Abstract
Two new species, Candida orthopsilosis and C. metapsilosis, are proposed to replace the existing designations of C. parapsilosis groups II and III, respectively. The species C. parapsilosis is retained for group I isolates. Attempts to construct a multilocus sequence typing scheme to differentiate individual strains of C. parapsilosis instead revealed fixed DNA sequence differences between pairs of subgroups in four genes: COX3, L1A1, SADH, and SYA1. PCR amplicons for sequencing were obtained for these four plus a further seven genes from 21 group I isolates. For nine group II isolates, PCR products were obtained from only 5 of the 11 genes, and for two group III isolates PCR products were obtained from a different set of 5 genes. Three of the PCR products from group II and III isolates differed in size from the group I products. Cluster analysis of sequence polymorphisms from COX3, SADH, and SYA1, which were common to the three groups, consistently separated the isolates into three distinct sets. All of these differences, together with DNA sequence similarities <90% in the ITS1 sequence, suggest the subgroups should be afforded species status. The near absence of DNA sequence variability among isolates of C. parapsilosis and relatively high levels of sequence variability among isolates of C. orthopsilosis suggest that the former species may have evolved very recently from the latter.
Although Candida albicans remains the most common Candida species encountered as a cause of human infections, other Candida species have been increasingly associated with disseminated disease since the 1990s. C. parapsilosis is the second most common yeast species isolated from the blood in Latin American countries and Asia (20, 23), and it is also found commonly in European surveys (19, 23). The species is particularly associated with bloodstream infections in neonates and also with catheter-associated candidemia and intravenous hyperalimentation (12).
Early work with restriction fragment polymorphisms showed that C. parapsilosis isolates were more genotypically heterogeneous than those of other Candida species (24). C. parapsilosis isolates can be divided into three groups distinguished on the basis of randomly amplified polymorphic DNA (RAPD) (16), multilocus enzyme electrophoresis (17), internal transcribed spacer (ITS) sequences of DNA encoding ribosomes (17), DNA relatedness (21), morphotyping (4), mitochondrial DNA sequence differences (18), DNA topoisomerase II gene sequences (11), and an oligonucleotide probe used for fingerprinting C. parapsilosis strains (7). In several of these studies, the authors stated that the extent of the differences between the subgroups of C. parapsilosis was sufficient to merit their designation as distinct species (4, 11, 17, 21). Kurtzman and Robnett (14, 15) considered the six nucleotide differences in the rRNA D1/D2 domain between the C. parapsilosis type strain and a group II isolate to be sufficient to justify regarding the latter as a new species.
As part of our ongoing research into multilocus sequence typing (MLST) of strains within pathogenic Candida species (3, 26, 27), we attempted to find genes with polymorphic loci that could differentiate a panel of C. parapsilosis isolates from diverse anatomic and geographic sources. Several of the genes we studied yielded PCR products only with isolates belonging to group I according to their RAPD profiles and ITS1 sequences (17). When isolates from C. parapsilosis groups I, II, and III gave products that could be sequenced, the sequences differed at a level characteristic for separate species. We describe here the MLST sequence diversity within these groups and formally designate C. parapsilosis groups II and III as the new species C. orthopsilosis and C. metapsilosis on the grounds that the level of genotypic differences between these taxa is too great for them to retain any longer their status as subgroups of a form species.
MATERIALS AND METHODS
C. parapsilosis isolates.
The 32 C. parapsilosis isolates used in the present study are listed in Table 1. They include the type strain of C. parapsilosis group I (CBS 604) and two representative isolates for group II (ATCC 96139 and ATCC 96141) and group III (ATCC 96143 and ATCC 96144). The remaining 27 isolates were chosen to represent presumed genetic and phenotypic diversity based on their date, anatomic site, and geographic source of isolation. The ability to assimilate a variety of carbohydrate source compounds was tested for all isolates with the API ID 32C (bioMérieux) according to the manufacturer's instructions. Colony morphology was analyzed after growth on Sabouraud agar (Oxoid, Basingstoke, United Kingdom) and CHROMagar Candida (M-Tech Diagnostic, Warrington, United Kingdom). For each isolate, one plate was incubated at 35, 37, and 42°C. The yeasts were maintained on Sabouraud agar slants.
TABLE 1.
Isolates and reference strains of C. parapsilosis used in this study
| Isolate | Site of isolation | Origin | RAPDa profile | Groupb |
|---|---|---|---|---|
| CBS 604 | Feces | Puerto Rico (type strain) | A | I |
| 711701 | Unknown | Aberdeen, United Kingdom | A | I |
| 74/046 | Aortic valve | Leeds, United Kingdom | A | I |
| 81/042 | Ear | London, United Kingdom | A | I |
| 73/114 | Anus | Leeds, United Kingdom | A | I |
| J930733 | Cat hair | Beerse, Belgium | A | I |
| J960578 | Nail | Hong Kong | A | I |
| J951066 | Nail | Korea | A | I |
| J961250 | Nail | Lisbon, Portugal | A | I |
| J931845 | Unknown | Japan | A | I |
| J931058 | Nail | Belgium | A | I |
| J950218 | Unknown | United States | A | I |
| J930631/1 | Cat hair | Africa | A | I |
| 81/041 | Vagina | Mayo Clinic, Minneapolis, Minn. | A | I |
| 81/253 | Nail | London, United Kingdom | A | I |
| 81/040 | Toe space | London, United Kingdom | A | I |
| 73/037 | Vagina | Leeds, United Kingdom | A | I |
| 103 | Anus | London, United Kingdom | A | I |
| 73/107 | Mouth | London, United Kingdom | A | I |
| 02-203 | Blood | Bergamo, Italy | A | I |
| 90-137 | Orbital tissue | San Jose, Calif | A | I |
| J981224 | Vagina | United States | B1 | II |
| 90-125 | Unknown | San Francisco, Calif. | B2 | II |
| 02-212 | Blood | Barcelona, Spain | B3 | II |
| 92-181 | Contaminated solution | Redwood City, Calif. | B1 | II |
| 02-201 | Blood | Italy | B2 | II |
| 82-33/698 | Unknown | San Jose, Calif. | B3 | II |
| J941221K | Nail | St. Niklaas, Belgium | B1 | II |
| ATCC 96139 | CVP catheterc | San Antonio, Tex. | B1 | II |
| ATCC 96141 | Blood | San Antonio, Tex. | B4 | II |
| ATCC 96143 | Unknown | Livermore, Calif. | C | III |
| ATCC 96144 | Hand | Tacoma, Wash. | C | III |
RAPD patterns were obtained with the primer RPO2.
Groups I, II, and III were differentiated on the basis of the ITS1 sequence (17).
That is, the tip of the catheter used for monitoring the central venous pressure.
DNA extraction.
