Abstract
Infection of calves with bovine herpesvirus 1 (BHV-1) results in transient immunosuppression that may lead to bacterium-induced pneumonia and, occasionally, death. Although sensory neurons in the trigeminal ganglia (TG) are the primary site of BHV-1 latency, viral genomes are detected in the tonsils of latently infected calves. Dexamethasone (DEX) consistently induces reactivation from latency, and viral gene expression is detected in TG and tonsils. In sensory neurons of latently infected calves, the latency-related (LR) gene is abundantly expressed and is required for reactivation from latency. In the present study, we compared the abilities of wild-type (wt) BHV-1 and a strain with a mutation in the LR gene (the LR mutant strain) to grow in the tonsils of infected calves and reactivate from latency. Lower levels of the LR mutant virus were detected in the tonsils of acutely infected calves. LR mutant viral DNA was consistently detected by PCR in the tonsils of latently infected calves, suggesting that the establishment of a latent or persistent infection occurred. Although the LR mutant did not reactivate from latency in vivo after DEX treatment, explantation of tonsil tissue from calves latently infected with the LR mutant yielded infectious virus. Relative to wt BHV-1, the LR mutant did not induce explant-induced reactivation as efficiently. These studies indicate that the LR gene promotes virus shedding from tonsil tissue during acute infection and reactivation from latency in tonsil tissue in vivo. We suggest that incorporation of the LR gene mutation into existing modified live vaccines would prevent reactivation from latency in neural and nonneural sites and would thus prevent transmission to other animals.
Bovine herpesvirus 1 (BHV-1), a member of the Alphaherpesvirinae subfamily, is responsible for a variety of clinical syndromes in cattle, including respiratory disorders, conjunctivitis, genital infections, encephalitis, abortions, and a multisystemic fatal disease in neonates. Acute infection of the respiratory tract by BHV-1 can induce immunosuppression, which predisposes cattle to secondary bacterial colonization, severe pneumonia, and even death (41).
Peripheral blood mononuclear cells prepared from calves infected with BHV-1 produce less interleukin 2 and have reduced mitogenic responses and lower natural cytotoxic activities (3). Furthermore, BHV-1 infects and induces at least 50 times more apoptosis in CD4+ T cells during acute infection relative to that induced in mock-infected cells (43, 46). BHV-1 antigens and apoptotic cells can be localized to germinal centers (GCs) of the pharyngeal tonsils in acutely or latently infected calves. BHV-1 induces apoptosis when cultured T cells, B lymphocytes, or monocytes are infected (14, 15). Glycoprotein D is important for the inducement of apoptosis during the infection process (12). Diverse mechanisms, such as induction of c-myc (13), p53 expression, and caspase activation (8), also stimulate apoptosis following BHV-1 infection. Finally, herpesviruses, including BHV-1, interfere with the major histocompatibility complex class I antigen presentation pathway and, consequently, interfere with cytotoxic lymphocyte-mediated killing of virus-infected cells (16, 31; for a review, see reference 9). Collectively, these studies indicate that the ability of BHV-1 to infect lymphoid cells and induce apoptosis is important for pathogenesis.
After acute infection, members of the Alphaherpesvirinae subfamily establish latent infections in their hosts, primarily in sensory neurons within trigeminal ganglia (TG) or dorsal root ganglia (23, 24). Latent virus can periodically reactivate from latency as a result of natural stressors or dexamethasone (DEX) treatment, which leads to virus shedding and spread to susceptible hosts (36, 44, 46). Although ganglionic neurons are the main site of latency for BHV-1 and other members of the Alphaherpesvirinae subfamily, latent or persistent infections can also occur in nonneural sites, for example, tonsils and lymph nodes. BHV-1 DNA is consistently detected in tonsils (46), peripheral blood cells (10), lymph nodes, and the spleen when infectious virus is not detected (30). Pseudorabies virus (4, 37), equine herpesvirus type 4 (2), and canine herpesvirus type 1 (28) DNA is also detected in lymphoid tissue during latency.
A small region of the BHV-1 genome, the latency-related (LR) gene, is abundantly transcribed in latently infected neurons (26). LR gene RNA is alternatively spliced (7, 17) and can be translated into more than one protein (22). LR gene proteins are expressed late in the productive infection cycle and are detected in a subset of latently infected neurons (17, 21). A BHV-1 strain with an LR gene mutation containing three stop codons at the beginning of the 5′ terminus of the LR gene transcript (the LR mutant) was recently constructed. The LR mutant expresses the LR gene transcript but not LR gene proteins (22). LR mutant and wild-type (wt) strains grow to similar endpoint titers in cultured bovine cells and in the nasal cavity of infected calves (18). However, calves infected with the LR mutant virus shed virus from the eye at lower titers and exhibit diminished clinical symptoms compared to the titers and symptoms in calves infected with wt BHV-1. In addition, lower levels of infectious virus are present in TG homogenates prepared from calves infected with the LR mutant relative to those in wt-infected calves, and calves latently infected with the LR mutant contain lower levels of viral DNA in TG (19). Finally, the LR mutant does not reactivate from latency following DEX treatment, in part, because the LR mutant induces higher levels of apoptosis in TG near the end of acute infection (27). Considering that BHV-1 modified live vaccines can reactivate from latency and are pathogenic in small calves (25), incorporation of the LR gene mutation into a modified live vaccine may prevent reactivation from latency. Thus, understanding of the pathogenic potential of the LR mutant and its ability to establish and reactivate from latency in the tonsil is important for determination of whether the LR gene mutation could be incorporated into a modified live vaccine strain.
In this study, we compared the abilities of LR mutant and wt viruses expressing LR gene products to grow in the tonsils of acutely infected calves and to establish and reactivate from latency. Expression of the LR gene by the wt virus stimulated the growth of BHV-1 in the tonsils of acutely infected calves. However, LR gene products do not have a major effect on apoptosis in the tonsils, perhaps because LR gene RNA is not abundantly expressed. Similar levels of viral DNA were detected in the tonsils of latently infected calves, regardless of which virus was used to infect the calves. Although we have no evidence that the LR mutant can reactivate from latency following DEX treatment of latently infected calves, the LR mutant virus can reactivate from latency when tonsil tissue is explanted.
MATERIALS AND METHODS
Viruses.
The Cooper strain of BHV-1 (wt virus) was obtained from the National Veterinary Services Laboratory, Animal and Plant Health Inspection Services, Ames, Iowa. To construct the LR mutant virus, 25 bp of the LR gene sequence that is near the first in-frame ATG of open reading frame ORF-2 was replaced with an oligonucleotide that contains a unique EcoRI restriction site and three stop codons to inhibit protein expression (18). This mutation was rescued by recombining the wt LR gene back into the LR gene locus of the LR mutant virus, and this strain was designated the LR gene rescued virus.
