BACTEC LYTIC/10 Anaerobic/F) blood cultures were obtained from each patient and incubated for 1 week in the BACTEC 9240 blood culture system (Becton Dickinson, Sparks, Md.). Valve tissue from patients was inoculated onto 5% blood agar (bioMérieux, Marcy-l'Etoile, France) and chocolate agar (bioMérieux) and incubated at 37°C in a 5% CO2 atmosphere for 10 days. Valve samples were also cultured on BCYE (bioMérieux) for 15 days and on Columbia media, where they were incubated under anaerobic conditions for 10 days. To isolate Bartonella spp., whole blood and valve tissue were inoculated onto both 5% blood agar (bioMérieux) and ECV 34 (20) and incubated at 37°C in a 5% CO2 atmosphere for up to 60 days. Isolates obtained from cell culture were identified by using species-specific mouse polyvalent antisera (26) and PCR (16, 39).
PCR amplification and sequencing.
Heart valve samples were stored at −80°C prior to DNA extraction, which was performed with the QIAmp kit (QIAGEN, Hilden, Germany) or the FastDNA kit (Bio 101, Carlsbad, Calif.). DNA was amplified using Taq DNA polymerase (Gibco BRL, Life Technologies) as previously described (12) but using the broad-range 16S rRNA gene primers 536f (5′-CAG CAG CCG CGG TAA TAC-3′) and 1050r (5′-CAC GAG CTG ACG ACA-3′). The sequences obtained from amplified DNA were compared with those available in GenBank using the BLASTN version 2.0 program through the website of the National Center for Biotechnology Information (2). PCR was also conducted on negative valve controls obtained from patients without IE, usually one control for every five patient samples. Positive amplification of any negative control caused the experiment to be considered unreliable. Only DNA corresponding to the bacterium responsible for the episode of IE was considered. Organisms identified as the recently reclassified Streptococcus equinus, Streptococcus gallolyticus, Streptococcus infantarius, Streptococcus lutetiensis, and Streptococcus alactolyticus (32, 41) were regarded as “Streptococcus bovis ” in our study.
Histopathology.
One histopathologist (H.L.) examined all hematoxylin-eosin-stained sections of the valves and classified lesions as (i) consistent with IE, (ii) having no signs of IE, or (iii) indeterminate according to criteria defined elsewhere (21). Specific stains, such as Gram and Whartin-Starry, were used when necessary (21).
Statistical analysis.
Demographic and clinical data, etiological agents (blood and valve culture results and serology), and duration of antibiotic treatments were compared using the χ2 or Mann-Whitney test according to PCR results. Univariate and multivariate logistic regressions were used to assess the situations more likely to benefit from systematic PCR amplification of the 16S rRNA gene and sequencing of the valves. The variables evaluated were age, sex, duration of antibiotic treatment, positive blood cultures, and histology. STATA software (version 7.0) was used for analysis.
RESULTS
A total of 147 patients who underwent 156 valve replacements were studied. Table 1 summarizes the demographic and clinical features of the patients at the time of valve replacement surgery. The patients were receiving antibiotic treatment for IE when 126 of the valve replacements were performed, and bacterial DNA was amplified more frequently in these patients (76 of 126; 60%) than in patients who had completed antibiotic treatment for IE at the time of valve replacement surgery (11 of 30; 37%; P = 0.02). The median ages of patients with positive and negative PCR results were 58 (interquartile range, 42 to 70) and 60 (interquartile range, 50 to 69) years, respectively. Seventy-one percent of patients with positive PCR results and 70% of patients with negative PCR results were males.
TABLE 1.
Characteristics of 156 episodes of valve replacement for IE according to PCR
| Parameter | Value for group
|
P value | |
|---|---|---|---|
| Positive PCR (n = 87) | Negative PCR (n = 69) | ||
| Demographic data | |||
| No. (%) male | 62 (71) | 49 (70) | 0.82 |
| Median (IQR) age (yr) | 58 (42-70) | 60 (50-69) | 0.27 |
| Histologic data | |||
| Histologic lesions in favor of IEa [no. (%)] | 64 (75) | 31 (48) | 0.001 |
| Presence of bacteria on Gram stainingb [no. (%)] | 43 (54) | 15 (24) | <0.001 |
| Etiologic diagnosis [no. (%)] | |||
| Streptococcus spp. | 46 (53) | 28 (41) | 0.13 |
| S. bovis | 22 (25) | 12 (17) | 0.24 |
| Oral Streptococcus | 19 (22) | 15 (22) | 0.99 |
| Enterococcus spp. | 10 (11) | 8 (12) | 0.99 |
| Enterococcus spp. and Streptococcus spp. | 56 (64) | 36 (51) | 0.12 |
| Staphylococcus spp. | 18 (21) | 23 (33) | 0.08 |
| CNS | 4 (5) | 9 (13) | 0.06 |
| S. aureus | 14 (16) | 14 (20) | 0.50 |
| Bartonella spp. | 7 (8) | 0 (0) | 0.02 |
| Othersc | 11 (13) | 11 (16) | 0.56 |
Six missing valve for histologic study.
