Abstract
We describe a simple genotyping method for Mycobacterium ulcerans based on PCR amplification of genomic regions between IS2404 and a frequently repeated GC-rich sequence. Application of this method to a global collection produced 10 M. ulcerans genotypes corresponding to their geographic origin.
Mycobacterium ulcerans is the causative agent of Buruli ulcer (BU), an emerging disease characterized by chronic and necrotizing skin ulcers. BU occurs mostly in rural communities in tropical regions and is the third most common mycobacterial disease in humans after tuberculosis and leprosy (1).
Presently, the mode of transmission and other key aspects of the epidemiology of BU are not fully understood, partly due to the apparent lack of genetic diversity in M. ulcerans (1-3, 6, 8-10). New typing tools with high resolution need to be explored for the study and better understanding of the epidemiology of BU disease. In this study, we exploited the high copy number, species specificity, and potential of the insertion sequence IS2404 to mediate genome rearrangements (8) and a frequently repeated GC-rich sequence (5′-CGG-CGG-CAA-CGG-CGG-CA-3′) in mycobacteria (7) to develop a simple PCR-based genotyping method for M. ulcerans.
The method, designated IS2404-Mtb2 PCR, involves the use of previously described outward-directed, IS2404-specific primers (MU3, 5′-CGC-GTG-GGT-CCC-TCG-GGT-CT-3′; and MU4, 5′-ATC-GCC-GAA-GCC-TGC-CGG-GAT-3′) (8) in combination with the oligonucleotide Mtb2 (5′-CGG-CGG-CAA-CGG-CGG-C-3′) targeting the repeated GC-rich motif to amplify DNA sequences located between adjacent and appropriately oriented copies of these elements. Previously, we used Mtb2 in combination with primers IS1 and IS2, directed at inverted repeats flanking IS6110, to develop a PCR-based epidemiological tool for the differentiation of Mycobacterium tuberculosis isolates (5).
We confirmed the presence of Mtb2 sequences in M. ulcerans by BLAST search of the M. ulcerans sequence database available on the BURULIST web server of the Institute Pasteur de Paris, Paris, France (http://genopole.pasteur.fr/Mulc/List.html).
Strains exhibiting differences in their genome architecture with respect to these elements show different banding patterns and are consequently differentiated. The M. ulcerans typing method 2426 PCR developed by Stinear et al. is based on this principle but amplifies regions between two insertion elements, IS2404 and IS2606 (9).
To investigate the IS2404-Mtb2 PCR, whole genomic DNA was prepared by the standardized method described by van Embden and coworkers (11). Briefly, suspensions of bacteria in Tris-EDTA buffer (pH 8.0) were digested with lysozyme (1 mg/ml). Suspensions were then treated with proteinase K (0.1 mg/ml) and sodium dodecyl sulfate (to 1%) and incubated with NaCl (0.6 M) and N-acetyl-N,N,N-trimethyl ammonium bromide. DNA was extracted with chloroform-isoamyl alcohol and precipitated with isopropanol. For the PCR amplification, each reaction mixture contained 3 μl of genomic DNA (about 10 ng/μl), 12.5 μl of water, 3 μl of PCR buffer with 1.5 mM MgCl2, a 20-pmol concentration of each primer, 5 mM deoxynucleotide triphosphate, 6 μl of Q solution, and 1 U of HotStarTaq DNA polymerase (QIAGEN, Hilden, Germany) in a final volume of 30 μl.
PCRs were run on a PTC 100 thermocycler (MJ Research, Waltham, Mass.) and consisted of an initial Taq activation and denaturation step of 95°C for 15 min, followed by 35 cycles of 94°C for 30 s, 65°C for 1 min, and 72°C for 1 min 30 s, and a final extension at 72°C for 10 min. The Agilent 2100 Bioanalyzer system (Agilent Technologies, Waldbronn, Germany) was used to electrophoretically separate 1 μl of PCR product. The resulting banding profiles were analyzed with the Bionumerics version 3.0 computer software (Sint-Martens-Latem, Belgium).
We tested this method with 32 M. ulcerans isolates representing a geographically diverse collection and 1 isolate (ITM 00-1026) classified as Mycobacterium marinum (2) but also harboring the M. ulcerans specific IS2404 element (4) and described previously as a “missing link” (4) (Table 1).
TABLE 1.