Animals.
BHV-1-free cross-bred calves (weight, ∼250 kg) were used for this study. Calves were inoculated with 107 PFU of the indicated virus into each nostril and eye for a total of 4 × 107 PFU/animal, as described previously (18, 19). Calves were housed under strict isolation and given antibiotics before and after BHV-1 infection to prevent secondary bacterial infection. Latently infected calves were injected intravenously with 100 mg of DEX at 60 days postinfection (dpi) to initiate reactivation from latency. Two additional intramuscular injections of DEX (25 mg) were given at 2 and 4 days after the initial injection of DEX to ensure that reactivation occurred. This protocol induces reactivation from latency in all calves latently infected with wt BHV-1 (19). All animal experiments were performed in accordance with the guidelines of the American Association of Laboratory Animal Care.
Tissue samples.
Tonsils and lymph nodes from calves infected with wt BHV-1, LR gene rescued virus, and the LR mutant were collected from acutely infected calves at 0, 2, 4, 6, 10, and 14 dpi. Tissue samples were also collected at 60 dpi (latency) and after DEX treatment to initiate reactivation (1, 2, 21, 28, 30, and 35 days). All tissue samples were stored at −100°C for nucleic acid extraction or were processed for histological methods.
Tonsil homogenates and virus titration.
Homogenates were prepared from 0.5 g of tonsil tissue with a glass tissue grinder as 10% solutions in minimal essential medium (MEM) containing antibiotics. Tonsil homogenates were centrifuged at 2,000 rpm for 30 min at 4°C (Beckman Avanti 30 centrifuge). The supernatant (125 μl) was added to 500 μl of medium (1:5 dilution), and then 1:5 serial dilutions were made. One hundred microliters of each dilution was added in quadruplicate to a 96-well plate. Each well contained 100 μl of MDBK cells (105 cells). After 4 days of incubation, the virus titer was measured by a 50% endpoint assay.
Tonsil explants and homogenates.
To prepare tonsil explants, tonsil tissue samples were minced into pieces of approximately 1 mm3 and washed in phosphate-buffered saline (PBS) containing antibiotics (penicillin and streptomycin). Five to six pieces were inoculated into petri dishes and overlaid with 4 ml of MEM containing 20% fetal bovine serum and antibiotics. MDBK cells were used as indicator cells for cocultivation of all explant cultures. The medium was changed every 3 to 4 days, and cultures were observed daily for a cytopathic effect.
Homogenates were prepared as a 10% solution in MEM containing antibiotics with glass tissue grinders. Tonsil homogenates were centrifuged at 2,000 rpm for 30 min at 4°C (Beckman Avanti 30 centrifuge). An aliquot of each supernatant (100 μl) was inoculated into MDBK cells in 24-well plates in duplicate. Cultures were observed daily for a cytopathic effect, and the supernatants were passaged every 3 days, for a total of four passages. All explant and homogenate cultures were tested for BHV-1 antigens by direct immunofluorescence with a BHV-1 polyclonal antibody (IBR FA conjugate; American BioResearch Lab). Cultures in 24-well plates were fixed in methanol-acetone (1:1) for 20 min at −20°C. FA conjugate diluted 1:50 was applied to each well. After incubation at 37°C for 45 min, the plates were washed three times in PBS and observed by immunofluorescence microscopy.
ISH.
DNA probes specific for the BHV-1 glycoprotein C (gC), ribonucleotide reductase, bICP0, and the LR gene sequences were synthesized and labeled by PCR in the presence of digoxigenin-dUTP (Boehringer Mannheim Corp., Indianapolis, Ind.), as described previously (43-46). Tissue sections were deparaffinized and rehydrated in a graded alcohol series. Deproteinization was carried out in 0.2 N HCl for 20 min at room temperature. Permeabilization with proteinase K (20 μg/ml; Boehringer Mannheim) was performed in PBS for 20 min at 37°C. After the tissue sections were rinsed with PBS (three times for 5 min each time), they were fixed with 4% paraformaldehyde in PBS for 5 min at room temperature. To reduce nonspecific binding of the probes, tissue sections were subjected to acetylation treatment in 0.1 M triethanolamine-HCl buffer (pH 8.0) with 0.25% acetic anhydride at room temperature. After 5 min of treatment, 0.25% acetic anhydride was added for an additional 5 min. The slides were then rinsed in 2× SSC (1× SSC is 150 mM NaCl plus 15 mM sodium citrate [pH 7.0]) for 5 min and then incubated for 1 h at 60°C in 200 μl of prehybridization mixture (in situ hybridization [ISH] mixture with 50% formamide; Amresco). Labeled probes were subsequently added to the prehybridization mixture, and hybridization was performed overnight at 60°C. The slides were then washed twice in 4× SSC for 5 min at room temperature, once in 2× SSC, and once in 0.5× SSC for 5 min at 40°C. The washes were then carried out at room temperature: twice in 2× SSC, twice in 0.5× SSC, and once in buffer I (100 mM maleic acid, 150 mM NaCl [pH 7.5]) for 5 min each time. The anti-digoxigenin-alkaline phosphatase conjugate (Boehringer Mannheim) was diluted in buffer II (1% blocking reagent in buffer I; Boehringer Mannheim), according to the recommendations of the manufacturer, and incubation was carried out for 1 h at room temperature. The slides were washed twice in buffer I and then once in buffer III (100 mM Tris-HCl, 100 mM NaCl, 50 mM MgCl2 [pH 9.5]) for 5 min each time. The slides were then incubated with color substrate solution consisting of 4-nitroblue tetrazolium chloride (Boerhinger Mannheim) and 5-bromo-4-chloro-3-indolylphosphate (X-phosphate; Boehringer Mannheim) in buffer III. The color reaction was stopped with TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]). The slides were counterstained with methyl green, and a coverslip was applied with Permount (Fisher).
Nucleic acid extraction.
Nucleic acid extraction was performed essentially as described by Chomczynski and Sacchi (5). Tonsil tissues were minced into small pieces, placed into 10 ml of solution D (4 M guanidine thiocyanate, 25 mM sodium citrate [pH 7.0], 0.5% sarcosyl, 14 mM β-mercaptoethanol), and homogenized. Two phenol extractions were performed, and the interface was saved for DNA extraction. The DNA was suspended in TEN buffer (50 mM Tris [pH 7.5], 100 mM NaCl, 10 mM EDTA) containing 0.5% sodium dodecyl sulfate and 0.3 mg of proteinase K/ml. This solution was digested overnight at 55°C. The DNA-containing solution was then extracted once with phenol-chloroform-isoamyl alcohol (25:24:1), precipitated with 2 volumes of ethanol, and suspended in Tris-EDTA to a final concentration of 1 μg/μl.