Fourteen missing valve for Gram-staining study.
Others: Actinobacillus actinomycetans (three cases), Corynebaterium jeikeium, Corynebacterium diphteriae, Proteus mirabilis, Haemophilus spp. (two cases), Streptococcus pneumoniae (two cases), Proprionobacterium acnes, Micrococcus spp., Enterobacter cloacae (two cases), E. coli (two cases), Streptococcus β haemolytic (three cases), Streptococcus agalactiae, Neisseria sicca, rastonia.
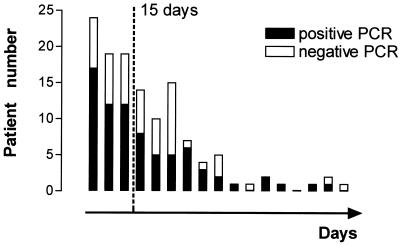
To determine the effect of current antibiotic treatment on positive PCR results, we analyzed the data on 126 patients with IE who were receiving antibiotic treatment at the time of surgery. The median duration of treatment was 19.5 days (range, 1 to 150) for patients with negative PCR results and 14 days (range, 1 to 79) for those with positive PCR results (P = 0.28). The median duration of antibiotic treatment for all of the patients was 15 days. Consequently, we analyzed as a whole and separately patients with >15 and ≤15 days of antibiotic therapy. PCRs were positive for 66% of patients receiving antibiotics for ≤15 days and for 55% of patients receiving >15 days of antibiotic treatment (P = 0.19) (Fig. 1).
FIG. 1.
Number of patients with a positive or negative PCR according to the delay between the beginning of antibiotic treatment and surgery (clusters of 5 days) among 126 episodes of valve replacement.
In this group of 126 patients with IE who were receiving antibiotic treatment at the time of surgery, S. bovis, oral viridans streptococci, and enterococci accounted for 21, 18, and 13% of cases of IE, respectively. Staphylococcus aureus and coagulase-negative staphylococci (CNS) represented 17 and 10% of the cases of IE, respectively. Bartonella spp. accounted for 5% of the cases of IE. Table 2 shows the PCR results for each of the species causing IE in the study. Valves infected with Streptococcus spp. were more likely to be positive by PCR than those infected with other species (P = 0.047). We detected bacterial DNA in all the valves of patients with Bartonella sp. IE (6 of 6). Valves infected with CNS were more likely to be negative by PCR than those infected with other species (P = 0.02). This association was confirmed by multivariate analysis adjusted for age, sex, duration of antibiotic treatment, and positive blood cultures (odds ratio = 0.26; 95% confidence interval, 0.07 to 0.95). Valves infected with streptococci were more likely to be positive by PCR (38 of 54) than those infected with CNS (4 of 13; P = 0.01) or with staphylococci and enterococci (26 of 51; P = 0.04).
TABLE 2.
Comparison of results of PCRs for one species versus all species performed on heart valves from 126 patients with IE with antibiotic treatment at the time of surgery
| Species | Positive PCR for species/total positive PCR (%) | Negative PCR for species/total negative PCR (%) | P value |
|---|---|---|---|
| Streptococcus spp. and enterococci | 47/76 (62) | 24/50 (48) | 0.13 |
| Streptococcus spp. | 38/76 (50) | 15/50 (30) | 0.047 |
| S. bovis | 18/76 (24) | 8/50 (16) | 0.30 |
| Oral streptococci | 16/76 (21) | 7/50 (14) | 0.32 |
| Enterococci | 9/76 (12) | 8/50 (16) | 0.50 |
| Staphylococcus spp. | 17/76 (22) | 17/50 (34) | 0.15 |
| S. aureus | 13/76 (17) | 8/50 (16) | 0.87 |
| CNS | 4/76 (5) | 9/50 (18) | 0.02 |
| Bartonella spp. | 6/76 (8) | 0 | 0.04 |
| Othersa | 10/76 (13) | 10/50 (20) | 0.30 |
Others: Actinobacillus actinomycetans (two cases), Corynebaterium jeikeium, Corynebacterium diphteriae, Proteus mirabilis, Haemophilus spp. (two cases), Streptococcus pneumoniae, Proprionobacterium acnes, Micrococcus spp., Enterobacter cloacae (two cases), E. coli (two cases), Streptococcus β haemolytic (three cases), Streptococcus agalactiae, Neisseria sicca, rastonia.