Genomic DNA was extracted from yeasts grown in a broth comprised of 2% glucose, 2% mycological peptone (Oxoid), and 1% yeast extract (Difco, Detroit, Mich.). Briefly, cells were harvested in stationary phase and lysed by vortexing the pellet for 3 min with 0.3-g glass beads (0.45 to 0.52 mm in diameter; Sigma, St. Louis, Mo.) in 200 μl of buffer (100 mM Tris-HCl [pH 8.0] containing 2% Triton X-100, 1% sodium dodecyl sulfate, 1 mM EDTA) and 200 μl of 1:1 (vol/vol) phenol-chloroform solution. After vortexing, 200 μl of TE (1 mM EDTA, 10 mM Tris-HCl [pH 8.0]) was added to the lysate; the mixture was microcentrifuged at full speed for 10 min, and the aqueous phase transferred to a new tube. DNA was precipitated by addition of 1 ml of ethanol to the supernatant. Samples were centrifuged, and the pellet was resuspended in 400 μl of TE containing 100 μl of RNase (10 mg/ml; Sigma). The mixture was incubated for 1 h at 37°C, and then DNA was precipitated with 2 volumes of isopropanol and 10 μl of 4 M ammonium acetate, dried, and redissolved in 50 μl of TE (pH 8.0).
RAPD-PCR.
C. parapsilosis isolates were screened by RAPD tests with the primer RPO2 (5′-GCGATCCCCA-3′) (22). Reaction volumes of 50 μl contained 100 ng of genomic DNA, 1.5 U of Taq polymerase (Promega, Madison, Wis.), 5 μl of 10× magnesium-free buffer, 3 μl of 25 mM MgCl2, a 200 μM concentration of deoxynucleoside triphosphates, and a 10 μM concentration of the primer. The PCR products were amplified in a Flexigene Thermocycler (Techne, Cambridge, United Kingdom) set up with a first cycle of denaturation for 2 min at 94°C, followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 36°C for 1 min, and elongation at 72°C for 75 s, with a final extension step of 10 min at 72°C. Amplified DNA products were separated by electrophoresis in a 1% agarose gel containing ethidium bromide (0.5 mg/ml); the running buffer was TAE (40 mM Tris acetate [pH 8.0], 1 mM EDTA), and a 100-bp DNA ladder was used as a molecular size marker (Promega). DNA bands were visualized by UV transillumination.
Choice of loci for C. parapsilosis MLST.
Eleven gene fragments were chosen in an attempt to set up an MLST scheme for C. parapsilosis (Table 2). One fragment was obtained with primers described for the C. albicans CaSYA1 gene encoding alanyl-tRNA synthetase (2), which showed cross-reactivity with other Candida species; the remainder were C. parapsilosis genes (GenBank), and they included one example of a mitochondrial gene (cytochrome oxidase subunit 3 [COX3]). Primers were designed to amplify gene fragments of 500 to 750 bp and are also described in Table 2, with the corresponding PCR product sizes.
TABLE 2.
List of gene fragments and primer details
| Gene | Derivation | GenBank accession no. | Primer sequencesd | Amplicon size (bp) | Sequence start
|
Sequenced fragment (bp) | PCR product obtained with group(s)a | |
|---|---|---|---|---|---|---|---|---|
| 5′ | 3′ | |||||||
| ACPL | Pro-acid protease | Z11918 | Fwd, 5′-CTCATTCAAGTCATTAGGCTC-3′ [A1F] | 660 | AGAGTTCTT | CCTTCGAC | 548 | I |
| Rev, 5′-GCAAGCATTGGCAGTACT-3′ [A1R] | ||||||||
| ACPR | Pro-acid protease | Z11919 | Fwd, 5′-GGAGCTGCTTTCACTATTCG-3′ [A2F] | 666 | AAGATACC | AAGATACC | 562 | I |
| Rev, 5′-GACAAGTACCATCACCATTT-3′ [A2R] | ||||||||
| COX3b | Cytochrome oxidase subunit 3 | X75679 | Fwd, 5′-AGGAGATCATACTATTGCAG-3′ [C1F] | 511 | TTTGTAGTTA | ATTATGCTTA | 380 | I, II, III |
| Rev, 5′-TAGTTGTTTCAGCACCA-3′ [C1R] | ||||||||
| GAL1 | Galactokinase | Y14704 | Fwd, 5′-CCCAAGTTTAACGATTTGTC-3′ [G1F] | 742 | TCGAATGGA | ACCTTGAC | 624 | I, III* |
| Rev, 5′-TAGTGGATCGGAGCCGTT-3′ [G1R] | ||||||||
| L1A1 | Cytochrome P450 demethylase gene | AF019902 | Fwd, 5′-TGCTCAATTGTATGCTGA-3′ [LIF] | 675 | TTTTGTTTTC | AGATTTTAC | 553 | I, II |
| Rev, 5′-CCATAATCAACTTCACCTGC-3′ [L1R] | ||||||||
| LIP2 | Lipase 2 | AJ320260 | Fwd, 5′-GGATTGGGGACGTTTCTT-3′ [L2F] | 618 | ACCTTGTGG | GCAAATAG | 514 | I |
| Rev, 5′-CGTGTTTTTGCAAGTTG-3′ [L2R] | ||||||||
| SADH | Secondary alcohol dehydrogenase | AB010636 | Fwd, 5′-GTTGATGCTGTTGGATTGT-3′ [S1F] | 716 | GGATTGTGGT | TTGATTT | 546 | I, II, III |
| Rev, 5′-CAATGCCAAATCTCCCAA-3′ [SIR] | ||||||||
| SAPP3 | Secreted aspartic protease 3 | AF339513 | Fwd, 5′-TAATTGCTGTCTTCACTGGA-3′ [S2F] | 537 | GTGTTTGCT | TACTAACC | 439 | I, II*, III* |
| Rev, 5′-AGACCCATGACCCCTTG-3′ [S2R] | ||||||||
| SYA1c | Putative alanyl-tRNA synthetase | Fwd, 5′-AGAAGAATTGTTGCTGTTACTG-3′ [S3F] | 543 | AATATGAACA | TTGATGC | 430 | I, II, III | |
| Rev, 5′-GTTACCTTTACCACCAGCTTT-3′ [S3R] | ||||||||
| TOP2 | Type II DNA topoisomerase | AB049144 | Fwd, 5′-GTGAATGCCGCTGATAAC-3′ [T1F] | 732 | TAACGTTAT | GCTCCCAA | 540 | I |
| Rev, 5′-GCCACTTCCCATCTTTC-3′ [T1R] | ||||||||
| URA3 | Orotidine-5′-phosphate decarboxylase | X99635 | Fwd, 5′-AGACTTGGGTATTACGTTGT-3′ [U1F] | 727 | ATTTTGGCT | GATTTAT | 587 | I |
| Rev, 5′-CAGGAGTCATGATTACCC-3′ [U1R] | ||||||||
Asterisked products differed in size between the groups.
Mitochondrial gene.
