Abstract
Four poliomyelitis outbreaks caused by vaccine-derived polioviruses have been reported recently, including one in Madagascar in 2002. In all cases, the viral strains involved were recombinant between poliovirus vaccine strains and nonpoliovirus strains, probably enterovirus species C. Nevertheless, little is known about the circulation and epidemiology of enteroviruses in the regions where these outbreaks occurred. To assess the circulation of enteroviruses (particularly enterovirus species C) in Madagascar, we genetically characterized 55 enterovirus strains isolated between 1994 and 2002. The strains were identified and compared by partially sequencing the region encoding the VP1 capsid protein. Phylogenetic analysis and pairwise comparison with prototype enterovirus strains distinguished two different species: 25 isolates belonged to human enterovirus B species, and 30 isolates were identified as coxsackievirus A13, A15, A17, A18, A20, A21, and A24, belonging to the human enterovirus species C. The relatively high frequency and the wide distribution of species C coxsackie A viruses in different regions of Madagascar suggest that they had been silently and widely circulating in the country during the whole study period. The circulation of coxsackie A viruses, combined with the low routine oral polio vaccine coverage, may have played a role in the emergence of the recent outbreak in Madagascar.
Human enteroviruses comprise pathogens that cause a wide spectrum of acute diseases, including the common cold syndrome, hand-foot-and-mouth disease, neonatal sepsis-like disease, aseptic meningitis, encephalitis, and in particular, poliomyelitis (18). These viruses were originally divided into 64 distinct serotypes: polioviruses 1 to 3; coxsackie A viruses (CAV) 1 to 22 and 24; coxsackie B viruses (CBV) 1 to 6; echoviruses (EV) 1 to 7, 9, 11 to 21, 24 to 27, and 29 to 33; and enteroviruses (ENV) 68 to 71 (18). Enteroviruses have recently been divided into five species on the bases of their molecular and pathogenetic characteristics: polioviruses and human enteroviruses A to D (HEV-A to -D) (16, 27). Species B includes all CBVs, all EVs, ENV69, and CAV9, whereas species C includes 11 CAV serotypes. Polioviruses and serotypes of the HEV-C species are closely related, according to the phylogenetic analysis of the entire genome (2, 13, 28).
The enterovirus genome is a single-stranded RNA molecule of positive polarity that is ∼7,500 nucleotides long. The most variable regions of the genome are the genes coding for the capsid proteins VP1 to VP4. The 5′ and 3′ untranslated regions are generally highly conserved. This genetic variability is caused by the high error rate of the viral RNA-dependent RNA polymerase, due to its lack of proofreading mechanisms during genome replication. Genetic recombination can also play a role in the evolution of enteroviruses and has been reported to occur between vaccine strains and/or wild polioviruses (7, 8, 10-12) and between nonpoliovirus enteroviruses belonging to the same or different serotypes (1, 24).
Poliomyelitis has been virtually eradicated worldwide, with only a few remaining countries of endemicity with reservoirs of wild polioviruses (6). However, poliomyelitis associated with mutated and pathogenic poliovirus strains derived from the oral polio vaccine is causing increasing concern. Outbreaks of poliomyelitis associated with vaccine-derived poliovirus recently occurred in Egypt, Hispaniola, and the Philippines (5, 15, 35). More recently, a similar outbreak occurred in Madagascar (29). The strains implicated in these four outbreaks were recombinant between vaccine polioviruses and unknown enteroviruses that could be either wild polioviruses or HEV-C. As wild polioviruses had not been detected in the affected regions for years (e.g., since 1998 in Madagascar), the unknown sequences present in the recombinants were probably derived from nonpoliovirus viruses belonging to HEV-C. Partial sequencing of the unknown parts of the two recombinant strains isolated in Madagascar was consistent with this hypothesis (29). However, little is known about the circulation and epidemiology of HEV-C in the regions where these outbreaks occurred, including Madagascar. The occurrence of recombinant vaccine-derived poliovirus stresses the importance of reliable and prompt diagnosis but also the need to improve our knowledge of HEV-C species epidemiology and circulation.
A study of 968 viral strains isolated between 1958 and 1977 in Madagascar revealed considerable circulation of enteroviruses, including polioviruses, coxsackieviruses, unspecified echoviruses, and enteroviruses (36). The present characteristics of circulating nonpoliovirus enteroviruses need to be clarified, as most strains isolated cannot be typed using the neutralization test, mainly because of the lack of corresponding antisera. Fortunately, molecular methods that allow enterovirus typing are now available (3, 4, 20-22, 31). These methods are based on the sequencing of part or all of the genomic region encoding the VP1 capsid protein, followed by comparison of these sequences with those of prototype enterovirus strains.
In this study, we used one of these molecular typing methods to characterize 55 enteroviruses isolated in Madagascar between 1994 and 2002. Forty-five of these strains could not be typed with the antiserum pools. A considerable number of coxsackie A virus strains belonging to the HEV-C species were discovered, indicating that this species has been continuously and widely circulating in Madagascar.
MATERIALS AND METHODS
Viruses.
Between 1994 and 2002, the Virology Unit of the Institut Pasteur de Madagascar isolated 436 enterovirus strains from 5,197 clinical samples (8.4%), mostly from healthy children. Due to the large number of isolates, we selected representative samples for further characterization. Since the annual number of samples was not constant and the annual rate of virus isolation varied from 5.0 to 19.2%, we selected all or some of the corresponding isolates, depending on the year considered. We tested 102 strains by using the neutralization assay: 24 (23.5%) were identified as coxsackie B viruses and different serotypes of echoviruses, whereas 78 (76.5%) could not be typed antigenically. Then, we randomly selected 55 viruses, including 10 isolates of known serotypes and 45 isolates that could not be typed with the antiserum pool. The panel included Malagasy viruses isolated during routine surveillance of enterovirus circulation among healthy children (n = 52) and clinical samples (acute flaccid paralysis and encephalitis; n = 3). All viruses were isolated from original specimens (stools, n = 54; throat swab, n = 1). They originated from four districts located in three provinces. Nine of the 55 viruses have already been described, and their sequences are available in nucleotide databases (accession no. AJ279179 to AJ279185, AJ279193, and AJ279195).