Thirty patients with a history of IE who had completed their antibiotic treatment had valve replacement surgery. The median time between surgery and antibiotic therapy was 1,210 days (range, 53 to 9,125) in patients with negative PCRs and 167 days (range, 45 to 2,920) in patients with positive PCRs (P = 0.01). In 11 patients in this group, bacterial DNA was found in valve tissue a long time after the end of antibiotic treatment. Seven patients did not have any histological signs of IE at the time of surgery (Table 3), while four had histological signs of IE. These four patients with histological signs of IE were likely to still have evolutive IE, since they all had biological markers of inflammation and two of them had persistent echographic signs of IE. Among the seven patients with no histological signs of IE, two patients (patients 1 and 3) did not have any clinical, biological, or echographic signs of active endocarditis. None of these patients developed signs of IE after valve replacement.
TABLE 3.
Long-term persistence of bacterial DNA in patients who had completed antibiotic treatment for IE at the time of surgerya
| Case/age/sexb | Bacterium | Involved valve | Delay between diagnosis and surgery |
|---|---|---|---|
| 1/55/M | Staphylococcus pneumoniae | Aortic bioprosthesis | 7 yr |
| 2/69/F | S. bovis | Aortic, native | 167 days |
| 3/80/M | S. bovis | Mitral, native | 730 days |
| 4/39/M | Enterococcus faecium | Aortic bioprosthesis | 850 days |
| 5/36/M | Streptococcus gordonii | Aortic, native | 45 days |
| 6/33/M | Bartonella quintana | Aortic homograft | 224 days |
| 7/70/F | Streptococcus sanguinis | Mitral bioprosthesis | 545 days |
All patients had no clinical or histological signs of IE, and valve cultures were sterile.
M, male; F, female.
Bacterial DNA was amplified by PCR in 67% (64 of 95) of valves with histological lesions indicating IE and in 38% (21 of 55) of valves that had no histological evidence of IE (P = 0.001). This significant correlation was confirmed by multivariate analysis adjusted for age and sex (odds ratio = 3.3; 95% confidence interval, 1.61 to 6.80). PCR amplification was positive in 74% (43 of 58) of valves in which bacteria were seen with Gram staining and in 44% (37 of 84) of valves that had no evidence of bacteria with Gram staining (P < 0.001). This significant correlation was also confirmed by multivariate analysis adjusted for age and sex.
DISCUSSION
In our study, we included only patients who had IE with known bacterial etiology and who underwent surgery while receiving antibiotic treatment or after successful antibiotic treatment of the condition.
We found no significant correlation between the duration of antibiotic treatment and PCR results in patients receiving antibiotics at the time of surgery. The use of PCR amplification to monitor treatment of infectious disease has become a subject of interest. In an experimental model of systemic candidiasis, PCR results correlated with the therapeutic efficacy of antifungal treatment (45). In a rabbit syphilis model, PCR amplification results suggested that in most instances the presence of Treponema pallidum DNA reflects living organisms (46). In infected mice, Borrelia burgdoferi DNA quickly disappeared from tissues that became culture negative after antibiotic treatment (24). These studies suggest that bacterial DNA is cleared from the tissue sites of animals after bacterial death but that the DNAs from different pathogens may have different kinetics at different sites. In human pathology, molecular methods based on DNA detection have been used to monitor the treatment of Whipple's disease (34), Lyme arthritis (33), and infections with Helicobacter pylori (42) and Chlamydia trachomatis (1). Our results show that even though bacterial DNA seems to be cleared over time in patients with IE, DNA detection should not be used for monitoring antibiotic efficacy in patients with ongoing IE.