Primers described for the C. albicans CaSYA1 gene, encoding alanyl-tRNA synthetase.
Fwd, forward; Rev, reverse. Laboratory reference names are given in brackets.
Amplification and nucleotide sequence determination.
PCRs were used to amplify the gene fragments listed in Table 2. Conditions were as described above, but PCR volumes contained 10 μM concentrations of each forward and reverse primer. The amplification conditions were as follows: a first cycle of denaturation for 7 min at 94°C, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 50°C for 1 min, and elongation at 74°C for 1 min, with a final extension step of 10 min at 74°C. The amplified products were precipitated in microdilution plates. Briefly, 60 μl of a 20% polyethylene glycol (PEG; Sigma)-2.5 M NaCl solution was added to each well containing 40 μl of PCR product. The microdilution plate was then sealed, vortexed, incubated at room temperature for 30 min, and centrifuged for 1 h at 2,250 × g (4°C). The supernatant was discarded, and the plate was inverted onto a piece of 3-mm chromatography paper and centrifuged again at 500 × g for 1 min to remove any residual PEG from the wells. Pellets were washed with 150 μl of 70% ethanol, precipitated as described above, and resuspended in 60 μl of sterile water. Both strands of purified gene fragments were sequenced on an ABI 3700 DNA analyzer (ABI, Foster City, Iowa) with a 2.5 μM concentration of the same primers that were used in the PCR step. The sequence data were coupled with DNASTAR software. Heterozygosities were defined by the presence of two coincident peaks in the forward and reverse sequence chromatograms. The one-letter code for nucleotides from the International Union of Pure and Applied Chemistry (IUPAC) nomenclature was used to define results.
BanI digestion of SADH fragment.
The SADH fragment was amplified in C. parapsilosis isolates, as well as in nine different Candida species (C. albicans, C. dubliniensis, C. famata, C. glabrata, C. guilliermondii, C. kefyr, C. krusei, C. lusitaniae, and C. tropicalis) and in Saccharomyces cerevisiae. PCR products were purified with a PCR purification kit (Qiagen), and 20 μl of PCR product obtained from C. parapsilosis isolates belonging to group I, II, and III were digested with BanI restriction enzyme. Sequence analysis had shown that the polymorphisms observed within the three groups in the SADH sequence result in the presence of one BanI restriction site in C. parapsilosis group I isolates, three cut sites in group III isolates, and none in group II isolates. The purified SADH fragments from the three groups were digested for 90 min with BanI (New England Biolabs) in a 40-μl reaction volume containing 20 μl of the PCR product, 4 ml of 10× buffer 4 (supplied with the enzyme), and 2 μl of the 20 U of BanI/μl. Digestion products were loaded onto a 2% agarose gel containing ethidium bromide (0.05 μg/ml), ΤΑΕ was used as the running buffer, and a 100-bp DNA ladder was used as a molecular size marker (Promega). DNA bands were visualized by UV transillumination.
Sequence analysis of ITS and D1-D2 large subunit rRNA regions.
Sequencing of the 5.8S RNA gene and the adjacent ITS1 and ITS2, as well as the variable D1-D2 region of the large ribosomal subunit, was carried out as described above. Primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) were used to amplify the ITS region as described by White et al. (28). A PCR product of 600 bp was obtained from C. parapsilosis isolates with the universal primers 11 (5′-GCATATCAATAAGCGGAGGAAAAG-3′) and 12 (5′-GGTCCGTGTTTCAAGACG-3′) specific for the D1-D2 large-subunit rRNA region (8). Amplification of both regions was carried out under the following conditions: denaturation at 94°C for 2 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 90 s, and elongation at 72°C for 75 s, with a final extension step of 10 min at 72°C. Amplified DNA was resolved by electrophoresis in a 1% agarose gel, stained with ethidium bromide, and purified and sequenced as described above. The D1-D2 large-subunit rRNA sequences were also determined for single isolates of C. albicans and C. tropicalis for purposes of comparison. ITS1 sequences from 15 other Candida species and S. cerevisiae (GenBank) were also compared to the ones obtained from C. parapsilosis groups I, II, and III.
Sequence analysis of other genes.
To determine whether the sequence differences between C. parapsilosis groups I, II, and III indicated positive or neutral selection, sequences of COX3, SADH, and SYA1 (for all three subgroups) and of L1A1 (for groups I and II) were assessed for the ratios of nonsynonymous to synonymous amino acid changes (DN/DS) by the modified Nei-Gojobori method with 500 bootstrap replicates, conducted with the MEGA 2.1 software package (13). The neighbor-joining method was used to generate a dendrogram based on sequence polymorphisms in COX3, SADH, and SYA1 for all isolates studied; the analysis, which utilized the Nei-Gojobori p-distance method with 1,000 bootstrap replicates, was also conducted with the MEGA 2.1 software package (13).
ITS1 sequences for 16 different yeast species were uploaded from GenBankdatabase (http://www.psc.edu/general/software/packages/GenBank/GenBank.html). These 16 sequences, together with those determined for C. parapsilosis groups I to III in the present study were submitted to the multiple alignment procedure with the on-line program CLUSTAL W (http://www.ebi.ac.uk/clustalw/#). The aligned sequences were then analyzed with the MEGA 2.1 software package (13). An evolutionary tree was generated based on p-distance with the neighbor-joining method. The validity of the branches was ascertained with 1,000 bootstrap replicates.
Nucleotide sequence accession numbers.
The sequences determined in the present study were submitted to GenBank. The accession numbers are as follows: COX3, C. orthopsilosis (AJ698046) and C. metapsilosis (AJ698116); ITS region, C. parapsilosis (AJ635316), C. orthopsilosis (AJ698048), and C. metapsilosis (AJ698049); L1A1, C. parapsilosis (AJ698042) and C. orthopsilosis (AJ698043); SADH, C. orthopsilosis (AJ698047) and C. metapsilosis (AJ698115); SYA1, C. parapsilosis (AJ635315), C. orthopsilosis (AJ698044), and C. metapsilosis (AJ698045).
RESULTS
Group assignment of test isolates.
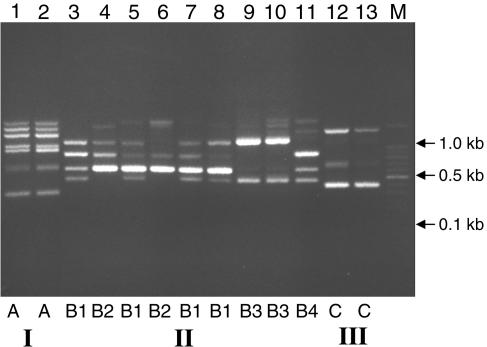
Of the 32 C. parapsilosis isolates used in the present study, 21 were assigned to the most commonly encountered subtype (group I), 9 were assigned to group II, and 2 were assigned to group III, as confirmed by ITS region sequencing (Table 1). All of the isolates were screened by RAPD testing, and the primer RPO2 was used under low-stringency conditions to amplify genomic DNA. The amplification profiles obtained showed a high genomic homogeneity within group I isolates, with just one RAPD pattern obtained from the 21 isolates (Fig. 1, profile A); the two group III isolates also gave an identical profile, as shown in Fig. 1 (profile C). In contrast, group II isolates exhibited a high level of genetic heterogeneity as shown by the variability observed in their RAPD patterns, with four different profiles identified within nine isolates (Fig. 1, profiles B1 to B4).