PCR.
An aliquot (2 μl) of DNA was used for each PCR with primers specific for the BHV-1 gC gene. The primers were GAGCAAAGCCCCGCCGAAGGA (forward primer) and TACGAACAGCAGCACGGGCGG (reverse primer). PCR was carried out in a 50-μl mixture containing 1× commercial PCR buffer, 200 μM each deoxynucleoside triphosphate, 1 μM each primer, and Taq polymerase. Amplification of the target was carried out for 40 cycles by denaturation at 95°C for 1 min, annealing at 65°C for 1 min, and extension at 72°C for 2 min. After the last cycle, the reaction mixtures were further incubated at 72°C for 7 min to ensure complete extension of the amplified product.
The amount of viral DNA in latently infected tonsils was assessed by semiquantitative PCR with BHV-1 gC gene-specific primers. The bovine growth hormone (GH) gene was amplified with the following primers (forward primer,GCTTTCGCCCTGCTCTGCC; reverse primer, TCCTGCCTCCCCACCCCTA). GH gene-specific primers were used to ensure that similar amounts of starting material were included in each reaction mixture. To demonstrate that the strategy used for the semiquantitative analysis was valid over a range of DNA concentrations, 10-fold dilutions of the positive control were amplified (30 cycles) in each assay to generate a standard curve. This ensured a linear relationship between the initial input DNA and the intensity of the product amplified from the gene for GH. The respective bands were captured by using a Bio-Rad FX Pro Plus device, and then the bands were quantified with Quantity One software (Bio-Rad).
In situ detection of apoptosis.
Tissue sections were deparaffinized in xylene for 10 min, rehydrated in a graded ethanol series, and treated with proteinase K (20 μg/ml; Gibco BRL). Proteinase K treatment was conducted with Tris buffer (100 mM Tris-HCl, 150 mM NaCl [pH 7.6]) for 20 min at 37°C. Nick end labeling of the DNA strand breaks in consecutive tissue sections was performed by the terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay, which uses alkaline phosphatase (Boehringer Mannheim). The slides were counterstained with methyl green, and then a coverslip was placed with Permount (Fisher). Apoptosis was evaluated in the following tonsillar compartments: GCs, crypts, epithelium, and interfollicular zones. The apoptotic cells observed in 10 different fields of each compartment were counted, and an intensity score was assigned on the basis of the average number of apoptotic cells counted for each tonsillar compartment. Scores were as follows: 0, no apoptotic cells; 1, 1 to 50 apoptotic cells; 2, 51 to 200 apoptotic cells; 3, 201 to 400 apoptotic cells; 4, 401 to 600 apoptotic cells; 5, 601 to 800 apoptotic cells; 6, ≥800 apoptotic cells. Statistical analysis was performed by the Kruskal-Wallis test for nonparametric data. The model statement included treatment (mock-infected, wt-infected, and LR mutant-infected calves), animal (treatment), day, tonsillar compartment, and their interactions (treatment-day, treatment-compartment, and treatment-day-compartment interactions). A 5% significance level was used. Means were compared by the Tukey-Kramer test at a 5% significance level.
RESULTS
Localization of viral DNA in tonsils by ISH.
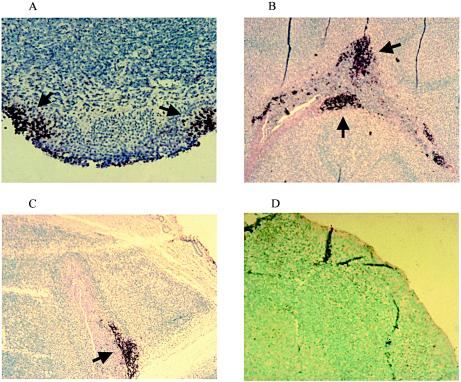
ISH was performed to localize the viral DNA in tonsil tissue during acute infection and latency following infection with the LR mutant or wt BHV-1. BHV-1-specific digoxigenin-labeled probes specific for four viral genes (the gC, bICP0, ribonucleotide reductase, and LR genes) were used for these studies. ISH-positive cells were consistently detected 2, 6, and 10 days after infection in the tonsils of calves infected with wt BHV-1 (Fig. 1A to C). Strong positive signals were primarily detected in areas adjacent to tonsillar crypts (Fig. 1B and C). Positive ISH signals were not readily detected in tonsil sections prepared from calves infected with the LR mutant during acute infection (data not shown). Viral DNA was not detected in the tonsils of mock-infected calves (Fig. 1D) or in the tonsils of calves latently infected with the LR mutant or wt BHV-1 (data not shown).
FIG. 1.
ISH of bovine tonsils after infection. Tonsil sections of calves at 2 (A), 6 (B), and 10 (C) dpi with wt BHV-1. (D) Representative tonsil sections from mock-infected calves. ISH was performed with the cocktail of viral probes described in Materials and Methods. The arrows denote the position of viral nucleic acid positive cells in the tonsil. Magnifications, ×1 (A and D) or ×40 (B and C).
Retropharyngeal and cervical lymph nodes, which drain from the initial site of infection, were also analyzed. Positive ISH signals were not readily detected with these tissues, regardless of the virus used to infect the calves (data not shown). A slight increase in the number of secondary lymphoid follicles was observed in the retropharyngeal lymph nodes but not in the cervical lymph nodes. Pathological differences in the lymph nodes of calves infected with the wt versus the LR mutant were not readily observed. In summary, these studies indicated that wt BHV-1 is readily detected by ISH in tonsils during acute infection, whereas LR mutant viral DNA is not.
Detection of viral DNA in tonsils of infected calves by PCR.
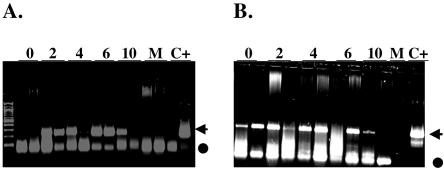
PCR assays were used to test whether viral DNA was present in the tonsils of calves infected with the LR mutant because the ISH results were negative. PCR was performed with gC-specific primers to detect viral DNA in pharyngeal tonsils at different times postinoculation. In wt-infected calves, the gC-specific amplification product was detected in the tonsils of both animals at 2 and 6 dpi and in one of two animals at 4 dpi (Fig. 2A). The expected 229-bp product was detected in the tonsils of both calves in the LR mutant-infected group at 2, 4, and 6 dpi (Fig. 2B). At 10 and 14 dpi, one of two calves contained detectable levels of viral DNA when the calves were infected with the LR mutant (Fig. 2B). Viral DNA was not readily detected in the tonsils at 14 dpi when the calves were infected with wt BHV-1 (Fig. 2A). Although amplified bands were detected in mock-infected MDBK cells (Fig. 2A), none of these bands was the specific amplified gC product. The ISH and PCR results suggest that lower levels of LR mutant viral DNA were present in the tonsil tissues of acutely infected calves.