When we considered bacterial species separately, all patients with endocarditis due to Bartonella spp. were positive by PCR. This may reflect greater amounts of DNA in infected valves or the fact that DNA of Bartonella spp. remains for longer periods in tissues than is the case with DNA of other bacteria. Patients with IE due to Streptococcus spp. were more likely to have a positive PCR. This shows that DNA of Streptococcus spp. persists longer than DNA of other species. On the other hand, patients infected with CNS were more likely to have a negative PCR, indicating that DNA of these species is less persistent than DNA of other species.
Our study showed that PCR was more likely to be positive in patients whose histology was indicative of IE and when bacteria were observed in histological preparations. These findings are in agreement with previous reports that molecular detection and sequencing were useful in cases of IE that were culture negative due to prior administration of antibiotics (4, 11, 18, 19, 28, 47), particularly when histopathology revealed the presence of bacteria (10, 47).
In previous studies, PCR was carried out only on valves from patients with acute IE and those with histological evidence of IE (4, 10). We were interested in patients under treatment for IE and those with a past history of IE who had completed their antibiotic treatment at the time of surgery. PCR amplification was positive despite the absence of histological signs of IE in 7 of 30 patients with a past history of IE. Two of these, one previously reported by Branger et al. (5) without clinical, laboratory, or echographic signs of active IE, had evidence of prolonged persistence of bacterial DNA in their heart valves. Both live and dead bacteria may result in positive PCR assays (17, 25). Despite negative bacterial cultures, the presence of bacterial DNA in clinical specimens has been shown for prolonged periods in septic arthritis (6, 44), pulmonary tuberculosis (14, 49), and leptospirosis (3, 27). The significance of the persistence of DNA for long periods after antibiotic treatment is uncertain (17, 25). Morris et al. found that in some patients with IE that had apparently been cured, valve cultures were sterile and had no histological signs of IE, although they were positive for organisms with Gram staining (29). Positive PCRs and Gram stains may represent results from previous episodes of IE that have resolved, and the results of both these tests must be interpreted with care. Gauduchon et al. reported a case of IE with blood cultures yielding Escherichia coli (9). Six months previously, however, the patient had been diagnosed with IE due to Streptococcus mutans. During this second episode of IE, the patient underwent valve replacement. Histology showed gram-positive cocci in the valve, and PCR amplification and sequencing detected S. mutans DNA. The authors considered the patient to have recurrent IE due to S. mutans rather than a new infection with E. coli. This interpretation is challenged by our finding that streptococcal DNA may persist long after IE is cured.
In conclusion, our results show that as the time between successful treatment of IE and valve surgery increases, the likelihood of bacterial DNA being demonstrated in heart valves decreases. This indicates that bacterial DNA is cleared over time but that this might be a very slow process. Since there is no correlation between the duration of antibiotic treatment and PCR results in patients still on antibiotic treatment at the time of surgery, DNA detection should not be used as a tool for monitoring treatment in IE. The persistence of DNA does not necessarily correspond to the persistence of viable bacteria and therefore to the successful treatment of the infection.
Acknowledgments
We thank Pat Kelly for critical reading of the manuscript.