FIG. 1.
RAPD patterns produced for 32 C. parapsilosis strains with the primer RPO2. Lanes 1 and 2, RAPD representative profiles obtained for all C. parapsilosis group I isolates (profile A); lanes 3 to 11, C. parapsilosis group II isolates (B1 to B4 profiles); lanes 12 and 13, C. parapsilosis group III isolates (profile C). M, 100-bp ladder.
PCR products and sequence variations for multiple gene fragments.
A total of 11 gene fragments, including one mitochondrial gene (COX3), were amplified in the course of our effort to distinguish the 32 C. parapsilosis isolates as individual strains by MLST (Table 2). Although all group I isolates gave a PCR product of the expected size with all 11 genes, no product could be obtained from group II isolates with the ACPL, ACPR, GAL1, LIP2, TOP2, and URA3 genes (Table 2). In addition, group II isolates yielded a PCR product of 1.5 kb for the SAPP3 gene, when the predicted size of the amplicon was 537 bp (Table 2). Both group III isolates gave no amplification product with ACPL, ACPR, L1A1, LIP2, TOP2, and URA3 genes. They gave larger amplicons with the GAL1 and SAPP3 genes (Table 2). Analysis of the sequences obtained for the 11 gene fragments from 21 group I isolates showed no base polymorphisms suitable for MLST, despite our sequencing of more than 7.5 kb of the C. parapsilosis genome. Only two polymorphic sites could be identified among the 21 aligned sequences of group I isolates: one in the L1A1 fragment (nucleotide position 498 [G or T]) and the other in the ACPL sequence (nucleotide position 428 [Y, C, or T]). This finding ruled out the possibility of creating a practical MLST scheme for C. parapsilosis group I isolates on the basis of the genes we sequenced. However, for three of the four gene fragments that could be amplified in more than one group (L1A1, SADH, and SYA1; Table 2), fixed sequence differences were detected at the group level between all isolates from each of the three groups. As shown in Table 3, intergroup DNA sequence similarities ranged from 82.5 to 86.6%, resulting in a large number of nonsynonymous changes at the amino acid level. Statistical analysis of the DN/DS ratios showed highly significant results in favor of neutral selection (P ≤ 0.006 for all paired sequences except for COX3, group II versus group III sequences, where P = 0.483).
TABLE 3.
Sequence similarity among four genes of C. parapsilosis groups I, II, and III
| Gene | Comparison | Sequence similarity (%)a | No. of gaps | No. of nucleotide polymorphisms/ total no. | No. of non- synonymous changes |
|---|---|---|---|---|---|
| COX3b | I vs II | 95.6 | 17/389 | 3 | |
| II vs III | 98.5 | 6/389 | 1 | ||
| I vs III | 97.2 | 11/389 | 2 | ||
| L1A1 | I vs II | 86.6 | 74/553 | 3 | |
| II vs III | |||||
| I vs III | |||||
| SADH | I vs II | 85.9 | 77/546 | 13 | |
| II vs III | 84.4 | 85/546 | 16 | ||
| I vs III | 82.8 | 94/546 | 23 | ||
| SYA1c | I vs II | 84.9 | 6 | 59/430 | 10 |
| II vs III | 84.4 | 3 | 64/424 | 10 | |
| I vs III | 82.5 | 9 | 66/428 | 13 |
That is, the number of matched bases in both sequences referred to the total number of bases in both sequences.
Mitochondrial gene.
Primers described for C. albicans CaSYA1 gene encoding alanyl-tRNA synthetase.
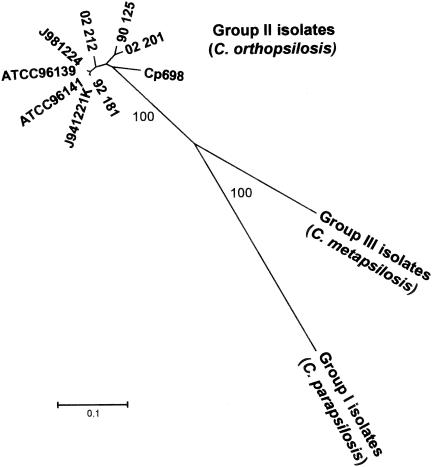
At a small number of sites in each of the three gene sequences the presence of three different bases could be detected in the same position between two or all three of the groups. In the L1A1 sequence, at nucleotide position 498, a G or a T was consistently found among group I isolates, whereas group II isolates had an A in the same position. In the SYA1 fragment, at least five of the variable sites showed the same phenomenon (positions 26 [A+C+T], 101 [A+C+T], 211 [A+C+T], 299 [A+T+C], and 311 [G+T+A]). The SADH sequence also revealed six polymorphic sites with three different nucleotides (position 72 [T+C+A], 109 [T+A+G], 127 [G+T+A], 169 [A+C+G], 268 [A+C+T], and 406 [C+G+T]). Fewer DNA sequence differences were detected among isolates from the three groups with the mitochondrial gene COX3, as reflected by sequence similarity values ranging from 95.6 to 97.5% (Table 3). However, even within the COX3 sequences, three different nucleotides were found at one polymorphic site (nucleotide position 117 [T+C+A]). In addition, a high number of changes at the protein level were detected between the amino acid sequence of group III isolates compared to group I or II sequences (from 1 to 23 nonsynonymous changes; Table 3). These findings, coupled with the absence of PCR product or amplicons of different size, indicate that differences between the three groups were at the level of different species rather than different groups in the same species. Apart from the mitochondrial gene COX1, only SADH and SYA1 gave sequence information for isolates of all three C. parapsilosis groups. In the SADH gene there were 17 alleles at which the sequence differed consistently between groups for all isolates in each group. Three alleles in SYA1 similarly showed group-specific sequence differences. The unrooted neighbor-joining tree (Fig. 2) shows the sequence differences clearly distinguished the three C. parapsilosis groups, with 100% bootstrap values. (Analysis of the same data by the unweighted pair group method with arithmetic mean [UPGMA] approach and by split-tree analysis generated dendrograms indistinguishable from Fig. 2.)
FIG. 2.
Unrooted radial tree showing nearest-neighbor clustering of sequence polymorphism data for COX3, SADH, and SYA1 genes. All 32 test isolates were sequenced, but sequences were identical for the 21 group I isolates and the 2 group III isolates, which are therefore shown as single entities. Bootstrap values are indicated at the nodes.