Cells and virus serotyping.
Human rhabdomyosarcoma (RD) and human larynx epidermoid carcinoma (HEp-2) cell lines were used for virus isolation. They were grown in tissue culture flasks with Dulbecco's modified Eagle's medium (Sigma-Aldrich, Inc., St. Louis, Mo.) supplemented with 5% fetal calf serum. Supernatants from infected cells were titrated using serial dilutions of viruses (from 10−1 to 10−8) in microplates. The 50% tissue culture infectious dose was then determined. The virus serotype was determined by a microseroneutralization test using 100 50% tissue culture infectious doses of virus and constant amounts of cross-reacting pools of antisera against enteroviruses (Rijksintituut voor Volksgezondheid en Milieuhygiene, Bilthoven, The Netherlands), according to the standard World Health Organization protocol (33). The amount of virus used for the test was systematically checked during the neutralization assay.
RNA extraction and reverse transcription-PCR.
Viral RNA was extracted from infected cell culture supernatants using Trizol LS (Invitrogen Life Technologies, Cergy Pontoise, France) according to the manufacturer's recommendations. A 1,452-bp gene fragment encompassing the 3′ third of the VP1 capsid region and the 2A, 2B, and part of the 2C coding regions was amplified by reverse transcription-PCR as previously described (3). Briefly, viral cDNA was synthesized using Moloney murine leukemia virus reverse transcriptase (Invitrogen Life Technologies). The resulting product was then amplified by PCR using Taq polymerase (Amersham Pharmacia Biotech, Inc., Piscataway, N.J.) in a GeneAmp PCR System 2400 thermal cycler (Perkin-Elmer Applied Biosystems, Foster City, Calif.). The PCR products were run on agarose gels, stained with ethidium bromide, and visualized under UV light.
Molecular typing of enteroviruses.
PCR products were purified from the gel with a QIAquick Gel Extraction kit (QIAGEN, Inc., Santa Clarita, Calif.) and directly sequenced using the PRISM BigDye Terminator Cycle Sequencing Reaction kit on an automated DNA sequencer (Perkin-Elmer Applied Biosystems). A fragment of 318 to 326 bp corresponding to the 3′ third of the VP1 capsid was compared with the corresponding region of available prototype sequences using the CLUSTAL W alignment program (32). The GenBank database was also screened for similar sequences using the FASTA program (26). Scores were established for each strain based on nucleotide identity and amino acid similarity with the closest prototype strains. The serotype of the field isolates was assumed to be that of the closest prototype strain according to the results of pairwise comparisons of nucleotide sequences, as previously described (3).
Phylogenetic analysis.
Phylogenetic analysis was performed using the NEIGHBOR program in PHYLIP (Phylogeny Inference Package) version 3.5 (9) and TREE-PUZZLE version 5.0 (30). Phylogenetic trees were constructed by the maximum-likelihood method of Kishino and Hasegawa (17) with PUZZLE, which uses quartet puzzling as the tree search algorithm. Distances were calculated by using the model of nucleotide substitutions described by Kishino and Hasegawa. The transition/transversion ratio was estimated directly from the data set. The reliability of the topology tree was estimated by the use of 1,000 puzzling steps. The trees were constructed using TREEVIEWX software version 0.4 (25). Alternatively, the DNADIST/NEIGHBOR method was used. The distance matrix (Kimura 2 or F84 model) was calculated by using a transition/transversion ratio of 8.0. The robustness of phylogenies constructed with NEIGHBOR was estimated by bootstrap analyses with 100 pseudoreplicate data sets generated with the SEQBOOT program.
Nucleotide sequence accession numbers.
The sequences reported here have been submitted to the EMBL Nucleotide Sequence Database under accession no. AJ567496 to AJ567533 and AJ584654 to AJ584661.
RESULTS
Molecular typing of isolates.
We studied 55 viruses isolated between 1994 and 2002 and recovered from HEp-2 and RD cell lines (Table 1). The neutralization test identified 10 of the isolates as CBV6 and EV7, -11, -13, -20, and -33. Molecular typing of these 10 isolates correlated well with the serotypes determined by the conventional method. For six EV7, -11, -13, -20, and -33 isolates, the highest scores with homotypic prototype strains were >75% for nucleotide sequences and >90% for amino acid sequences, compatible with the sequence homology criteria previously defined (3). Four CBV6 strains presented slightly lower nucleotide identity scores (73.6 to 74.8% nucleotide identity) with the prototype strain CBV6 Schmitt but higher nucleotide identity scores (79.0 to 96.0%) when compared with recently isolated CBV6 strains reported in GenBank.
TABLE 1.