REFERENCES
- 1.Adair, C. D., M. Gunter, T. G. Stovall, G. McElroy, J. C. Veille, and J. M. Ernest. 1998. Chlamydia in pregnancy: a randomized trial of azithromycin and erythromycin. Obstet. Gynecol. 91:165-168. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, X., F. Stephen, L. Thomas, X. Madden, A. Alejandro, X. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bal, A. E., C. Gravekamp, R. A. Hartskeerl, J. De Meza-Brewster, H. Korver, and W. J. Terpstra. 1994. Detection of leptospires in urine by PCR for early diagnosis of leptospirosis. J. Clin. Microbiol. 32:1894-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosshard, P. P., A. Kronenberg, R. Zbinden, C. Cuef, E. C. Böttberg, and M. Altwegg. 2003. Etiologic diagnosis of infective endocarditis by broad-range polymerase chain reaction: a 3-year experience. Clin. Infect. Dis. 37:167-172. [DOI] [PubMed] [Google Scholar]
- 5.Branger, S., J. P. Casalta, G. Habib, F. Collart, and D. Raoult. 2003. Streptococcus pneumoniae endocarditis: persistence of DNA on heart valve material 7 years after infectious episode. J. Clin. Microbiol. 41:4435-4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canvin, J. M. G., S. C. Goutcher, M. Hagig, C. G. Gemmell, and R. D. Sturrock. 1997. Persistence of Staphylococcus aureus as detected by polymerase chain reaction in the synovial fluid of a patient with septic arthritis. Br. J. Rheumatol. 36:203-206. [DOI] [PubMed] [Google Scholar]
- 7.Dupray, E., M. P. Caprais, A. Derrien, and P. Fach. 1997. Salmonella DNA persistence in natural seawaters using PCR analysis. J. Appl. Microbiol. 82:507-510. [DOI] [PubMed] [Google Scholar]
- 8.Fournier, P. E., and D. Raoult. 1999. Nonculture laboratory methods for the diagnosis of infectious endocarditis. Curr. Infect. Dis. Rep. 1:136-141. [DOI] [PubMed] [Google Scholar]
- 9.Gauduchon, V., Y. Benito, M. Celard, et al. 2001. Molecular diagnosis of recurrent Streptococcus mutans endocarditis by PCR amplification and sequencing. Clin. Microbiol. Infect. 7:36-37. [PubMed] [Google Scholar]
- 10.Gauduchon, V., L. Chalabreysse, J. Etienne, M. Célard, Y. Benito, H. Lepidi, F. Thivlet-Béjui, and F. Vandenesch. 2003. Molecular diagnosis of infective endocarditis by PCR amplification and direct sequencing of DNA from valvular tissue. J. Clin. Microbiol. 41:763-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldenberger, D., A. Künzli, P. Vogt, R. Zbinden, and M. Altwegg. 1997. Molecular diagnosis of bacterial endocarditis by broad-range PCR amplification and direct sequencing. J. Clin. Microbiol. 35:2733-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greub, G., and D. Raoult. 2002. “Actinobaculum massilae”, a new species causing chronic urinary tract infection. J. Clin. Microbiol. 40:3938-3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grijalva, M., R. Horvath, M. Dendis, J. Cerny, and J. Benedik. 2003. Molecular diagnosis of culture negative infective endocarditis: clinical validation in a group of surgically treated patients. Heart 89:263-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hellyer, T. J., T. W. Fletcher, J. H. Bates, W. W. Stead, G. L. Templeton, M. D. Cave, and K. D. Eisenach. 1996. Strand displacement amplification and the polymerase chain reaction for monitoring response to treatment in patients with pulmonary tuberculosis. J. Infect. Dis. 173:934-941. [DOI] [PubMed] [Google Scholar]
- 15.Houpikian, P., and D. Raoult. 2002. Diagnostic methods: current best practices and guidelines for identification of difficult-to-culture pathogens. Infect. Dis. Clin. N. Am. 16:377-392. [DOI] [PubMed] [Google Scholar]
- 16.Joblet, C., V. Roux, M. Drancourt, J. Gouvernet, and D. Raoult. 1995. Identification of Bartonella (Rochalimea) species among fastidious gram-negative bacteria based on partial sequence of the citrate-synthetase gene. J. Clin. Microbiol. 33:1879-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Josephson, K. L., C. P. Gerba, and I. L. Pepper. 1993. Polymerase chain reaction detection of nonviable bacterial pathogens. Appl. Environ. Microbiol. 59:3513-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamas, C. C., and S. J. Eykyn. 2003. Blood culture negative endocarditis: analysis of 63 cases presenting over 25 years. Heart 89:258-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lang, S., R. W. Watkin, P. A. Lambert, R. S. Ronser, W. A. Littler, and T. S. J. Elliott. 2004. Evaluation of PCR in the molecular diagnosis of endocarditis. J. Infect. 48:269-275. [DOI] [PubMed] [Google Scholar]