FIG. 2.
PCR of DNA from tonsils at different times after infection. Calves were infected with wt BHV-1 (A) or the LR mutant strain (B) as described in the Materials and Methods. Data representative of those for two calves are shown for each day postinfection (as indicated by the numbers above the lanes). PCR was performed as described in Materials and Methods with gC BHV-1-specific primers that yield a 229-bp product (arrows). The gC band was excised and sequenced to confirm that it was the amplified product. PCR products were electrophoresed on 2% agarose gels, and the DNA was stained with ethidium bromide. The first (unlabeled) lane contains a 100-bp ladder (New England Biolabs). Mock-infected MDBK cells (lanes M) and MDBK cells infected with BHV-1 Cooper strain (lanes C+) were included in all PCRs as negative and positive controls, respectively. Closed circle, positions of primer dimers or bands resulting from mispriming.
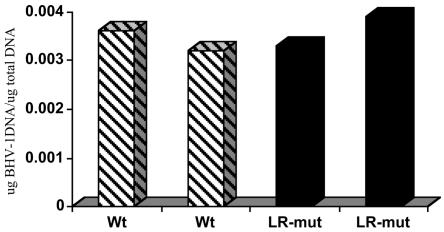
Previous studies demonstrated that lower levels of viral DNA were present in TG of calves latently infected with the LR mutant relative to the levels in calves latently infected with wt BHV-1 (19). Thus, it was of interest to test whether this was the case in the tonsils of calves latently infected with the two virus strains. We initially attempted real-time PCR, but the background levels of fluorescence in mock-infected samples and the control with no template were too high (data not shown). Consequently, we performed semiquantitative PCR using the gC-specific primers and bovine GH-specific primers to normalize the samples. The GH-specific primers were used to establish a standard curve for estimation of the levels of gC in tonsil tissue. This approach had an advantage, in that we knew the sizes of the amplified bands and were able to compare the intensities of the bands in the respective samples. In contrast to the results obtained with TG (19), similar levels of viral DNA were present in tonsils during latency (60 dpi) (Fig. 3). In summary, these studies demonstrated that the LR mutant viral DNA was present in the tonsils of latently infected calves and that similar levels of viral DNA were present in the tonsils during latency.
FIG. 3.
Quantitation of viral DNA in the pharyngeal tonsils of latently infected cattle. Semiquantitative PCR was performed with the virus-specific gC and the bovine GH gene-specific primers described in the Materials and Methods. A standard curve was used to measure the intensities of the GH amplification products and was generated with 10-fold serial dilutions of DNA. Under the PCR conditions used, the intensity of the GH gene band was linear when 0.6 to 0.00006 μg of total DNA from tonsil was used. The amount of total DNA used to detect the level of the gC-amplified product in the respective samples was within the linear range of PCR amplification. Two calves infected with the LR mutant (LR-mut) and two calves infected with wt BHV-1 were then analyzed at 60 dpi (latency). The gC-specific primers do not amplify the 229-bp gC-specific band with tonsil tissue samples from mock-infected animals (Fig. 2). The images of the ethidium bromide-stained gels were captured with a Bio-Rad FX Pro Plus device, and the bands were quantified and then compared to the standard curve generated with the GH gene. The values presented are expressed as the amount of viral DNA/micrograms total DNA present in the tonsil.
Isolation and titration of virus present in tonsils of acutely infected cattle.
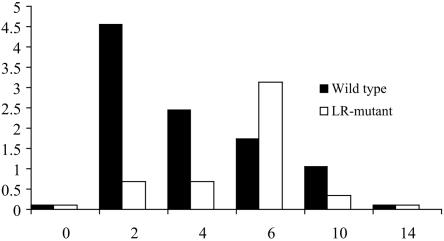
Infectious virus was isolated from two of two tonsils from calves infected with wt BHV-1 or the LR gene rescued virus at 2, 4, 6, and 10 dpi. The LR mutant strain was isolated from two of two tonsils at 2 and 6 dpi but from just one of two tonsils at 4 and 10 dpi. No infectious virus was recovered at 14 dpi, regardless of whether the calves were infected with wt BHV-1, the LR gene rescued virus, or the LR mutant. As expected, samples prepared from mock-infected calves were negative for virus isolation.
In general, 2 to 3 log units less virus was detected in homogenates prepared from calves infected with the LR mutant strain than from calves infected with the wt virus (Fig. 4). The exception to this was at 6 dpi, when virus titers from tonsil homogenates prepared from calves infected with the LR mutant were slightly higher than those from calves infected with wt BHV-1. This result suggests that virus infection was delayed in the tonsils of calves infected with the LR mutant and that, in general, the LR mutant grew less efficiently in the tonsils of infected calves. The decreased growth of the LR mutant in the tonsils was similar to the results obtained with TG and the eye (18, 19).
FIG. 4.
Isolation and titration of virus present in tonsils during acute infection. Virus was isolated from tonsils of mock-infected (day 0), LR mutant-infected, or wt-infected calves at different times after infection, as described in the Material and Methods. Virus titration was performed with MDBK cells, and the data represent the average for each group. Two calves in each group were used for this study. Negative results are represented by a value of 0.2 in order to visualize the bars.
Analysis of apoptosis in tonsil tissue.
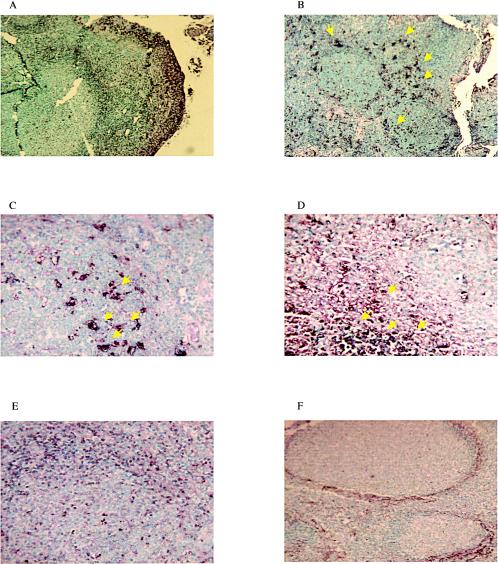
A previous study demonstrated that calves infected with BHV-1 contain numerous TUNEL assay-positive cells in their pharyngeal tonsils (43). In agreement with this finding, apoptosis was detected in the tonsils of the infected (Fig. 5A to E) and the noninfected (Fig. 5F) calves used for this study. Apoptotic cells were observed in all tonsillar compartments analyzed. Although apoptosis was detected in calves infected with the LR mutant (Fig. 5C and D), during the course of acute infection there appeared to be reduced levels in these calves compared to the levels in calves infected with wt BHV-1 or the LR gene rescued virus.