M. ulcerans strains investigated in this study
| Isolate | Origina | Source (other strain designation)b | 2426 PCR typec | IS2404-Mtb2 type |
|---|---|---|---|---|
| ITM 5142 | Australia | ATCC 19423 | Victorian | Victorian |
| ITM 94-1326 | Australia | L.S., 93160339 | Victorian | Victorian |
| ITM 94-1329 | Australia | F.P., 144727 | Victorian | Victorian |
| ITM 9550 | Australia | D.D., 17679 | NT | Victorian |
| ITM 9540 | Australia | D.D., 11098/70 | NT | Queensland |
| ITM 94-1324 | Australia | F.P., 176862 | NT | Queensland |
| ITM 9537 | PNG | K.J., 11878/70 | PNG I | PNG I |
| ITM 94-1331 | PNG | K.J., 186395 | PNG II | PNG II |
| ITM 03-524 | PNG | F.P. | NT | PNG I |
| ITM 94-1328 | Malaysia | K.J., 186510 | Malaysian | Malaysian |
| ITM 7922 | French Guyana | V.V., 141090018 | NT | South American |
| ITM 842 | Surinam | V.K., 701357 | Surinam | South American |
| ITM 5114 | Mexico | P.L. | Mexican | Mexican |
| ITM 5143 | Mexico | P.L. | Mexican | Mexican |
| ITM 8756 | Japan | ATCC 33728 | Asian | Japanese |
| ITM 98-912 | China | F.P. | Asian | Chinese |
| ITM 5150 | D.R. Congo | F.P. | NT | African |
| ITM 5152 | D.R. Congo | F.P. | African | African |
| ITM 5155 | D.R. Congo | F.P. | African | African |
| ITM 94-662 | Ivory Coast | F.P. | NT | African |
| ITM 96-657 | Angola | F.P. | NT | African |
| ITM 96-658 | Angola | F.P. | African | African |
| ITM 97-321 | Ghana | F.P. | NT | African |
| ITM 97-483 | Ghana | F.P. | NT | African |
| ITM 97-610 | Ghana | F.P. | African | African |
| ITM 94-856 | Benin | F.P. | African | African |
| ITM 97-111 | Benin | F.P. | African | African |
| ITM 98-239 | Benin | F.P. | NT | African |
| ITM 00-0040 | Benin | F.P. | NT | African |
| ITM 00-1213 | Benin | F.P. | NT | African |
| ITM 00-1441 | Benin | F.P. | NT | African |
| ITM 02-279 | Cameroon | F.P. | NT | African |
| ITM 00-1026 | France | F.P. | NT | M. marinumd |
D.R. Congo, Democratic Republic of Congo.
F. P., F. Portaels, Institute of Tropical Medicine, Antwerp, Belgium; L.S., L. Stanford, School of Pathology, London, United Kingdom; D.D., D. Dawson, Laboratory of Microbiology and Pathology, Queensland Health, Brisbane, Australia; V.K., P. Van Keulen, Academic Medical Centre, Amsterdam, The Netherlands; P.L., P. Lavalle, Centro Dermatologico Pascua, Mexico, Mexico; K.J., K. Jackson, Victorian Infectious Diseases Reference Laboratory, Victoria, Australia; V.V., V. Vincent, Institute Pasteur de Paris, Paris, France.
Genotype designation as determined by 2426 PCR (9). NT, not tested.
M. marinum genotype designation as determined by amplified fragment length polymorphism and phenotypic tests (4).
All the M. ulcerans isolates with the exception of isolate ITM 00-1441 were obtained from humans. Isolate ITM 00-1441 was recovered from an aquatic insect (Gerris sp.) in Benin (Portaels et al., unpublished results).