- 20.La Scola, B., and D. Raoult. 1999. Culture of Bartonella quintana and Bartonella henselae from human samples: a 5-year experience (1993 to 1998). J. Clin. Microbiol. 37:1899-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lepidi, H., D. T. Durack, and D. Raoult. 2002. Diagnostic methods: current best practices and guidelines for histologic evaluation in infective endocarditis. Infect. Dis. Clin. N. Am. 16:339-361. [DOI] [PubMed] [Google Scholar]
- 22.Lepidi, H., P. Houpikian, Z. Liang, and D. Raoult. 2003. The heart valves of patients with Q fever endocarditis: microbiological, molecular and histological studies. J. Infect. Dis. 187:1097-1106. [DOI] [PubMed] [Google Scholar]
- 23.Li, J. S., D. J. Sexton, N. Mick, R. Nettles, V. G. J. Fowler, T. Ryan, T. Bashore, and G. R. Corey. 2000. Proposed modifications to the Duke criteria for the diagnosis of infective endocarditis. Clin. Infect. Dis. 30:633-638. [DOI] [PubMed] [Google Scholar]
- 24.Malawista, S. E., S. W. Barthold, and D. H. Persing. 1994. Fate of Borrelia burgdoferi DNA in tissues of infected mice after antibiotic treatment. J. Infect. Dis. 170:1312-1316. [DOI] [PubMed] [Google Scholar]
- 25.Masters, C. I., J. A. Schallcross, and B. M. Mackey. 1994. Effect of stress treatments on the detection of Listeria monocytogenes and enterotoxigenic Escherichia coli by the polymerase chain reaction. J. Appl. Bacteriol. 77:73-79. [DOI] [PubMed] [Google Scholar]
- 26.Maurin, M., V. Roux, A. Stein, F. Ferrier, R. Viraben, and D. Raoult. 1994. Isolation and characterization by immunofluorescence, SDS-PAGE, Western blot, RFLP-PCR, 16S rRNA gene sequencing and pulsed-field gel electrophoresis of Rochalimea quintana from a French patient with bacillary angiomatosis. J. Clin. Microbiol. 32:1166-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merien, F., G. Baranton, and P. Perolat. 1995. Comparison of polymerase chain reaction with microagglutination test and culture for diagnosis of leptospirosis. J. Infect. Dis. 172:281-285. [DOI] [PubMed] [Google Scholar]
- 28.Millar, B. M. J., P. Mallon, J. Xu, M. Crowe, R. McClurg, D. Raoult, J. Earle, R. Hone, and P. Murphy. 2001. Molecular diagnosis of infective endocarditis—a new Duke's criterion. Scand. J. Infect. Dis. 33:673-680. [DOI] [PubMed] [Google Scholar]
- 29.Morris, A. J., D. Drinkovic, S. Pottumarthy, M. G. Strickett, D. MacCulloch, N. Lambie, and A. R. Kerr. 2003. Gram stain, culture, and histopathlogical examination findings for heart valves removed because of infective endocarditis. Clin. Infect. Dis. 36:697-704. [DOI] [PubMed] [Google Scholar]
- 30.Ohara-Nemoto, Y., S. Tajika, M. Sasaki, and M. Kaneko. 1997. Identification of Abiotrophia adiacens and Abiotrophia defectiva by 16S rRNA gene PCR and restriction fragment length polymorphism analysis. J. Clin. Microbiol. 35:2458-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paabo, S. G., J. A. Gifford, and A. C. Wilson. 1988. Mitochondrial DNA sequences from a 7000 year old brain. Nucleic Acids Res. 16:9775-9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poyart, C., G. Quesne, and P. Trieu-Cuot. 2002. Taxonomic dissection of the Streptococcus bovis group by analysis of manganese-dependent superoxide dismutase gene (sodA) sequences: reclassification of ‘Streptococcus infantarius subsp. coli’ as Streptococcus lutetiensis sp. nov. and of Streptococcus bovis biotype II.2 as Streptococcus pasteurianus sp. nov. Int. J. Syst. Evol. Microbiol. 52:1247-1255. [DOI] [PubMed] [Google Scholar]
- 33.Priem, S., G. R. Burmester, T. Kamradt, K. Wolbart, M. G. Rittig, and A. Krause. 1998. Detection of Borrelia burgdorferi by polymerase chain reaction in synovial membrane but not in synovial fluid from patients with persisting Lyme arthritis after antibiotic therapy. Ann. Rheum. Dis. 57:118-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pron, B., C. Poyart, E. Abachin, T. Fest, C. Belanger, C. Bonnet, P. Capelle, J. F. Bretagne, A. Fabianek, L. Girard, H. Hagège, and P. Berche. 1999. Diagnosis and follow-up of Whipple's disease by amplification of the 16S rRNA gene of Tropheryma whippelii. Eur. J. Clin. Microbiol. Infect. Dis. 18:62-65. [DOI] [PubMed] [Google Scholar]