FIG. 5.
TUNEL assay with tonsil tissues of calves during acute infection. The TUNEL assay was performed as described in the Materials and Methods. Shown are tonsil tissue sections from wt BHV-1-infected calves at 2 (A), 4 (B), and 14 (E) dpi and from LR mutant-infected calves at 4 (C) and 6 (D) dpi. (F) Results for a mock-infected calf. The arrows denote areas of TUNEL assay-positive cells.
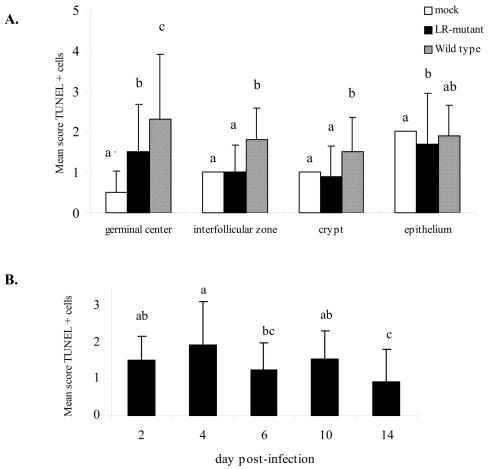
To determine the frequency of apoptosis in the tonsils of infected calves, the number of apoptotic cells in the tonsils of calves infected with wt BHV-1 was counted, and these results were compared to the apoptotic frequency in calves infected with the LR mutant or mock-infected calves. Significant differences (P < 0.01) in the number of apoptotic cells were detected in the different regions of the tonsils during acute infection (Fig. 6A). Analysis of apoptosis in the different tonsillar compartments revealed that the highest mean score of apoptotic cells was observed in tonsils from calves acutely infected with wt BHV-1. The highest score of TUNEL assay-positive cells for wt infection was observed in GCs. Statistically significant differences (P < 0.05) in the scores for GCs of calves infected with wt BHV-1 compared to the scores for the tonsils of calves infected with the LR mutant strain or the control group were detected (Fig. 6A).
FIG. 6.
Analysis of apoptosis in tonsils of calves infected with the LR mutant-, wt BHV-1-, or mock-infected calves. (A) Mean scores for apoptotic cells in different compartments of the tonsils of mock-, LR mutant-, or wt-BHV-1-infected calves. The results of the treatment-tonsil compartment interaction (P < 0.01) are shown. The number of apoptotic cells in consecutive tonsil tissue sections that stained positive by the TUNEL assay was counted as described in the Materials and Methods. The intensity score represents the average number of apoptotic cells counted in each tonsil compartment. The number of apoptotic cells in the epithelium infected with the wt was similar to the number in the epithelia infected with the LR mutant and mock infected, and this is denoted ab. Columns with different letters within each group are significantly different (P ≤ 0.05). (B) Comparison of the frequencies of apoptosis in the tonsils of calves infected with BHV-1. Overall differences in the scores of apoptotic cells in the tonsils at different times postinfection were not detected between wt BHV-1- and LR mutant-infected calves (P > 0.05). Therefore, the scores for apoptotic cells were analyzed independently of those for the infected group. The score at each time represents the average number of apoptotic cells counted on each day. Columns with different letters denote significant differences (P < 0.05) in the scores for apoptotic cells at different times after infection.
For the interfollicular zone and tonsillar crypts, significant differences were not detected (P > 0.05) when mock-infected calves were compared to calves infected with the LR mutant. A high score for apoptotic cells was observed in the epithelia of the tonsils from mock-infected calves, but this score was not significantly different (P > 0.05) from the score for calves infected with wt BHV-1 (Fig. 6A).
The effect of time after infection on the apoptotic score was analyzed independently of the treatment (Fig. 6B). These results demonstrated that wt BHV-1 and the LR mutant strain followed the same pattern with respect to the induction of apoptosis in tonsils during acute infection. The highest score for apoptotic cells was detected at 4 dpi, although it was not statistically different (P > 0.05) from the scores detected at 2 and 10 dpi. By 14 dpi, when latency was established (23, 24), a reduction in the scores for TUNEL assay-positive cells was detected. The mean score at day 14 was statistically different (P < 0.05) from those at day 2, 4, and 10 but not from that at day 6 (P < 0.05). In summary, these studies confirmed that apoptosis occurs in the tonsils of infected calves. With respect to GCs, higher frequencies of apoptosis were detected in calves infected with wt BHV-1, which correlated with the higher titers of virus in the tonsils of calves infected with wt BHV-1.
The LR mutant can reactivate in explanted tonsil tissue.
In our previous studies (19), we were not able to detect infectious virus from calves latently infected with the LR mutant following DEX treatment. In contrast, in all calves infected with wt BHV-1 or the LR mutant rescued virus, virus was reactivated from latency because infectious virus was readily detected and the BHV-1-specific antibody titers increased after DEX treatment. When the wt LR gene (32) was inserted into a herpes simplex virus type 1 (HSV-1) LAT null mutant, the resulting recombinant virus was capable of high levels of spontaneous reactivation in a rabbit ocular model. In contrast, the LAT null mutant exhibits low levels of spontaneous reactivation from latency (29, 32-35). When the LR gene containing the stop codons used to generate the LR mutant virus strain was inserted into the LAT null mutant, this recombinant virus behaved like the LAT null HSV-1 mutant (29). These studies indicate that the LR gene plays a crucial role during the latency-reactivation cycle.
It is possible to take TGs from mice or rabbits that are latently infected with HSV-1 and to explant the tissue and generate infectious virus (29, 32, 33, 39). In this assay, HSV-1 LAT is also important for the promotion of explant-induced reactivation. Consequently, we tested whether infectious virus could be detected when tonsil tissue from latently infected calves or calves that were latently infected and then treated with DEX were explanted. Infectious virus was detected when tonsil tissue from calves latently infected with LR gene-positive BHV-1 strains (the wt or the LR gene rescued virus) was explanted by 10 days after explantation (15 of 18 calves; Table 1). When tonsil tissues from calves latently infected with the LR mutant were explanted, 6 of 13 animals yielded infectious virus, but virus was not generally detected until 15 to 18 days after explantation (Table 2). As expected, infectious virus was not detected when tonsil tissue from mock-infected calves was explanted (Table 1 and 2). These studies indicate that the LR mutant virus is capable of undergoing explant-induced reactivation in the tonsils of latently infected calves but that the explant-induced reactivation is not as efficient as that obtained with wt BHV-1.