Occasional, unequivocal heterozygosities at some sites in the group I and II sequences pointed to these isolates having a diploid (or aneuploid) genome, such as C. albicans and C. tropicalis. For example, one of the rare polymorphic sites found for group I isolates showed heterozygosity at position 428 in the ACPL gene, and eight heterozygous sites were found in SYA1 for group II isolates (all polymorphic sites). The two group III isolates always showed identical sequences for all of the fragments that could be amplified. For SYA1 the group III sequences contained 15 heterozygous loci.
Genetic diversity of group II isolates at multiple loci.
The genetic variability exhibited by group II isolates when screened by RAPD was confirmed by differences in their DNA sequences. Two polymorphic sites were detected in the L1A1 sequence of nine group II isolates, which led to the identification of three different genotypes (Table 4). For SADH 11 polymorphic sites defined four genotypes, for SYA1 10 variable sites defined four genotypes, and COX3 gave three different genotypes with only two polymorphic sites. The combination of the genotypes illustrates that, with just four gene fragments sequenced, it was possible to distinguish eight out of the nine group II isolates as unique diploid sequence types (Table 4). It is worth noting that the two isolates that were indistinguishable by MLST also shared a unique RAPD profile (Fig. 1, profile B3, and Table 1). Although group II isolates could be differentiated as individual isolates on the basis of nucleotide polymorphisms, these isolates nevertheless clustered together as a group well differentiated from group I and group III isolates by neighbor-joining analysis (Fig. 2).
TABLE 4.
Polymorphic nucleotides in gene fragments COX3, L1A1, SADH and SYA1 for C. parapsilosis group II strainsa
| Strain no. | Nucleotide(s) and genotype in:
|
|||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
COX3
|
L1A1
|
SADH
|
SYA1
|
|||||||||||||||||||||||||||
| 117 | 360 | G | 206 | 496 | G | 72 | 79 | 109 | 127 | 169 | 250 | 268 | 295 | 352 | 406 | 472 | G | 26 | 48 | 59 | 92 | 158 | 211 | 215 | 239 | 287 | 323 | G | DSTb | |
| J981224 | A | A | 1 | T | T | 1 | C | T | T | T | A | C | C | T | G | G | G | 1 | Y | R | Y | R | Y | T | Y | C | Y | M | 1 | 1 |
| 90-125 | - | - | 1 | - | - | 1 | A | C | G | A | G | T | T | C | A | T | A | 2 | T | A | C | A | T | - | C | C | T | A | 2 | 2 |
| 02-212 | C | T | 2 | C | C | 2 | - | C | - | - | - | - | - | - | - | - | - | 3 | C | G | T | A | T | A | C | T | T | A | 3 | 3 |
| 92-181 | C | - | 3 | - | - | 1 | - | C | - | - | - | - | - | - | - | - | - | 3 | - | - | - | - | - | - | - | - | - | - | 1 | 4 |
| 02-201 | C | - | 3 | C | - | 3 | A | C | G | A | G | T | C | T | A | T | A | 4 | T | A | C | A | T | A | C | T | T | A | 4 | 5 |
| 82-33/698 | C | T | 2 | C | C | 2 | - | C | - | - | - | - | - | - | - | - | - | 3 | C | G | T | A | T | A | C | T | T | A | 3 | 3 |
| J941221K | - | - | 1 | - | - | 1 | - | C | - | - | - | - | - | - | - | - | - | 3 | - | - | - | - | - | - | - | - | - | - | 1 | 6 |
| ATCC 96139 | C | A | 3 | - | - | 1 | - | - | - | - | - | - | - | - | - | - | - | 1 | - | - | - | - | - | - | - | - | - | - | 1 | 7 |
| ATCC 96141 | C | A | 3 | C | - | 3 | - | C | - | - | - | - | - | - | - | - | - | 3 | C | - | - | - | - | - | - | - | - | - | 5 | 8 |
Numbers in column subheadings indicate the positions of each polymorphic site in the DNA sequence. The first sequence shown (strain J981224) is the complete sequence for MLST diploid sequence type (DST) 1. For the remainder of the table, only sites that differ from those of DST 1 are shown; sites that share the same nucleotide as DST1 are indicated by dashes. IUPAC one-letter codes for single and paired nucleotides are used throughout. G, genotype.
DST, diploid sequence type, i.e., The composite of four genotypes.
Restriction analysis of SADH PCR product.
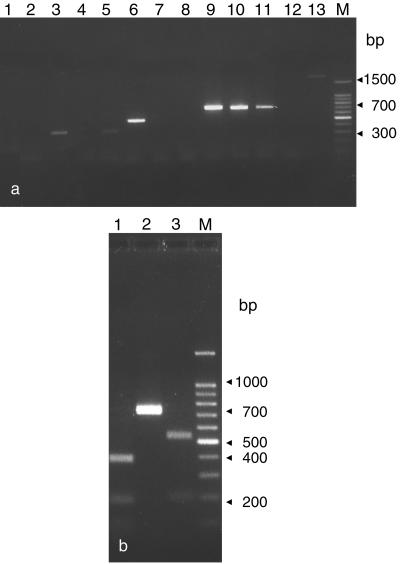
No amplification was obtained with SADH primers (Table 2) with five of the nine Candida species tested (C. dubliniensis, C. glabrata, C. krusei, C. lusitaniae, and C. tropicalis; Fig. 3a). C. albicans gave two faint bands of approximately 300 and 600 bp and a PCR product different from the predicted 716 bp size was also obtained with C. famata (350 bp), C. guilliermondii (350 bp), C. kefyr (500 bp) and S. cerevisiae (1,600 bp) (Fig. 3a). Only C. parapsilosis isolates belonging to groups I, II, and III gave a PCR product of 716 bp (Fig. 3a). Sequence analysis showed that the SADH DNA from the three groups had different restriction maps of the amplified fragment. Although SADH sequence from group I isolates had one predicted BanI site in position 196, three BanI cut sites were found in the sequence obtained from group III isolates (positions 96, 469, and 529), whereas no BanI site was found in SADH sequence from group II isolates. As shown in Fig. 3b, electrophoresis of the BanI digest from group I isolates revealed two bands (521 and 196 bp) as expected, since only one BanI site was found in the SADH sequence. The presence of three restriction sites in the PCR product obtained from group III isolates gave four bands of 370, 188, 93, and 60 bp (Fig. 3b), whereas just one band, the uncut PCR product, was obtained from the digestion of SADH amplicon obtained from group II isolates, where no BanI sites had been previously identified (Fig. 3b).
FIG. 3.