- 35.Qin, X., and K. B. Urdahl. 2001. PCR and sequencing of independent genetic targets for the diagnosis of culture negative bacterial endocarditis. Diagn. Microbiol. Infect. Dis. 40:145-149. [DOI] [PubMed] [Google Scholar]
- 36.Rafi, A., M. Spigelman, J. Standford, E. Lemma, H. Donoghue, and J. Zias. 1994. Mycobacterium leprae DNA from ancient bone detected by PCR. Lancet 343:1360-1361. [PubMed] [Google Scholar]
- 37.Raoult, D., G. Abdouharam, E. Crubézy, G. Larrouy, B. Ludes, and M. Drancourt. 2000. Molecular identification by “suicide PCR” of Yersinia pestis as the agent of medieval black death. Proc. Natl. Acad. Sci. USA 97:12800-12803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raoult, D., P. E. Fournier, F. Vandenesch, J. L. Mainardi, S. Eykyn, E. Nash, E. James, and T. J. Marrie. 2003. Outcome and treatment of Bartonella endocarditis. Arch. Intern. Med. 163:226-230. [DOI] [PubMed] [Google Scholar]
- 39.Roux, V., and D. Raoult. 1995. Inter- and intraspecies identification of Bartonella (Rochalimea) species. J. Clin. Microbiol. 33:1573-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rubinstein, E., and C. Carbon. 1998. Staphylococcal endocarditis: recommendations for therapy. Clin. Microbiol. Infect. 4:S27-S33. [PubMed] [Google Scholar]
- 41.Schlegel, L., F. Grimont, E. Ageron, P. A. D. Grimont, and A. Bouvet. 2003. Reappraisal of the taxonomy of the Streptococcus bovis/Streptococcus equinus complex and related species: description of Streptococcus gallolyticus subsp. gallolyticus subsp. nov., S. gallolyticus subsp. macedonicus subsp. nov. and S. gallolyticus subsp. pasteurianus subsp. nov. Int. J. Syst. Evol. Microbiol. 53:631-645. [DOI] [PubMed] [Google Scholar]
- 42.Shuber, A. P., J. J. Ascano, K. A. Boynton, A. Mitchell, H. F. Frierson, W. El-Rifai, and S. M. Powell. 2002. Accurate, noninvasive detection of Helicobacter pylori DNA from stool samples: potential usefulness for monitoring treatment. J. Clin. Microbiol. 40:262-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor, G. M., M. Goyal, A. J. Legge, R. J. Shaw, and D. Young. 1999. Genotypic analysis of Mycobacterium tuberculosis from medieval human remains. Microbiology 145:899-904. [DOI] [PubMed] [Google Scholar]
- 44.Van der Heijden, I. M., B. Wilbrink, A. E. M. Vije, L. M. Schouls, F. C. Breedveld, and P. P. Tak. 1999. Detection of bacterial DNA in serial synovial samples obtained during antibiotic treatment from patients in septic arthritis. Arthritis Rheum. 42:2198-2203. [DOI] [PubMed] [Google Scholar]
- 45.Van Deventer, A. J. M., W. H. F. Goessens, A. Van Belkum, E. W. M. Van Etten, H. J. A. Van Vliet, and H. A. Verbrugh. 1996. PCR monitoring of response to liposomal amphotericin B treatment of systemic candidiasis in neutropenic mice. J. Clin. Microbiol. 34:25-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wicher, K., F. Abbruscato, V. Wicher, D. N. Collins, I. Auger, and H. W. Horowitz. 1998. Identification of persistent infection in experimental syphilis by PCR. Infect. Immun. 66:2309-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilck, M. B., Y. Wu, J. G. Howe, J. Y. Crouch, and S. Edberg. 2001. Endocarditis caused by culture-negative organisms visible by Brown and Brenn staining: utility of PCR and DNA sequencing for diagnosis. J. Clin. Microbiol. 39:2025-2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson, W. R., and the Endocarditis Working Group of the International Society for Chemotherapy. 1998. Antibiotic treatment of infective endocarditis due to viridans streptococci, enterococci, and other streptococci. Clin. Microbiol. Infect. 4:S17-S26. [PubMed] [Google Scholar]
- 49.Yuen, K. Y., K. S. Chan, C. M. Chan, B. Ho, L. Dai, P. Chau, and M. Ng. 1993. Use of PCR in routine diagnosis of treated and untreated pulmonary tuberculosis. J. Clin. Pathol. 46:318-322. [DOI] [PMC free article] [PubMed] [Google Scholar]