TABLE 1.
Results for wt BHV-1 and LR gene rescued virus from tonsil explants and homogenates during latencya
| Parameter | Mock infected | Latently infected (wt) | DEX, 24 h (wt) | DEX, 48 h (wt) | DEX, 21 days (wt) | DEX, 28 days (wt) | DEX, 35 days (wt) | DEX, 30 days (LR-R) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Calf no. | 44 | 52 | 42 | 63 | 228 | 245 | 71 | 72 | 50 | 44 | 3196 | 3198 | 51 | 54 | 54 | 57 | 65 | 1 | 2 | 124 | 142 |
| Explant infection result | − | − | + | + | + | + | − | + | + | + | + | + | − | + | − | + | − | + | + | + | + |
| Homogenate infection result | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
Tonsils from calves that were latently infected (60 dpi) with wt BHV-1 (wt) or the LR rescued virus (LR-R) were used to examine cell-free homogenates for infectious virus or to perform explant-induced reactivation. Samples were also obtained from latently infected calves after they were treated with DEX, as described in the Materials and Methods, to initiate reactivation from latency. The times after DEX initiated reactivation, in hours or days, are indicated. The procedures used to examine explant-induced reactivation and prepare the homogenates to identify cell-free virus are described in the Materials and Methods. The calf identification number is indicated because these numbers were arbitrarily assigned when the animals were purchased.
TABLE 2.
Results for LR mutant virus from tonsil explants and homogenates during latencya
| Parameter | Mock infected | Latently infected | DEX, 24 h | DEX, 48 h | DEX, 21 days | DEX, 28 days | DEX, 35 days | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Calf no. | 33 | 44 | 45 | 49 | 74 | 233 | 230 | 231 | 234 | 43 | 47 | 1001 | 3197 | 52 | 63 |
| Explant infection result | − | − | + | − | − | − | − | − | + | − | + | + | + | − | + |
| Homogenate infection result | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
Tonsils from calves that were latently infected (60 dpi) with the LR mutant virus were used to examine cell-free homogenates for infectious virus or to perform explant-induced reactivation. Samples were also obtained from latently infected calves after they were treated with DEX, as described in the Materials and Methods, to initiate reactivation from latency. The times after DEX initiated reactivation, in hours or days, are indicated. The procedures used to examine explant-induced reactivation and prepare the homogenates to identify cell-free virus are described in the Materials and Methods. The calf identification number is indicated because these numbers were arbitrarily assigned when the animals were purchased.
DISCUSSION
In this study, we compared the ability of wt BHV-1 and the LR mutant virus to grow in the tonsils of acutely infected calves and to establish and reactivate from latency in the tonsils. Previous studies with the LR mutant strain demonstrated that LR gene products promote virus growth in certain cell types during acute infection of cattle, for example, the eye and TG (18, 19). The present study demonstrated that the LR gene stimulated growth in tonsil tissue. Although the LR mutant was not consistently detected in the tonsils by ISH, sensitive PCR studies demonstrated that LR mutant DNA was present in the tonsils of acutely and latently infected calves. The finding that similar levels of viral DNA were detected in the tonsils of calves latently infected with the LR mutant or wt BHV-1 implies that small amounts of cells in the tonsil and/or circulating blood survive infection. The LR mutant established a persistent or latent infection in the tonsils but was not able to reactivate from latency in vivo after DEX treatment.
Our studies suggest that the LR gene stimulates virus growth in the tonsils because there is reduced growth of the LR mutant in the tonsils. Since the LR mutant grows in the nasal cavity to titers similar to those of wt BHV-1 (18), we suggest that similar levels of the respective viruses reached the tonsil. Although there was less virus growth during acute infection, similar levels of LR mutant and wt BHV-1 DNA were present in the tonsils of latently infected calves, indicating that the LR mutant establishes a persistent or latent infection in the tonsils. Our previous studies (19) demonstrated that the LR mutant did not reactivate from latency following DEX treatment, which led us to conclude that the LR gene is necessary for in vivo reactivation. In this study, we found that the LR mutant virus was capable of explant-induced reactivation, indicating that the LR mutant DNA is infectious following the establishment of latency in calves. Explant-induced reactivation is an ex vivo process, and thus, the immune response is not a significant limiting factor for virus growth and spread. Our results suggest that expression of LR gene products interfered with immune recognition of latently infected cells undergoing reactivation from latency. Furthermore, the ability of the LR gene to inhibit apoptosis (6) may allow maximal virus shedding during reactivation from latency in vivo, which is crucial for a successful reactivation episode in cattle. Finally, LR gene products may promote productive infection following a stress response in vivo because DEX was used to initiate reactivation from latency in cattle.
A previous study (27) demonstrated that the LR mutant induces higher levels of apoptosis near the end of acute infection, despite reduced virus shedding in TG. In contrast to the results obtained in TG, the LR gene did not play a dramatic role in regulating apoptosis in the tonsil, in part because it is not abundantly expressed. Although LR gene RNA is abundantly expressed in TG of infected calves (23, 24), LR gene RNA is not abundantly expressed in tonsil tissue prepared from latently infected calves (46; S. Perez; unpublished data), suggesting that proteins encoded by the LR gene are not abundantly expressed. Neuron-specific splicing of the LR gene transcript also occurs (7, 17), suggesting that proper splicing does not occur in the tonsil, and thus, LR gene proteins may not have potent antiapoptotic activity.
Apoptotic cells were detected in all tonsillar compartments examined, regardless of the virus that was used to infect calves. However, the highest level of apoptosis was detected in GCs and, to a lesser extent, interfollicular zones. GCs provide a specialized environment where B cells initiate the maturation program after the primary encounter with the antigen in the extrafollicular T-cell-rich zone (11). Other viruses that infect lymph nodes or lymphoid cells, such as human immunodeficiency virus (1), human herpesvirus 6 (20), and human herpesvirus 7 (38), induce the apoptosis of surrounding uninfected cells (bystander effect) because many T cells express Fas and the Fas ligand. The ability of viruses to induce apoptosis in a direct manner and by the bystander effect is an important mechanism for inducing immunosuppression (47). The ability of BHV-1 to induce apoptosis in CD4+ T cells in a direct manner (43) and by the bystander effect would contribute to transient immunosuppression during acute infection.