(a) SADH fragment amplification (expected PCR product of 716 bp) in different Candida species: lane 1, C. albicans; lane 2, C. dubliniensis; lane 3, C. famata; lane 4, C. glabrata; lane 5, C. guilliermondii; lane 6, C. kefyr; lane 7, C. krusei; lane 8, C. lusitaniae; lane 9, C. metapsilosis; lane 10, C. orthopsilosis; lane 11, C. parapsilosis; lane 12, C. tropicalis. Lane 13, S. cerevisiae; lane M, 100-bp ladder. (b) BanI restriction digestion of SADH-PCR product obtained from representative C. metapsilosis (lane 1), C. orthopsilosis (lane 2), and C. parapsilosis (lane 3) strain types. Lane M, 100-bp ladder.
Analysis of rRNA and ITS regions.
All isolates within each group (I, II, and III) gave an identical sequence at the variable region (D1-D2) of the large ribosomal subunit. The variable sequences differed between isolates in groups I, II, and III, but the differences were minor, with percent similarities all higher than 98% (99.0% [group I versus group II], 98.8% [group I versus group III], and 99.2% [group II versus group III]). The D1-D2 large subunit rRNA sequences for C. parapsilosis group I isolates was 99.0% similar to the C. albicans sequence and 95.0% similar to the C. tropicalis sequence.
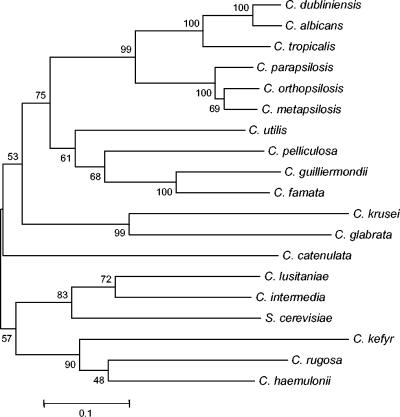
The 5.8S ribosomal gene DNA sequence of C. parapsilosis isolates from all three groups was 100% identical, and only minor differences were observed in ITS2 sequence between the three groups (98.1% [group I versus group II], 95.6% [group I versus group III], and 97.5% [group II versus group III]). However, considerable dissimilarities were found in the ITS1 sequences, as has been previously observed (17). Our data showed ITS1 sequence similarity of 82.5% (group I versus group III), 88.1% (group I versus group II), and 86.1% (group II versus group III). No ITS1 sequence differences were found between isolates within each group. For comparison, ITS1 sequences from GenBank for 15 Candida other species and S. cerevisiae were used to compile a genetic distance matrix. An evolutionary tree was generated based on the genetic distance matrix with the neighbor-joining method (Fig. 4), showing that group II and III isolates clustered separately from C. parapsilosis. The level of difference between the ITS1 sequences for the three C. parapsilosis groups was similar to that between C. albicans and C. dubliniensis. The high bootstrap values for the ITS1 aligned sequence differences validate the strong separation of C. parapsilosis groups I to III from each other and from other yeast species (Fig. 4).
FIG. 4.
Phylogenetic neighbor-joining tree generated from a genetic similarity matrix based on comparison of ITS1 sequences. Numbers at each node indicate similarity percentage.
Based on the extensive differences between the three subgroups of C. parapsilosis, we propose that the name C. parapsilosis be reserved for group I isolates only, and we propose two new species in place of the former groups II and III as follows.
Candida orthopsilosis (Tavanti, Davidson, Gow, Maiden, et Odds, sp. nov.).
Haec species, olim appellata C. parapsilosis Grex II (16, 17), a C. parapsilosi et C. metapsilosi distinguenda non forma sed nucleotidis. Infra illa nucleotida describuntur secundum genum, positionem in geno, et classem in hac specie institutam. Synthetasum, ut videtur, ARN translationis alanylici: 101, C; 299, T; 311, T. Dehydrogenasum alcoholis secundarium: 10, A; 28, C; 58, T; 76, T; 128, T; 149, A; 170, A; 197, T; 260, T; 275, T; 341, A; 350, A; 422, T; 431, G; 512, G; 518, C; 524, G. Opus nullum efficit reactio polymerasi serialis usu parium ordinum nucleotidorum initialium A1F et A1R, A2F et A2R, G1F et G1R, L2F et L2R, T1F et T1R, et U1F et U1R.
(i) Description.
Formerly known as C. parapsilosis group II (16, 17), C. orthopsilosis (orthopsilosis, indicating affinity with C. parapsilosis and C. metapsilosis) is morphologically indistinguishable from C. parapsilosis and C. metapsilosis. The species is diagnosed based on the following nucleotide characteristics, given as the gene, the nucleotide position in the gene, and the nucleotide fixed in C. orthopsilosis: SYA1 (putative alanyl-tRNA synthetase), 101, C; 299, T; and 311, T; and SADH (secondary alcohol dehydrogenase), 10, A; 28, C; 58, T; 76, T; 128, T; 149, A; 170, A; 197, T; 260, T; 275, T; 341, A; 350, A; 422, T; 431, G; 512, G; 518, C; and 524, G. No PCR product formed with primer pairs A1F and A1R, A2F and A2R, G1F and G1R, L2F and L2R, T1F and T1R, and U1F and U1R.
(ii) Holotype.
Strain ATCC 96139 has all of the above-mentioned characteristics. It was originally isolated from the tip of a catheter used for monitoring central venous pressure in San Antonio.
Candida metapsilosis (Tavanti, Davidson, Gow, Maiden et Odds, sp. nov.).
Haec species, olim appellata C. parapsilosis Grex III (16, 17), a C. parapsilosi et C. orthopsilosi distinguenda non forma sed nucleotidis. Infra illa nucleotida describuntur secundum genum, positionem in geno, et classem in hac specie institutam. Synthetasum, ut videtur, ARN translationis alanylici: 101, T; 299, C; 311, A. Dehydrogenasum alcoholis secundarium: 10, G; 28, T; 58, A; 76, A; 128, C; 149, T; 170, G; 197, C; 260, C; 275, G; 341, T; 350, G; 422, C; 431, C; 512, A; 518, A; 524, T. Opus nullum efficit reactio polymerasi serialis usu parium ordinum nucleotidorum initialium A1F et A1R, A2F et A2R, L1F et L1R, L2F et L2R, T1F et T1R, U1F et U1R.
(i) Description.
Formerly known as C. parapsilosis group III (16, 17), C. metapsilosis (metapsilosis, indicating affinity with C. parapsilosis and C. orthopsilosis) is morphologically indistinguishable from C. parapsilosis and C. orthopsilosis. The species is diagnosed on the basis of the following DNA-based characteristics, given as nucleotide position and base specific to C. metapsilosis: SYA1 (putative alanyl tRNA synthetase), 101, T; 299, C; and 311, A; and SADH (secondary alcohol dehydrogenase), 10, G; 28, T; 58, A; 76, A; 128, C; 149, T; 170, G; 197, C; 260, C; 275, G; 341, T; 350, G; 422, C; 431, C; 512, A; 518, and A; 524, T. No PCR product formed with primer pairs A1F and A1R, A2F and A2R, L1F and L1R, L2F and L2R, T1F and T1R, and U1F and U1R.