Serotyping and partial VP1 sequencing of isolates
| Isolatea | Serotype by neutralization testb | Highest identity score (%)c
|
Second-highest identity score (%)c
|
Delta score (%)d | Accession no. (reference) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Type | nt | aa | Type | nt | Type | aa | ||||
| TN-374/94 | CBV6 | CBV6 | 74.8 | 89.8 | EV30 | 69.3 | EV11 | 69.2 | 5.5 | AJ567515 |
| IH-001/97 | CBV6 | CBV6 | 74.5 | 89.8 | ENV69 | 69.3 | EV11 | 72.9 | 5.2 | AJ567516 |
| IH-003/97 | CBV6 | CBV6 | 74.8 | 89.8 | EV30 | 69.1 | EV11 | 72.9 | 5.7 | AJ567517 |
| IH-004/97 | CBV6 | CBV6 | 73.6 | 87.1 | EV11 | 68.9 | EV13 | 69.8 | 4.7 | AJ567518 |
| MG-451/94 | UT | CBV6 | 77.9 | 92.6 | ENV69 | 70.6 | EV11 | 72.9 | 7.3 | AJ279183 (3) |
| MG-44381/98 | EV7 | EV7 | 77.5 | 95.0 | EV2 | 72.4 | EV11 | 76.2 | 5.1 | AJ279195 (3) |
| MG-41094/97 | EV11 | EV11 | 79.1 | 91.8 | EV33 | 72.8 | EV19 | 83.7 | 6.3 | AJ279193 (3) |
| MI-006/97 | EV13 | EV13 | 79.1 | 96.3 | ENV69 | 72.4 | ENV69 | 87.1 | 6.7 | AJ567520 |
| MI-007/97 | EV20 | EV20 | 81.9 | 96.3 | EV33 | 69.0 | EV33 | 75.0 | 12.9 | AJ567521 |
| IH-008/97 | UT | EV20 | 81.9 | 96.3 | EV33 | 69.1 | EV33 | 75.0 | 12.8 | AJ567522 |
| MI-009/97 | UT | EV20 | 81.6 | 95.4 | EV33 | 69.0 | EV33 | 74.0 | 12.6 | AJ567523 |
| MI-010/97 | UT | EV20 | 80.7 | 92.6 | EV33 | 67.8 | EV33 | 72.2 | 12.9 | AJ567524 |
| MI-011/97 | UT | EV20 | 81.3 | 96.3 | EV33 | 68.7 | EV33 | 75.0 | 12.6 | AJ567525 |
| MI-012/97 | UT | EV20 | 82.2 | 96.3 | EV33 | 68.4 | EV33 | 75.0 | 13.8 | AJ567526 |
| MI-013/97 | UT | EV20 | 82.5 | 97.2 | EV33 | 68.7 | EV33 | 75.9 | 13.8 | AJ567527 |
| MI-020/97 | UT | EV20 | 81.0 | 94.5 | EV33 | 69.0 | EV33 | 74.1 | 13.0 | AJ567528 |
| MI-021/97 | UT | EV20 | 82.5 | 97.2 | EV33 | 68.7 | EV33 | 75.9 | 13.8 | AJ567529 |
| MI-014/97 | EV33 | EV33 | 77.0 | 95.4 | EV19 | 69.0 | EV19 | 77.4 | 8.0 | AJ567530 |
| MI-015/97 | EV33 | EV33 | 77.0 | 95.4 | EV19 | 69.0 | EV19 | 77.4 | 8.0 | AJ567531 |
| MI-017/97 | UT | EV33 | 76.1 | 93.5 | EV19 | 68.4 | EV19 | 76.4 | 7.7 | AJ567532 |
| MI-019/97 | UT | EV33 | 76.0 | 93.5 | EV19 | 68.3 | EV19 | 76.4 | 7.7 | AJ567533 |
| TL-66984/02 | UT | EV19 | 81.0 | 96.3 | EV11 | 78.5 | EV11 | 86.1 | 2.5 | AJ584658 |
| TL-66995/02 | UT | EV19 | 80.4 | 95.4 | EV11 | 77.8 | EV11 | 85.2 | 2.6 | AJ584661 |
| TL-68143/02 | UT | EV25 | 81.3 | 96.3 | EV21 | 71.5 | EV21 | 83.3 | 9.8 | AJ584659 |
| TN-RD-1/00 | UT | EV27 | 75.1 | 79.4 | EV6 | 72.9 | EV17 | 69.8 | 2.2 | AJ567519 |
| IH-024/97 | UT | CAV13 | 75.2 | 90.7 | CAV18 | 72.8 | CAV18 | 87.9 | 2.4 | AJ567499 |
| TN-444/94 | UT | CAV13 | 76.9 | 89.7 | CAV18 | 76.0 | CAV18 | 87.9 | 0.9 | AJ567500 |
| TN-445/94 | UT | CAV13 | 76.9 | 89.7 | CAV18 | 76.0 | CAV18 | 87.9 | 0.9 | AJ567501 |
| TN-446/94 | UT | CAV13 | 76.9 | 89.7 | CAV18 | 76.0 | CAV18 | 87.9 | 0.9 | AJ567502 |
| TN-456/94 | UT | CAV13 | 77.6 | 89.7 | CAV18 | 75.9 | CAV18 | 88.8 | 1.7 | AJ567503 |
| MG-354/94 | UT | CAV13 | 77.9 | 90.6 | CAV18 | 76.3 | CAV18 | 88.8 | 1.6 | AJ279179 (3) |
| MG-356/94 | UT | CAV13 | 77.4 | 90.6 | CAV18 | 75.4 | CAV18 | 88.8 | 2.0 | AJ279180 (3) |
| MG-404/94 | UT | CAV13 | 80.2 | 92.5 | CAV18 | 78.0 | CAV18 | 90.6 | 2.2 | AJ279181 (3) |
| MG-498/94 | UT | CAV13 | 78.7 | 93.5 | CAV18 | 77.3 | CAV18 | 91.6 | 1.4 | AJ279185 (3) |
| TL-67001/02 | UT | CAV13 | 77.2 | 86.9 | CAV18 | 75.9 | CAV13 | 86.0 | 1.3 | AJ584655 |
| TL-66025/02 | UT | CAV18 | 76.9 | 87.9 | CAV13 | 76.3 | CAV13 | 88.8 | 0.6 | AJ584654 |
| MI-025/97 | UT | CAV18 | 77.9 | 89.7 | CAV13 | 73.5 | CAV13 | 90.7 | 4.4 | AJ567504 |
| MI-026/97 | UT | CAV18 | 76.6 | 88.8 | CAV13 | 72.9 | CAV13 | 89.7 | 3.7 | AJ567505 |
| MI-027/97 | UT | CAV18 | 77.0 | 88.8 | CAV13 | 72.9 | CAV13 | 89.7 | 4.1 | AJ567506 |
| MI-029/97 | UT | CAV18 | 76.3 | 86.0 | CAV17 | 72.6 | CAV13 | 86.9 | 3.7 | AJ567507 |