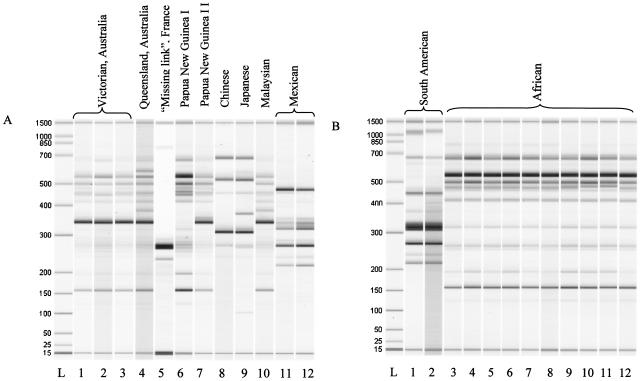
Comparison of IS2404-Mtb2 banding profiles revealed 10 different patterns, corresponding to the geographic origin of the isolates (Fig. 1). All of the 16 isolates originating from six different African countries produced an identical profile. The environmental isolate ITM 00-1441 also had the same profile as that of isolates from human origin. This homogeneous pattern is designated the African genotype. Two different genotypes were identified in east Asia (Japan and China genotypes), Australia (Victorian and Queensland genotypes corresponding to southeastern and northern Australia, respectively) and in Papua New Guinea (PNG I and II genotypes). The Malaysian genotype was nearly identical to the Queensland genotype, differing by the absence of one band in its profile. Similarly, there was one band difference between the profiles of the PNG I and Victorian genotypes. A South American genotype comprising identical profiles of two isolates from Surinam and French Guyana was shown to be different from the unique profiles of the Mexican and the missing link genotypes.
FIG. 1.
IS2404-Mtb2 PCR analysis of M. ulcerans isolates from different geographic regions and isolate ITM 00-1026, the missing link. (A) Lanes: L, DNA ladder; 1, ITM 5142; 2, ITM 1326; 3, ITM 1329; 4, ITM 9540; 5, ITM 00-1026; 6, ITM 9537; 7, ITM 94-1331; 8, ITM 98-912; 9, ITM 8756; 10, ITM 94-1328; 11, ITM 5143; 12, ITM 5114. (B) Lanes: L, DNA ladder; 1, ITM 842; 2, ITM 7922; 3, ITM 5150; 4, ITM 5155; 5, ITM 96-658; 6, ITM 96-657; 7, ITM 97-610; 8, ITM 02-279; 9, ITM 00-0040; 10, ITM 00-1213; 11, ITM 00-1441; 12, ITM 94-662. The 15- and 1,500-bp bands in all lanes represent lower and upper internal markers.
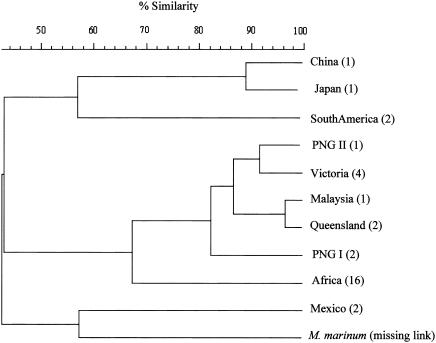
A dendrogram comparing the relationship among the genotypes was constructed with the similarity scores of the banding profiles and shows clustering of isolates from the same geographic region (Fig. 2). Among the geographic groupings, the Southeast Asian cluster (comprising the Malaysian, PNG I and II, Victorian, and Queensland genotypes) and the African genotype display about 68% similarity and represent the most closely related M. ulcerans genotypes originating from different geographic regions. The Mexican genotype and the missing link constituted a separate cluster least related to the other M. ulcerans genotypes. The link between these two genotypes may be consistent with the observation that both isolates contain comparatively fewer IS2404 elements (3).
FIG. 2.
Dendrogram showing relationships among the 33 isolates. Cluster analysis was performed by the unweighted pair group method, using the arithmetic average algorithm with the Bionumerics version 3.0 computer software (Sint-Martens-Latem, Belgium). Percent similarity was calculated with the band-based Dice correlation coefficient. The numbers in parentheses represent the number of isolates tested.
Overall, the results of the IS2404-Mtb2 PCR method corroborate previous M. ulcerans typing data (2, 3, 6, 8, 9, 10) in revealing a clonal population structure in M. ulcerans within a given geographic region. More interestingly, both 2426 PCR (9) and IS2404-Mtb2 PCR revealed two different M. ulcerans genotypes in Australia and Papua New Guinea.
2426 PCR analysis of a set of overlapping isolates used in this study produced nine M. ulcerans genotypes and has until now been the most discriminatory method for M. ulcerans. However, using the IS2404-Mtb2 PCR method, we could for the first time identify two different genotypes (i.e., Chinese and Japanese) in east Asia and have consequently been able to resolve 10 different M. ulcerans genotypes globally. In addition to the increased resolution, this method is reproducible and technically easy to perform. The results of this work allow for the speculation that limited IS2404-mediated genome reshuffling may be responsible for the apparent lack of genotype diversity in M. ulcerans, especially among African isolates. Variation in intensities of bands that may lead to inaccuracies in the assignment of genotypes could be a limitation of this approach. Here, this concern was addressed by using approximately equal concentrations of template DNA and maintaining uniform amplification conditions. High annealing temperature (65°C) was also used to ensure specific binding of primers to target sequences. While IS2404-Mtb2 PCR, like previous M. ulcerans genotyping methods, may not provide the requisite level of discrimination necessary for local epidemiological investigations, it could, however, be considered as a simple and rapid means of differentiating among M. ulcerans from different geographic regions.