Although several commercially modified live vaccines directed against BHV-1 are available (42), these vaccine strains can establish latency and reactivate from latency (25, 40). Reactivation from latency does not frequently lead to recurrent disease, but it is the major means of BHV-1 transmission in the field. Since the existing modified live vaccines can be pathogenic in small calves and can cause abortion (42), it would be desirable to generate a vaccine strain of BHV-1 that does not reactivate from latency. Introduction of the LR gene mutation into the strain included in the present modified live vaccines that are commercially available may offer protection from challenge with a field strain, but the virus in the modified live vaccine would not reactivate from latency.
Acknowledgments
This study was supported by grants from USDA (grants 2002-02450 and 2003-02213) and the Public Health Service (grant 1P20RR15635).
REFERENCES
- 1.Badley, A., A. Pilon, A. Landay, and D. Lynch. 2000. Mechanisms of HIV associates lympocyte apoptosis. Blood 96:2951-2964. [PubMed] [Google Scholar]
- 2.Borchers, K., U. Wolfinger, and H. Ludwig. 1999. Latency-associated transcript of equine herpesvirus 4 in trigeminal ganglia of naturally infected horses. J. Gen. Virol. 80:2165-2171. [DOI] [PubMed] [Google Scholar]
- 3.Carter, J. J., A. D. Weinberg, A. Pollard, R. Reeves, J. A. Magnuson, and N. S. Magnuson. 1989. Inhibition of T-lymphocyte mitogenic responses and effects on cell functions by bovine herpesvirus 1. J. Virol. 63:1525-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung, A. K. 1995. Investigation of pseudorabies virus DNA and RNA in trigeminal ganglia and tonsil tissues of latently infected swine. Am. J. Vet. Res. 56:45-50. [PubMed] [Google Scholar]
- 5.Chomczynski, P., and N. Sacchi. 1987. Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 6.Ciacci-Zanella, J., M. Stone, G. Henderson, and C. Jones. 1999. The latency-related gene of bovine herpesvirus 1 inhibits programmed cell death. J. Virol. 73:9734-9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devireddy, L. R., and C. Jones. 1998. Alternative splicing of the latency-related transcript of bovine herpesvirus 1 yields RNAs containing unique open reading frames. J. Virol. 72:7294-7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devireddy, L. R., and C. J. Jones. 1999. Activation of caspases and p53 by bovine herpesvirus 1 infection results in programmed cell death and efficient virus release. J. Virol. 73:3778-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Favoreel, H., H. Nauwynck, and M. Pensaert. 1999. Immunological hiding of herpesvirus-infected cells. Arch. Virol. 145:1269-1290. [DOI] [PubMed] [Google Scholar]
- 10.Fuchs, M., P. Hubert, J. Detterer, and H. J. Rziha. 1999. Detection of bovine herpesvirus type 1 in blood from naturally infected cattle by using a sensitive PCR that discriminates between wild-type virus and virus lacking glycoprotein E. J. Clin. Microbiol. 37:2498-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guzman-Rojas, L., J. C. Sims-Mourtada, R. Rangel, and H. Martinez-Valdez. 2002. Life and death within germinal centers: a double-edged sword. Immunology 107:167-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanon, E., G. Keil, S. van Drunen Little-van den Hurk, P. Griebel, A. Vanderplasschen, F. A. Rijsewijk, L. Babiuk, and P. P. Pastoret. 1999. Bovine herpesvirus 1-induced apoptotic cell death: role of glycoprotein D. Virology 257:191-197. [DOI] [PubMed] [Google Scholar]
- 13.Hanon, E., S. Hoornaert, F. Dequiedt, A. Vanderplasschen, J. Lyaku, L. Willems, and P. P. Pastoret. 1997. Bovine herpesvirus 1-induced apoptosis occurs at the G0/G1 phase of the cell cycle. Virology 232:351-358. [DOI] [PubMed] [Google Scholar]
- 14.Hanon, E., G. Meyer, A. Vanderplasschen, C. Dessy-Doize, E. Thiry, and P. P. Pastoret. 1998. Attachment but not penetration of bovine herpesvirus 1 is necessary to induce apoptosis in target cells. J. Virol. 72:7638-7641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanon, E., A. Vanderplasschen, S. Lyaku, G. Keil, M. Denis, and P. P. Pastoret. 1996. Inactivated bovine herpesvirus 1 induces apoptotic cell death of mitogen-stimulated bovine peripheral blood mononuclear cells. J. Virol. 70:4116-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hariharan, M. J., C. Nataraj, and S. Srikumaran. 1993. Down regulation of murine MHC class I expression by bovine herpesvirus 1. Viral Immunol. 6:273-284. [DOI] [PubMed] [Google Scholar]
- 17.Hossain, A., L. M. Schang, and C. Jones. 1995. Identification of gene products encoded by the latency-related gene of bovine herpesvirus 1. J. Virol. 69:5345-5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inman, M., L. Lovato, A. Doster, and C. Jones. 2001. A mutation in the latency-related gene of bovine herpesvirus 1 leads to impaired ocular shedding in acutely infected calves. J. Virol. 75:8507-8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inman, M., L. Lovato, A. Doster, and C. Jones. 2002. A mutation in the latency related gene of bovine herpesvirus 1 interferes with the latency-reactivation cycle of latency in calves. J. Virol. 76:6771-6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inoue, Y., M. Yasukawa, and S. Fujita. 1997. Induction of T cell apoptosis by human herpesvirus 6. J. Virol. 71:3751-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang, Y., A. Hossain, M. T. Winkler, T. Holt, A. Doster, and C. Jones. 1998. A protein encoded by the latency-related gene of bovine herpesvirus 1 is expressed in trigeminal ganglionic neurons of latently infected cattle and interacts with cyclin-dependent kinase 2 during productive infection. J. Virol. 72:8133-8142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang, Y., M. Inman, Y. Zhang, N. A. Posadas, and C. Jones. 2004. A mutation in the latency related gene of bovine herpesvirus 1 (BHV-1) inhibits protein expression of a protein from open reading frame 2 (ORF-2) and an adjacent reading frame during productive infection. J. Virol. 78:3184-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones, C. 1998. Alphaherpesvirus latency: its role in disease and survival of the virus in nature. Adv. Virus Res. 51:81-133. [DOI] [PubMed] [Google Scholar]