| IH-034/97 | UT | CAV18 | 76.1 | 88.8 | CAV13 | 74.1 | CAV13 | 89.7 | 2.0 | AJ567508 |
| IH-035/97 | UT | CAV18 | 76.0 | 88.8 | CAV13 | 74.8 | CAV13 | 89.7 | 1.2 | AJ567509 |
| TN-439/94 | UT | CAV18 | 77.9 | 90.7 | CAV13 | 76.1 | CAV13 | 91.6 | 1.8 | AJ567510 |
| MI-018/97 | UT | CAV15 | 83.5 | 98.1 | CAV11 | 78.5 | CAV11 | 97.2 | 5.0 | AJ567511 |
| MI-031/97 | UT | CAV15 | 83.2 | 97.2 | CAV11 | 78.2 | CAV11 | 96.3 | 5.0 | AJ567512 |
| TL-66990/02 | UT | CAV15 | 83.2 | 96.3 | CAV11 | 79.7 | CAV11 | 95.3 | 3.5 | AJ584657 |
| TL-67004/02 | UT | CAV15 | 82.5 | 94.4 | CAV11 | 78.4 | CAV11 | 93.4 | 4.1 | AJ584660 |
| MI-032/97 | UT | CAV17 | 77.0 | 92.5 | CAV13 | 72.3 | CAV13 | 81.3 | 4.7 | AJ567513 |
| MI-033/97 | UT | CAV17 | 76.6 | 91.5 | CAV13 | 71.9 | CAV13 | 80.4 | 4.7 | AJ567514 |
| TN-D05/01 | UT | CAV21 | 90.7 | 99.1 | CAV24 | 67.5 | CAV24 | 73.8 | 23.2 | AJ567496 |
| IH-022/97 | UT | CAV24 | 80.1 | 92.5 | CAV17 | 71.3 | CAV15 | 78.0 | 8.8 | AJ567497 |
| IH-023/97 | UT | CAV24 | 80.1 | 92.5 | CAV17 | 71.3 | CAV15 | 78.0 | 8.8 | AJ567498 |
| TL-68269/02 | UT | CAV24 | 75.0 | 86.0 | CAV21 | 70.6 | CAV21 | 77.6 | 4.4 | AJ584656 |
| TN-448/94 | UT | CAV20 | 74.8 | 92.5 | CAV17 | 71.9 | CAV13 | 82.3 | 2.9 | AJ279182 (3) |
| TN-423/94 | UT | CAV20 | 75.0 | 91.6 | CAV13 | 72.3 | CAV13 | 81.3 | 2.7 | AJ279184 (3) |
Isolates are identified by geographical location (two-letter code: TN, Antananarivo; IH, Ihosy; MI, Miantsoarivo; TL, Tolagnaro; MG; Madagascar), internal laboratory code, and year of isolation (e.g., 94, 1994).
Results obtained with the standard neutralization test; UT, untypeable.
nt, nucleotide; aa, amino acid.
Difference between highest and second-highest nucleotide scores.
The remaining 45 isolates that could not be typed using standard antisera (untypeable isolates) were characterized only by the molecular method. All sequences showed ≥74.8% nucleotide sequence identity and ≥86.0% amino acid sequence similarity with one of the enterovirus prototype sequences available in the database (Table 1). These results allowed us to assign a serotype to each of these untypeable isolates. In all cases, the second-highest scores were obtained with a heterotypic strain that belonged to the same species as the homotypic strain giving the highest scores. Comparisons of the partial VP1 sequences of the 55 isolates allowed us to distinguish two different species: 25 strains belonged to HEV-B species, and surprisingly, 30 strains were coxsackie A viruses of the HEV-C species.
Strains belonging to HEV-B species.
The group of strains belonging to the HEV-B species comprises the 10 isolates previously identified by the neutralization test (described above) and 15 untypeable viruses (Table 1). Most of them were isolated from the RD cell line. On the basis of sequence data, the untypeable isolates were identified as CBV6 and EV19, -20, -25, -27, and -33 strains.
However, for three recent untypeable isolates, the two highest scores differed by only 2.2 to 2.6%. The sequences of strains TL-66995/02 and TL-66984/02 showed 80.4 and 81.0% nucleotide identity with the prototype strain EV19 (Burke), respectively, and were thus classified as EV19 strains (Table 1). However, the second-highest nucleotide identity scores were 77.8 and 78.5% with the EV11 prototype strain (Silva). A search of the GenBank database indicated that these two strains were more similar (>80.0% nucleotide identity) to other field EV19 strains than to field EV11 strains (<77.5% nucleotide identity).
The nucleotide sequence of strain TN-RD-1/00 was 75.1% identical to that of the prototype strain EV27 (Bacon). It was also similar (72.9% nucleotide identity) to that of strain EV6 (d'Amori). Unfortunately, no other EV27 field strain sequences were present in GenBank to confirm the serotype of TN-RD-1/00.