Acknowledgments
This work was partly supported by a grant from the Fund for Scientific Research, Flanders (Belgium) (F.W.O-Vlaanderen. contract no. G.0301.01). A.A was supported by a grant from the Damien Foundation (Brussels, Belgium).
We are grateful to Juan Carlos Palomino, Karim Chemlal, Leen Rigouts, Anandi Martin, and Pieter Stragier for assistance and advice. We also thank Pim de Rijk, Krista Fissette, and Æecile Uwizeye for excellent technical work.
REFERENCES
- 1.Asiedu, K., R. Scherpbier, and M. Raviglione. 2000. Buruli ulcer infection: Mycobacterium ulcerans infection, p. 1-4. (W.H.O./CDS/CPE/GBUI/2000.1) World Health Organization, Geneva, Switzerland.
- 2.Chemlal, K., G. Huys, P.-A. Fonteyne, V. Vincent, A. G. Lopez, L. Rigouts, J. Swings, W. M. Meyers, and F. Portaels. 2001. Evaluation of PCR-restriction profile analysis and IS2404 restriction fragment length polymorphism and amplified fragment length polymorphism fingerprinting for identification and typing of Mycobacterium ulcerans and M. marinum. J. Clin. Microbiol. 39:3272-3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chemlal, K., K. de Ridder, P. A. Fonteyne, W. M. Meyers, J. Swings, and F. Portaels. 2001. The use of IS2404 restriction fragment length polymorphisms suggests the diversity of Mycobacterium ulcerans from different geographic areas. Am. J. Trop. Med. Hyg. 64:270-273. [DOI] [PubMed] [Google Scholar]
- 4.Chemlal, K., G. Huys, F. Laval, V. Vincent, C. Savage, C. Gutierrez, M.-A. Laneelle, J. Swings, W. M. Meyers, M. Daffe, and F. Portaels. 2002. Characterization of an unusual mycobacterium: a possible missing link between Mycobacterium marinum and Mycobacterium ulcerans. J. Clin. Microbiol. 40:2370-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kotlowski, R., I. Chola Shamputa, N. A. El Aila, A. Sajduda, L. Rigouts, A. van Deun, and F. Portaels. 2004. PCR-based genotyping of Mycobacterium tuberculosis with new GC-rich repeat sequences and IS6110 inverted repeats used as primers. J. Clin. Microbiol. 42:372-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Portaels, F., P. A. Fonteyne, H. de Beenhouwer, P. de Rijk, A. Guedenon, J. Hayman, and W. M. Meyers. 1996. Variability in 3′ end of 16S rRNA sequence of Mycobacterium ulcerans is related to geographic origin of isolates. J. Clin. Microbiol. 34:962-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poulet, S., and S. T. Cole. 1995. Repeated DNA sequences in mycobacteria. Arch. Microbiol. 163:79-86. [DOI] [PubMed] [Google Scholar]
- 8.Stinear, T., B. C. Ross, J. K. Davies, L. Marino, R. M. Robins-Brown, F. Oppedisano, A. Sievers, and P. D. R. Johnson. 1999. Identification and characterization of IS2404 and IS2606: two distinct repeated sequences for detection of Mycobacterium ulcerans by PCR. J. Clin. Microbiol. 37:1018-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stinear, T., J. K. Davies, G. A. Jenkin, F. Portaels, B. C. Ross, F. Oppedisano, M. Purcell, J. A. Hayman, and P. D. R. Johnson. 2000. A simple PCR method for rapid genotype analysis of Mycobacterium ulcerans. J. Clin. Microbiol. 38:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stinear, T. P., G. A. Jenkin, P. D. R. Johnson, and J. K. Davies. 2000. Comparative genetic analysis of Mycobacterium ulcerans and Mycobacterium marinum reveals evidence of recent divergence. J. Bacteriol. 182:6322-6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Embden, J. D. A., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, and P. M. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]