(ii) Holotype.
Strain ATCC 96144 has all of the above-mentioned characteristics. It was originally isolated from a hand sample in Tacoma, Wash. It is now part of the American Type Culture Collection.
Phenotypic tests.
Several approaches to phenotypic characterization of the 32 isolates in the present study revealed no clear-cut differences between the three species. Sugar assimilation profiles evaluated by API 32 C were essentially the same for all of the isolates studied. Isolates of all groups formed white to pale colonies on CHROMagar Candida, and isolates from all species grew equally well at 30 and 37°C but failed to grow at 42°C. Antifungal susceptibility testing with amphotericin B, flucytosine, fluconazole, itraconazole, ketoconazole, and miconazole revealed no gross differences between the species: all isolates were susceptible to the agents tested. Tests for biofilm formation in vitro also showed no statistically significant differences between the three species (details not shown).
DISCUSSION
The dividing line for microorganisms that demarcates “species” is, and always has been, a controversial matter. Molecular phylogenetic analyses based on DNA sequence data have greatly altered many traditional concepts of species delineation. Among fungal pathogens, molecular phylogenies form the basis for differentiating C. dubliniensis from C. albicans (25) and Coccidioides posadasii from Coccidioides immitis (9). In the latter case, phenotypic differentiation of the species has not yet been achieved and, in the former, it has taken many years to establish reliable phenotypic tests to identify the species (6).
We consider the differences in DNA sequence between several genes in C. parapsilosis groups I, II, and III to be too great for these groups to continue to be designated as subspecies, and we propose the names C. orthopsilosis and C. metapsilosis, respectively, for groups II and group III. The term “psilosis” means “sprue” (tropical diarrhea), and the species epithet “parapsilosis” was coined by Ashford in 1928 to describe the novel yeast he isolated from diarrhea in Puerto Rico (1). We are proposing the new species names in an ironic parallel to the manner in which the prefixes “ortho” (originally meaning “right” or “correct”) and “meta” (originally meaning “with” or “after”) are used by chemists, along with “para” to denote different positions in the same six-membered ring. We have refrained from proposing more individualized species names in recognition of the phenotypic indistinguishability of the three species in the tests we have used.
That C. metapsilosis, C. orthopsilosis, and C. parapsilosis merit species status at the genomic level seems to us to be unequivocally proved. The level of dissimilarity in the ITS1 sequences is of the same order as that between C. albicans and C. dubliniensis (Fig. 4). Our ITS1 sequence data for the three species match the earlier results of Lin et al. (17), who previously considered the differences were compatible with the delineation of species rather than subgroups. Although SADH and SYA1 are not highly conserved genes, the finding of consistent sequence similarities well below 90% between these genes from C. parapsilosis, C. metapsilosis, and C. orthopsilosis (Table 3) confirms a fundamental lack of genomic conformity appropriate to a single species, and the unrooted UPGMA tree analysis (Fig. 2) reinforces the considerable intergroup diversity. The fact that for 7 of 11 genes we attempted to sequence we obtained no PCR product, or a product of a consistently different size, from C. metapsilosis and C. orthopsilosis with primers from C. parapsilosis lends further weight to the considerable dissimilarities between the genomes of the three species. C. metapsilosis and C. orthopsilosis as here defined react less strongly with oligonucleotide repetitive element Cp13 (7). DNA reassociation studies by Roy and Meyer indicated species-level differences in DNA relatedness between the three “C. parapsilosis” groups (21). Kurtzman and Robnett considered a mere six nucleotide sequence difference in the rRNA gene to be sufficient to demarcate a group II isolate as a new species (14, 15). Although the linearity versus circularity of mitochondrial DNA (mtDNA) is not a taxonomic criterion, it is interesting that most group II isolates have a circular mtDNA, whereas the mtDNA is linear in most isolates belonging to groups I and III (22). Our own data are a clear addition to previous suggestions that the “C. parapsilosis complex” requires redelineation. There are too many differences between the genes of these yeasts for them to be regarded as mere variants.
Evolutionary significance of the three related species.
We embarked on the present study to devise an MLST system for C. parapsilosis and not to produce evidence for novel yeast species. For C. parapsilosis (as here redefined to refer only to group I isolates) the lack of sequence variation appears to make it unlikely that strains can be typed by sequence differences. We found only two polymorphic nucleotide sites among approximately 7.5 kb sequenced. Fundyga et al. recently reported a similar finding: a search for single nucleotide polymorphisms revealed just 4 among more than 36 kb sequenced (10). Although C. parapsilosis isolates can be distinguished by differences in numbers and positions of the repetitive element Cp13 (7), the lack of nucleotide sequence diversity strongly suggests that C. parapsilosis is highly clonal and therefore has emerged as a species very recently on the scale of evolutionary time, probably within the past one million years (10). It is not inconceivable that the species has coevolved with the migrations of early humans. Its global distribution and the much higher frequency of its isolation from clinical material than C. orthopsilosis or C. metapsilosis (17) indicate a rapid geographic spread of C. parapsilosis, perhaps because it is more easily transmitted from person to person or better adapted to a human commensal environment than its two relatives. C. parapsilosis has recently been shown to be predominantly aneuploid rather than strictly haploid or diploid (10). Our finding of occasional heterozygosities in C. parapsilosis gene sequences is compatible with this conclusion. The prevalence of heterozygosity in C. orthopsilosis and C. metapsilosis sequences was much higher than in C. parapsilosis and may make a prima facie case for diploidy in these species.
For C. orthopsilosis we found considerable nucleotide sequence diversity among the nine isolates we studied (Table 4 and Fig. 2). These results provide a measure of the genetic diversity at multiple loci within a small number of epidemiologically unrelated isolates and suggest that an MLST-like approach is suitable for application to C. orthopsilosis isolates. Since we studied only two C. metapsilosis isolates, we cannot yet comment on the possible level of genetic diversity within this species. However, the sequence diversity between C. orthopsilosis isolates and its near absence in C. parapsilosis generates a hypothesis in which C. orthopsilosis is a possible ancestor of C. parapsilosis. The ancestral position of C. metapsilosis in the evolution of the three species is as yet unknown, but its extreme rarity among clinical isolates, evidenced by the paucity of C. parapsilosis group III isolates in culture collections and the comment of Clark et al. that the species has not yet been isolated from clinical material at the Centers for Disease Control and Prevention (5) raises the possibility that C. metapsilosis might be the progenitor, associated with a nonmammalian environment, of both the related species C. orthopsilosis and C. parapsilosis, which evolved by adaptation to commensalism in human or other mammalian niches.
Acknowledgments
This study was supported by grant 069615 from the Wellcome Trust.
We are grateful to Mark A. Garland for the Latin translation of the species descriptions.