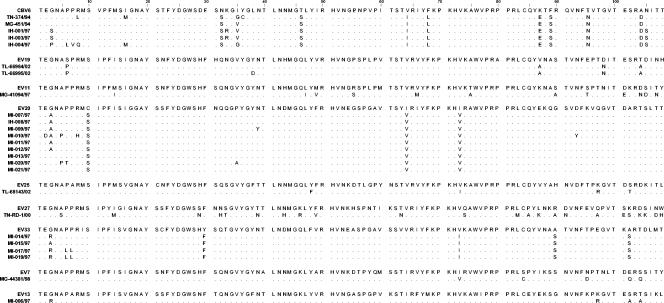
Alignment of the amino acid sequences of HEV-B strains with their respective prototype strains revealed just a few amino acid changes in most cases (4 to 15 differences for 108-amino-acid segments) (Fig. 1). Most of these changes were identical in all Madagascar HEV-B strains of the same serotype, indicating that the analyzed strains were derived from common ancestors that were somewhat different from (or more recent than) the prototype strains. Strain EV27 TN-RD-1/00 appeared to be more different from its prototype strain than the other serotypes (23 amino acid differences).
FIG. 1.
Comparisons of amino acid sequences in the 3′ third of the VP1 proteins of the HEV-B species isolates and their prototype strains. Dots indicate amino acid similarity with the VP1 sequence of each prototype strain.
Strains belonging to HEV-C species.
The group of 30 strains belonging to the HEV-C species included only untypeable strains. All were isolated in the HEp-2 cell line, with the exception of the CAV21 strain, which was recovered from the RD cell line. Nucleotide sequences clearly confirmed that all these strains were indeed HEV-C strains. The highest nucleotide identity scores were with the CAV13, -15, -17, -18, -20, -21, and -24 prototype strains (Table 1).
However, for 22 nucleotide sequences, the two highest scores with prototype strains differed by only 0.6 to 5.0% and the second-highest nucleotide identity score was relatively high, between 72.6 and 79.7%. Therefore, some ambiguity remains as to the true identities of certain strains. This group of strains includes, in particular, the pairs of closely related strains CAV13-CAV18 and CAV11-CAV15, as previously described (2, 22). For the remaining nucleotide sequences, their identities as CAV17, CAV20, CAV21, or CAV24 sequences were strengthened with greater certainty by the presence in data banks of sequences of closely related field isolates of the corresponding serotypes.
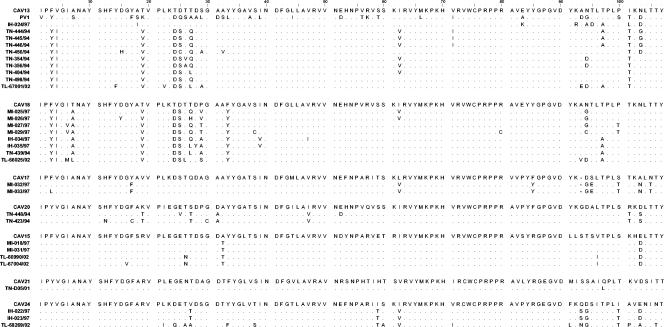
The amino acid sequences corresponding to the analyzed nucleotide sequences encoding the 3′ third of the VP1 capsid of field isolates were aligned and compared with those of the corresponding prototype strains (Fig. 2). In most cases, a few amino acid changes were found (2 to 17 differences for 107-amino-acid segments). As for the HEV-B field strains described above, most of these changes were identical in all field strains belonging to the same serotype. Surprisingly, a common core of conserved residues was found at positions 3 to 4 (YI), 7 (A), 19 (V), 24 to 25 (DS), 33 (Y), and 101 (T) in most of the CAV13 and CAV18 isolates but not in one or both prototype strains. Hence, both types of field strains are derived from a common ancestor.
FIG. 2.
Comparisons of amino acid sequences in the 3′ third of the VP1 proteins of the HEV-C species isolates and their prototype strains. Dots indicate amino acid similarity with the VP1 sequences of each prototype strain, while dashes indicate gaps.
Phylogenetic clustering of enteroviruses.
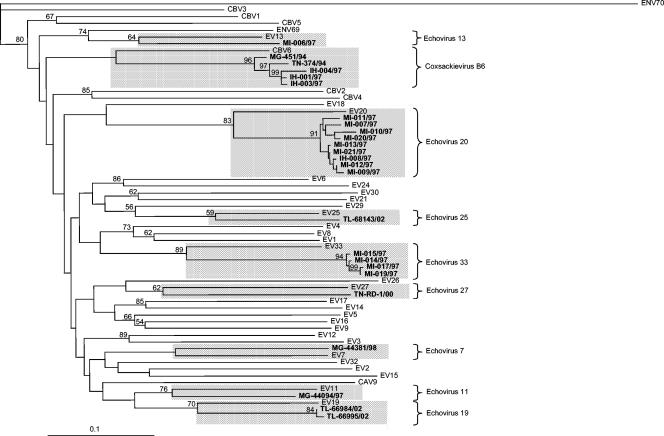
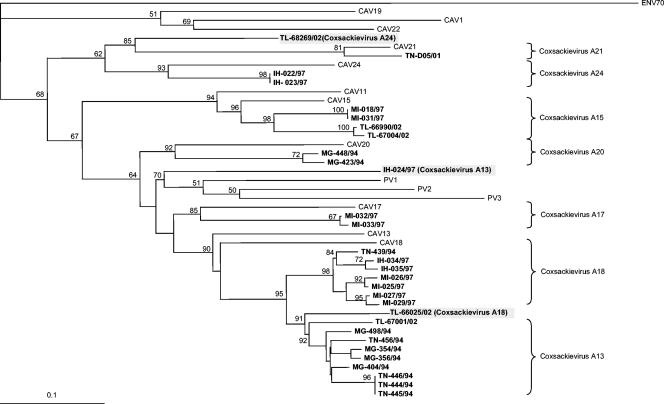
Phylogenetic trees were constructed for each defined species based on the sequence of the 3′ third of the VP1 capsid. The phylogenetic relationships between the field isolates and the HEV-B prototype strains are shown in Fig. 3. All HEV-B field isolates formed a monophyletic group with their respective homotypic prototype strains. This finding was supported by bootstrap values of 59 to 89%. The phylogenetic relationships between the field isolates and the HEV-C prototype strains are shown in Fig. 4. The HEV-C species segregated into four subgroups as previously described (22). Most field isolates were monophyletic with respect to their homotypic prototype strains, and clusters were supported by high bootstrap values (≥81%). However, some variations in the branching order of two isolates were observed. Isolates IH-024/97 and TL-68269/02, identified as CAV13 and CAV24, respectively, were rather far from their supposed prototype strains in the phylogenetic tree. The use of a different method for constructing the phylogenetic tree indicated that the positions of these strains were not stable in the tree and that in most cases they do not cluster with their putative prototype strains. Although these strains clearly belong to HEV-C, their true serotypes need to be confirmed by the use of longer nucleotide sequences and/or specific antisera.