- 24.Jones, C. 2003. Herpes simplex virus type 1 and bovine herpesvirus 1 latency. Clin. Microbiol. Rev. 16:79-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, C., T. J. Newby, T. Holt, A. Doster, M. Stone, J. Ciacci-Zanella, C. J. Webster, and M. W. Jackwood. 2000. Analysis of latency in cattle after inoculation with a temperature sensitive mutant of bovine herpesvirus 1 (RLB106). Vaccine 18:3185-3195. [DOI] [PubMed] [Google Scholar]
- 26.Kutish, G., T. Mainprize, and D. Rock. 1990. Characterization of the latency-related transcriptionally active region of the bovine herpesvirus 1 genome. J. Virol. 64:5730-5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lovato, L., M. Inman, G. Henderson, A. Doster, and C. Jones. 2003. Infection of cattle with a bovine herpesvirus 1 (BHV-1) strain that contains a mutation in the latency related gene leads to increased apoptosis in trigeminal ganglia during the transition from acute infection to latency. J. Virol. 77:4848-4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyoshi, M., Y. Ishii, M. Takiguchi, A. Takada, J. Yasuda, A. Hashimoto, K. Okasaki, and H. Kida. 1999. Detection of canine herpesvirus DNA in ganglionic neurons and the lymph node lymphocytes of latently infected dogs. J. Vet. Med. Sci. 61:375-379. [DOI] [PubMed] [Google Scholar]
- 29.Mott, K., N. Osorio, L. Jin, D. Brick, J. Naito, J. Cooper, G. Henderson, M. Inman, C. Jones, S. L. Wechsler, and G.-C. Perng. 2003. The bovine herpesvirus 1 LR ORF2 is crucial for this gene's ability to restore the high reactivation phenotype to a herpes simplex virus-1 LAT null mutant. J. Gen. Virol. 84:2975-2985. [DOI] [PubMed] [Google Scholar]
- 30.Mweene, A. S., K. Okazaki, and H. Kida. 1996. Detection of viral genome in non-neural tissues of cattle experimentally infected with bovine herpesvirus 1. Jpn J. Vet. Res. 44:165-174. [PubMed] [Google Scholar]
- 31.Nataraj, C., S. Eidmann, M. J. Hariharan, J. H. Sur, G. A. Perry, and S. Srikumaran. 1997. Bovine herpesvirus 1 downregulates the expression of bovine MHC class I molecules. Viral Immunol. 10:21-34. [DOI] [PubMed] [Google Scholar]
- 32.Perng, G.-C., B. Maguen, L. Jin, K. R. Mott, N. Osorio, S. M. Slanina, A. Yukht, H. Ghiasi, A. B. Nesburn, M. Inman, G. Henderson, C. Jones, and S. L. Wechsler. 2002. A gene capable of blocking apoptosis can substitute for the herpes simplex virus type 1 latency-associated transcript gene and restore wild-type reactivation levels. J. Virol. 76:1224-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perng, G.-C., D. Esmail, S. Slanina, A. Yukht, H. Ghiasi, N. Osorio, K. R. Mott, B. Maguen, L. Jin, A. B. Nesburn, and S. L. Wechsler. 2001. Three herpes simplex virus type 1 latency-associated transcript mutants with distinct and asymmetric effects on virulence in mice compared with rabbits. J. Virol. 75:9018-9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perng, G. C., E. C. Dunkel, P. A. Geary, S. M. Slanina, H. Ghiasi, R. Kaiwar, A. B. Nesburn, and S. L. Wechsler. 1994. The latency-associated transcript gene of herpes simplex virus type 1 (HSV-1) is required for efficient in vivo spontaneous reactivation of HSV-1 from latency. J. Virol. 68:8045-8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perng, G. C., H. Ghiasi, S. M. Slanina, A. B. Nesburn, and S. L. Wechsler. 1996. The spontaneous reactivation function of the herpes simplex virus type 1 LAT gene resides completely within the first 1.5 kilobases of the 8.3-kilobase primary transcript. J. Virol. 70:976-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rock, D., J. Lokensgard, T. Lewis, and G. Kutish. 1992. Characterization of dexamethasone-induced reactivation of latent bovine herpesvirus 1. J. Virol. 66:2484-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sabo, A., and J. Rajcani. 1976. Latent pseudorabies virus infection in pigs. Acta Virol. 20:208-214. [PubMed] [Google Scholar]
- 38.Secchiero, P., L. Flamand, D. Gibellini, E. Flacieri, I. Robuffo, S. Capitani, R. Gallo, and G. Zauli. 1997. Human herpesvirus 7 induces CD4+ T cell death by two distinct mechanisms: necrotic lysis in productively infected cells and apoptosis in uninfected or non productively infected cells. Blood 90:4502-4512. [PubMed] [Google Scholar]
- 39.Stevens, J. G., and M. L. Cook. 1971. Latent herpes simplex virus in spinal ganglia of mice. Science 173:843-845. [DOI] [PubMed] [Google Scholar]
- 40.Thiry, E., B. Brochier, J. Saliki, M. Pirak, and P. P. Pastoret. 1985. Excretion and reexcretion of thermosensitive and wild-type strains of infectious bovine rhinotracheitis virus after co-infection or two successive infections. Vet. Microbiol. 10:371-380. [DOI] [PubMed] [Google Scholar]
- 41.Tikoo, S. K., M. Campos, and L. A. Babiuk. 1995. Bovine herpesvirus 1 (BHV-1): biology, pathogenesis, and control. Adv. Virus Res. 45:191-223. [DOI] [PubMed] [Google Scholar]
- 42.van Drunen Littel-van den Hurk, S., S. K. Tikoo, X. Liang, and L. A. Babiuk. 1993. Bovine herpesvirus-1 vaccines. Immunol. Cell Biol. 71:405-420. [DOI] [PubMed] [Google Scholar]
- 43.Winkler, M. T., A. Doster, and C. Jones. 1999. Bovine herpesvirus 1 can infect CD4+ T lymphocytes and induce programmed cell death during acute infection of cattle. J. Virol. 73:8657-8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winkler, M. T., A. Doster, J. H. Sur, and C. Jones. 2002. Analysis of bovine trigeminal ganglia following infection with bovine herpesvirus 1. Vet. Microbiol. 86:139-155. [DOI] [PubMed] [Google Scholar]
- 45.Winkler, M. T., L. S. Schang, A. Doster, T. Holt, and C. Jones. 2000. Analysis of cyclins in trigeminal ganglia of calves infected with bovine herpesvirus-1. J. Gen. Virol. 81(Pt 12):2993-2998. [DOI] [PubMed] [Google Scholar]
- 46.Winkler, M. T. C., A. Doster, and C. Jones. 2000. Persistence and reactivation of bovine herpesvirus 1 in the tonsils of latently infected calves. J. Virol. 74:5337-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zaronski, C. C., J. M. McNally, B. L. Loman, K. A. Daniels, and R. M. Welsh. 2000. Bystander sensitization to activation-induced cell death as a mechanism of virus-induced suppression. J. Virol. 74:3650-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]