REFERENCES
- 1.Ashford, B. K. 1928. Certain conditions of the gastrointestinal tract in Puerto Rico and their relation to tropical sprue. Am. J. Trop. Med. Hyg. 8:507-538. [Google Scholar]
- 2.Bougnoux, M.-E., S. Morand, and C. d'Enfert. 2002. Usefulness of multilocus sequence typing for characterization of clinical isolates of Candida albicans. J. Clin. Microbiol. 40:1290-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bougnoux, M. E., A. Tavanti, C. Bouchier, N. A. R. Gow, A. Magnier, A. D. Davidson, M. C. J. Maiden, C. d'Enfert, and F. C. Odds. 2003. Collaborative consensus for optimized multilocus sequence typing of Candida albicans. J. Clin. Microbiol. 41:5265-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cassone, A., F. De Bernardis, E. Pontieri, G. Carruba, C. Girmenia, P. Martino, M. Fernandez-Rodriguez, G. Quindos, and J. Ponton. 1995. Biotype diversity of Candida parapsilosis and its relationship to the clinical source and experimental pathogenicity. J. Infect. Dis. 171:967-975. [DOI] [PubMed] [Google Scholar]
- 5.Clark, T. A., J. Morgan, M. Brandt, T. Lott, S. Slavinski, S. Taylor, H. Flowers, S. Fridkin, and R. Hajjeh. 2002. Hospital outbreak of Candida parapsilosis bloodstream infections-Mississippi, 2001. Program Abstr. 42nd Intersci. Conf. Antimicrob. Agents Chemother., abstr. M-880.
- 6.Ellepola, A. N. B., S. F. Hurst, C. M. Elie, and C. J. Morrison. 2003. Rapid and unequivocal differentiation of Candida dubliniensis from other Candida species using species-specific DNA probes: comparison with phenotypic identification methods. Oral Microbiol. Immunol. 18:379-388. [DOI] [PubMed] [Google Scholar]
- 7.Enger, L., S. Joly, C. Pujol, P. Simonson, M. Pfaller, and D. R. Soll. 2001. Cloning and characterization of a complex DNA fingerprinting probe for Candida parapsilosis. J. Clin. Microbiol. 39:658-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fell, J. W. 1993. Rapid identification of yeast species using three primers in a polymerase chain reaction. Mol. Mar. Biol. Biotechnol. 2:174-180. [PubMed] [Google Scholar]
- 9.Fisher, M. C., G. L. Koenig, T. J. White, and J. T. Taylor. 2002. Molecular and phenotypic description of Coccidioides posadasii sp. nov., previously recognized as the non-California population of Coccidioides immitis. Mycologia 94:73-84. [PubMed] [Google Scholar]
- 10.Fundyga, R. E., R. J. Kuykendall, W. Lee-Yang, and T. J. Lott. 2004. Evidence for aneuploidy and recombination in the human commensal yeast Candida parapsilosis. Infect. Genet. Evol. 4:37-43. [DOI] [PubMed] [Google Scholar]
- 11.Kato, M., M. Ozeki, A. Kikuchi, and T. Kanbe. 2001. Phylogenetic relationship and mode of evolution of yeast DNA topoisomerase II gene in the pathogenic Candida species. Gene 272:275-281. [DOI] [PubMed] [Google Scholar]
- 12.Krcmery, V., and A. J. Barnes. 2002. Non-albicans Candida spp. causing fungaemia: pathogenicity and antifungal resistance. J. Hosp. Infect. 50:243-260. [DOI] [PubMed] [Google Scholar]
- 13.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 14.Kurtzman, C. P., and C. J. Robnett. 1998. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Leeuwenhoek 73:331-371. [DOI] [PubMed] [Google Scholar]
- 15.Kurtzman, C. P., and C. J. Robnett. 1997. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J. Clin. Microbiol. 35:1216-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehmann, P. F., D. M. Lin, and B. A. Lasker. 1992. Genotypic Identification and characterization of species and strains within the genus Candida by using random amplified polymorphic DNA. J. Clin. Microbiol. 30:3249-3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin, D. M., L. C. Wu, M. G. Rinaldi, and P. F. Lehmann. 1995. Three distinct genotypes within Candida parapsilosis from clinical sources. J. Clin. Microbiol. 33:1815-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nosek, J., L. Tomaska, A. Rycovska, and H. Fukuhara. 2002. Mitochondrial telomeres as molecular markers for identification of the opportunistic yeast pathogen Candida parapsilosis. J. Clin. Microbiol. 40:1283-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfaller, M. A., R. N. Jones, G. V. Doern, A. C. Fluit, J. Verhoef, H. S. Sader, S. A. Messer, A. Houston, S. Coffman, R. J. Hollis, et al. 1999. International surveillance of blood stream infections due to Candida species in the European SENTRY program: species distribution and antifungal susceptibility including the investigational triazole and echinocandin agents. Diagn. Microbiol. Infect. Dis. 35:19-25. [DOI] [PubMed] [Google Scholar]
- 20.Pfaller, M. A., R. N. Jones, G. V. Doern, H. S. Sader, S. A. Messer, A. Houston, S. Coffman, and R. J. Hollis. 2000. Bloodstream infections due to Candida species: SENTRY Antimicrobial Surveillance Program in North America and Latin America, 1997-1998. Antimicrob. Agents Chemother. 44:747-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roy, B., and S. A. Meyer. 1998. Confirmation of the distinct genotype groups within the form species Candida parapsilosis. J. Clin. Microbiol. 36:216-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rycovska, A., M. Valach, L. Tomaska, M. Bolotin-Fukuhara, and J. Nosek. 2004. Linear versus circular mitochondrial genomes: intraspecies variability of mitochondrial genome architecture in Candida parapsilosis. Microbiology 150:1571-1580. [DOI] [PubMed] [Google Scholar]
- 23.Sandven, P. 2000. Epidemiology of candidemia. Rev. Iberoamer. Micol. 17:73-81. [PubMed] [Google Scholar]
- 24.Scherer, S., and D. A. Stevens. 1987. Application of DNA typing methods to epidemiology and taxonomy of Candida species. J. Clin. Microbiol. 25:675-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sullivan, D. J., T. J. Westerneng, K. A. Haynes, D. E. Bennett, and D. C. Coleman. 1995. Candida dubliniensis sp. nov: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology 141:1507-1521. [DOI] [PubMed] [Google Scholar]
- 26.Tavanti, A., N. A. R. Gow, M. C. J. Maiden, F. C. Odds, and D. J. Shaw. 2004. Genetic evidence for recombination in Candida albicans based on haplotype analysis. Fungal Genet. Biol. 41:553-562. [DOI] [PubMed] [Google Scholar]
- 27.Tavanti, A., N. A. R. Gow, S. Senesi, M. C. J. Maiden, and F. C. Odds. 2003. Optimization and validation of multilocus sequence typing for Candida albicans. J. Clin. Microbiol. 41:3765-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White, T. J., T. D. Bruns, S. B. Lee, and J. W. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, Inc., San Diego, Calif.