FIG. 3.
Phylogenetic tree showing genetic relationships between human enterovirus species B. This neighbor-joining tree was based on nucleotide sequence alignments for the 3′ third of the VP1 coding regions. Branch lengths were calculated using the maximum-likelihood quartet-puzzling method (PUZZLE). Numbers at nodes correspond to the percentage of 1,000 puzzle steps supporting the distal cluster. The genetic distance is indicated (bar). The nucleotide sequences of enterovirus 70 were used as an outgroup.
FIG. 4.
Phylogenetic tree depicting genetic relationships among human enterovirus species C strains. This tree was constructed as described in the legend to Fig. 3.
DISCUSSION
Since the discovery of circulating vaccine-derived polioviruses bearing nonpoliovirus sequences thought to be derived from HEV-C, interest in the epidemiology and circulation of these enterovirus species has increased tremendously. However, HEV-C isolates have been poorly studied due to the inability of certain serotypes to grow in common cell lines and to the lack of specific antisera. Molecular methods based on the correlation between the nucleotide sequence of the gene encoding the VP1 capsid protein and those of prototype enterovirus strains allowed us to search for such HEV-C isolates. We used one of these molecular methods to identify untypeable enterovirus isolates collected in Madagascar from 1994 to 2002. We discovered that 30 of the 45 untypeable viruses belonged to HEV-C. These viruses also belonged to different serotypes, including CAV13, -15, -17, -18, -20, -21, and -24. Given that 76.5% (annual range, 70 to 90%) of the enterovirus strains isolated in Madagascar in this period were untypeable, these results suggest that ∼50% of these enteroviruses were HEV-C. This is the first comprehensive study of enterovirus circulation showing a high proportion of circulating HEV-C species among healthy children living in a given country.
The number of enterovirus-positive samples and the proportion of these enteroviruses that were HEV-C may have been biased by the cell lines used for isolation (RD and HEp-2 cell lines). Some circulating enteroviruses that cannot grow in these cell lines may have been missed, and some specific serotypes may have been favored. Nevertheless, the CAV strains analyzed in this study represent seven of the eight HEV-C serotypes known to grow in cell culture. The other three HEV-C serotypes (CAV1, -19, and -22) display genetic features different from those of most other HEV-C isolates and are rarely, if ever, isolated in cell culture (2). For example, the only way to isolate these three CAV phenotypes is to inoculate newborn mice with fecal samples (18).
The molecular method also identified some untypeable isolates as serotypes of the HEV-B species. The fact that these strains were not neutralized with the standard neutralizing antiserum pools used in the classical identification method is possibly due to antigenic variation compared to the prototype strains used to immunize animals. It is less easy to obtain positive results with this test if the virus is in a mixture or an aggregate or if an inadequate amount of virus is available. Furthermore, the fact that the conventional and molecular techniques gave the same results for the HEV-B isolates previously typed by neutralization test shows that our method is accurate (3). However, some ambiguities remain concerning the true identities of some HEV-B and HEV-C isolates. The use of monospecific antisera could clarify the results. Nevertheless, genetic features clearly indicated that these isolates were members of the HEV-B or HEV-C species.
Our data showed that CAV13 and CAV18 field isolates are closely genetically related. This is consistent with previous studies of the complete VP1 sequences of the respective prototype strains, which suggested that CAV18 isolates are (and could be classified as) antigenic variants of the CAV13 serotype (2, 22). The close relationship between the CAV13 and CAV18 serotypes is also supported by the common core of highly conserved amino acid residues found in both CAV13 and CAV18 Madagascar field isolates but not in their respective prototype strains. This strongly suggests that the CAV18-CAV13 field isolates in Madagascar were derived from a common CAV18-CAV13 ancestor strain.
Coxsackie A viruses of the HEV-C species are human pathogens that are often responsible for common cold symptoms, like rhinoviruses (18). Most HEV-C isolates and the major group of rhinoviruses use ICAM-1 (intercellular adhesion molecule 1) as a cellular receptor (19, 34). Coxsackievirus A24 strains have been involved in several outbreaks of acute hemorrhagic conjunctivitis worldwide (14, 23), and coxsackieviruses A1 and A22 are known to cause herpangina (18). However, little is known about the implication of members of HEV-C in other diseases. In Madagascar during the period surveyed, the only coxsackievirus A21 strain isolated was recovered from a clinical case. The virus was isolated from the throat of a patient with encephalitis. The disease association was confirmed by directly detecting the genome in a blood sample. No other clinical cases of coxsackie A viruses have been reported. However, this may be due to inadequate investigation of suspected cases. In fact, many of the strains isolated from stool samples from symptomatic children in Madagascar were “by-products” of poliomyelitis surveillance, which is essentially based on the surveillance of acute flaccid paralysis. Little attention has been paid to nonpoliovirus enteroviruses and to other related diseases in tropical countries.
During the 9-year study period, several HEV-C serotypes were isolated during the same year (e.g., CAV15, -17, -18, and -24 in 1997), and some of them were isolated in several years (e.g., CAV13 in 1994 and 2002 or CAV15 in 1997 and 2002). Moreover phylogenetic clusters of CAV13 and CAV18 strains suggest long-term circulation from 1994 to 2002 and from 1994 to 1997, respectively (Fig. 4). However, many isolates were not isolated in more than 1 or 2 years (e.g., CAV17, CAV20, and CAV21). From these results, therefore, it is difficult to distinguish between continuous circulation of indigenous strains and periodic introduction of serotypes from outside the country. Nevertheless, this study clearly indicates that HEV-C isolates were common during the period considered. Interestingly, different serotypes of coxsackie A viruses were identified in different geographical locations, including the region where an outbreak of recombinant vaccine-derived poliovirus occurred (29).
Studies of vaccine-derived poliovirus outbreaks, including the one that recently occurred in Madagascar, have shown that all of the implicated strains resulted from recombination between vaccine poliovirus and unidentified wild polioviruses or HEV-C (5, 15, 29, 35). Wild polioviruses have not been detected in the affected regions for years, and nothing was known about the circulation of HEV-C. We provide the temporal evidence for the circulation of coxsackie A viruses in a country in which a vaccine-derived poliovirus outbreak has occurred. This is the first step toward identifying unknown putative parental strains that recombined with polioviruses. This will improve our understanding of the evolution of enteroviruses and, in particular, of interspecific genetic exchanges between poliovirus and HEV-C. The role of recombination in the appearance of such vaccine-derived epidemic strains is unclear, but it is possible that the implicated recombinants acquired phenotypic characteristics (pathogenicity and transmissibility) of wild polioviruses via recombination. A more detailed study is under way to assess the impacts of the genetic exchanges between poliovirus and HEV-C species on the characteristics of the recombinant viruses. From a practical point of view, genetic recombination can also be used as a virological marker of vaccine-derived poliovirus circulation.
As we approach the final stage of poliomyelitis eradication, one of the remaining challenges is to stop immunization with the oral polio vaccine. However, more detailed studies of the natural evolution of enteroviruses are needed before this is possible without causing the reappearance of poliovirus. Wide circulation of HEV-C (as in Madagascar) and low polio vaccine coverage (as in the regions where the four vaccine-derived outbreaks occurred) may be the two major factors or indicators leading to the emergence of vaccine-derived epidemic polioviruses.
Acknowledgments
This study was partly supported by grants from the Transverse Research Programs (PTR120) and from the DVPI (Virus ARN) of the Pasteur Institute and from the French Ministry of Foreign Affairs (FSP 2001-168).
REFERENCES
- 1.Andersson, P., K. Edman, and A. M. Lindberg. 2002. Molecular analysis of the echovirus 18 prototype: evidence of interserotypic recombination with echovirus 9. Virus Res. 85:71-83. [DOI] [PubMed] [Google Scholar]
- 2.Brown, B., M. S. Oberste, K. Maher, and M. A. Pallansch. 2003. Complete genomic sequencing shows that polioviruses and members of human enterovirus species C are closely related in the noncapsid coding region. J. Virol. 77:8973-8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caro, V., S. Guillot, F. Delpeyroux, and R. Crainic. 2001. Molecular strategy for ‘serotyping’ of human enteroviruses. J. Gen. Virol. 82:79-91. [DOI] [PubMed] [Google Scholar]
- 4.Casas, I., G. F. Palacios, G. Trallero, D. Cisterna, M. C. Freire, and A. Tenorio. 2001. Molecular characterization of human enteroviruses in clinical samples: comparison between VP2, VP1, and RNA polymerase regions using RT nested PCR assays and direct sequencing of products. J. Med. Virol. 65:138-148. [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2001. Acute flaccid paralysis associated with circulating vaccine-derived poliovirus—Philippines, 2001. Morb. Mortal. Wkly. Rep. 50:874-875. [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2004. Global polio eradication initiative strategic plan, 2004. Morb. Mortal. Wkly. Rep. 53:107-108. [PubMed] [Google Scholar]
- 7.Cuervo, N. S., S. Guillot, N. Romanenkova, M. Combiescu, A. Aubert-Combiescu, M. Seghier, V. Caro, R. Crainic, and F. Delpeyroux. 2001. Genomic features of intertypic recombinant Sabin poliovirus strains excreted by primary vaccinees. J. Virol. 75:5740-5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dahourou, G., S. Guillot, O. Le Gall, and R. Crainic. 2002. Genetic recombination in wild-type poliovirus. J. Gen. Virol. 83:3103-3110. [DOI] [PubMed] [Google Scholar]
- 9.Felsenstein, J. 1993. PHYLIP: Phylogeny Inference Package, version 3.5c. Department of Genetics, University of Washington, Seattle.
- 10.Georgescu, M. M., F. Delpeyroux, and R. Crainic. 1995. Tripartite genome organization of a natural type 2 vaccine/nonvaccine recombinant poliovirus. J. Gen. Virol. 76:2343-2348. [DOI] [PubMed] [Google Scholar]
- 11.Georgescu, M. M., F. Delpeyroux, M. Tardy-Panit, J. Balanant, M. Combiescu, A. A. Combiescu, S. Guillot, and R. Crainic. 1994. High diversity of poliovirus strains isolated from the central nervous system from patients with vaccine-associated paralytic poliomyelitis. J. Virol. 68:8089-8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guillot, S., V. Caro, N. Cuervo, E. Korotkova, M. Combiescu, A. Persu, A. Aubert-Combiescu, F. Delpeyroux, and R. Crainic. 2000. Natural genetic exchanges between vaccine and wild poliovirus strains in humans. J. Virol. 74:8434-8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes, P. J., C. North, P. D. Minor, and G. Stanway. 1989. The complete nucleotide sequence of coxsackievirus A21. J. Gen. Virol. 70:2943-2952. [DOI] [PubMed] [Google Scholar]
- 14.Ishiko, H., N. Takeda, K. Miyamura, N. Kato, M. Tanimura, K. H. Lin, M. Yin-Murphy, J. S. Tam, G. F. Mu, and S. Yamazaki. 1992. Phylogenetic analysis of a coxsackievirus A24 variant: the most recent worldwide pandemic was caused by progenies of a virus prevalent around 1981. Virology 187:748-759. [DOI] [PubMed] [Google Scholar]
- 15.Kew, O., V. Morris-Glasgow, M. Landaverde, C. Burns, J. Shaw, Z. Garib, J. Andre, E. Blackman, C. J. Freeman, J. Jorba, R. Sutter, G. Tambini, L. Venczel, C. Pedreira, F. Laender, H. Shimizu, T. Yoneyama, T. Miyamura, H. van Der Avoort, M. S. Oberste, D. Kilpatrick, S. Cochi, M. Pallansch, and C. deq Uadros. 2002. Outbreak of poliomyelitis in Hispaniola associated with circulating type 1 vaccine-derived poliovirus. Science 296:356-359. [DOI] [PubMed] [Google Scholar]
- 16.King, A. M. Q., F. Brown, P. Christian, T. Hovi, T. Hyypiä, N. J. Knowles, S. M. Lemon, P. D. Minor, A. C. Palmenberg, T. Skern, and G. Stanway. 2000. Picornaviridae, p. 657-673. In M. H. V. Van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Seventh report of the International Committee for the Taxonomy of Viruses. Academic Press, Inc., San Diego, Calif.
- 17.Kishino, H., and M. Hasegawa. 1989. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J. Mol. Evol. 29:170-179. [DOI] [PubMed] [Google Scholar]
- 18.Melnick, J. L. 1996. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses, p. 655-712. In B. N. Fields, D. M. Knipe, P. M. Howley, R. M. Channock, J. L. Melnick, T. P. Monath, B. Roizman, and S. E. Straus (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 19.Newcombe, N. G., P. Andersson, E. S. Johansson, G. G. Au, A. M. Lindberg, R. D. Barry, and D. R. Shafren. 2003. Cellular receptor interactions of C-cluster human group A coxsackieviruses. J. Gen. Virol. 84:3041-3050. [DOI] [PubMed] [Google Scholar]
- 20.Norder, H., L. Bjerregaard, and L. O. Magnius. 2001. Homotypic echoviruses share aminoterminal VP1 sequence homology applicable for typing. J. Med. Virol. 63:35-44. [PubMed] [Google Scholar]
- 21.Oberste, M. S., K. Maher, D. R. Kilpatrick, M. R. Flemister, B. A. Brown, and M. A. Pallansch. 1999. Typing of human enteroviruses by partial sequencing of VP1. J. Clin. Microbiol. 37:1288-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oberste, M. S., K. Maher, D. R. Kilpatrick, and M. A. Pallansch. 1999. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J. Virol. 73:1941-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oh, M. D., S. Park, Y. Choi, H. Kim, K. Lee, W. Park, Y. Yoo, E. C. Kim, and K. Choe. 2003. Acute hemorrhagic conjunctivitis caused by coxsackievirus A24 variant, South Korea, 2002. Emerg. Infect. Dis. 9:1010-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oprisan, G., M. Combiescu, S. Guillot, V. Caro, A. Combiescu, F. Delpeyroux, and R. Crainic. 2002. Natural genetic recombination between co-circulating heterotypic enteroviruses. J. Gen. Virol. 83:2193-2200. [DOI] [PubMed] [Google Scholar]
- 25.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 26.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pringle, C. R. 1999. Virus taxonomy at the XIth International Congress of Virology, Sydney, Australia, 1999. Arch. Virol. 144:2065-2070. [DOI] [PubMed] [Google Scholar]
- 28.Pulli, T., P. Koskimies, and T. Hyypia. 1995. Molecular comparison of coxsackie A virus serotypes. Virology 212:30-38. [DOI] [PubMed] [Google Scholar]
- 29.Rousset, D., M. Rakoto-Andrianarivelo, R. Razafindratsimandresy, B. Randriamanalina, S. Guillot, J. Balanant, P. Mauclère, and F. Delpeyroux. 2003. Recombinant vaccine-derived poliovirus in Madagascar. Emerg. Infect. Dis. 9:885-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strimmer, K., and A. von Haeseler. 1996. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13:964-969. [Google Scholar]
- 31.Thoelen, I., P. Lemey, I. Van Der Donck, K. Beuselinck, A. M. Lindberg, and M. Van Ranst. 2003. Molecular typing and epidemiology of enteroviruses identified from an outbreak of aseptic meningitis in Belgium during the summer of 2000. J. Med. Virol. 70:420-429. [DOI] [PubMed] [Google Scholar]
- 32.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.World Health Organization. 2001. Polio laboratory manual. Department of Vaccines and Biologicals. World Health Organization, Geneva, Switzerland.
- 34.Xiao, C., C. M. Bator, V. D. Bowman, E. Rieder, Y. He, B. Hebert, J. Bella, T. S. Baker, E. Wimmer, R. J. Kuhn, and M. G. Rossmann. 2001. Interaction of coxsackievirus A21 with its cellular receptor, ICAM-1. J. Virol. 75:2444-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang, C. F., T. Naguib, S. J. Yang, E. Nasr, J. Jorba, N. Ahmed, R. Campagnoli, H. van der Avoort, H. Shimizu, T. Yoneyama, T. Miyamura, M. Pallansch, and O. Kew. 2003. Circulation of endemic type 2 vaccine-derived poliovirus in Egypt from 1983 to 1993. J. Virol. 77:8366-8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeller, H., and P. Coulanges. 1979. Les entérovirus à Madagascar. Bilan de 20 ans d'activité du laboratoire de virologie de l'Institut Pasteur de Madagascar. Arch. Inst. Pasteur Madagascar 47:121-169. [PubMed] [Google Scholar]