Abstract
Nontoxigenic strains of Corynebacterium diphtheriae represent a potential reservoir for the emergence of toxigenic C. diphtheriae strains if they possessed functional diphtheria toxin repressor (dtxR) genes. We studied the predominant strain of nontoxigenic C. diphtheriae circulating in the United Kingdom to see if they possessed dtxR genes and ascertain whether they were functional. A total of 26 nontoxigenic C. diphtheriae strains isolated in the United Kingdom during 1995 and 4 nontoxigenic strains isolated in other countries were analyzed by PCR and direct sequencing to determine the presence and intactness of the dtxR genes. The functionality of the DtxR proteins was assayed by testing for the production of siderophore in medium containing high and low concentrations of iron. PCR amplification and sequence analysis of the dtxR genes revealed four variants of the predicted DtxR protein among the nontoxigenic strains isolated in the United Kingdom. Production of siderophore in medium containing a low concentration of iron and repression of siderophore production in medium containing a high concentration of iron demonstrated that in all the strains the dtxR genes were functional. These findings demonstrate that, if lysogenised by a bacteriophage, nontoxigenic strains circulating in the United Kingdom could produce toxin and therefore represent a potential reservoir for toxigenic C. diphtheriae.
The production of diphtheria toxin by toxigenic strains of Corynebacterium diphtheriae is regulated in an iron-dependent manner by the diphtheria toxin repressor protein (DtxR) (13, 16). DtxR is the prototype of a family of metal-dependant regulatory proteins that have been identified in numerous gram-positive and gram-negative bacteria (14, 22, 25, 32). The physiological role of DtxR in C. diphtheriae is similar to that of the ferric uptake regulator, Fur. However, the two proteins have very little amino acid homology (3, 12, 29). The DtxR protein is known to function as a global regulator of metabolism in C. diphtheriae. It is not only involved in the regulation of expression of diphtheria toxin but also responsible for regulating the synthesis and production of the C. diphtheriae siderophore, corynebactin, as well as at least seven other promoters (15, 23, 27, 29, 31, 33, 39). These include iron-regulated promoters (IRPs) designated IRP1 through IRP5, as well as the promoters for the tox gene (36) and hmuO gene (the latter encodes a heme oxygenase that is essential for the acquisition of iron by C. diphtheriae from heme and hemoglobin) (27, 28).
Iron is required for the growth of virtually all living cells, primarily as a cofactor for essential enzymatic reactions. Fe3+ has very low solubility under physiological conditions, and most of the iron in body fluids is removed by high-affinity iron-binding proteins such as transferrin (serum) and lactoferrin (secretions) (38). The concentration of free iron in living tissues is estimated to be 10−12 μM, which is much lower than that required for bacterial growth (4). Invasive pathogenic bacteria overcome this problem by using inducible, high-affinity iron uptake systems, many of which involve siderophores, to scavenge iron from the host (1, 19). Siderophores are low-molecular-weight high-affinity ferric-ion-chelating agents, which are synthesized and secreted by many microorganisms growing under iron deprivation (20). These compounds solubilize and bind iron and transport it back into the bacterial cell, through specific membrane receptors (19).
In 2000, Reacher et al. (26) reported that a predominant strain of nontoxigenic C. diphtheriae is circulating within England and Wales. The discovery by Freeman in 1951 (7) that Tox− C. diphtheriae could be converted to the Tox+ state by the β corynephage demonstrated that nontoxigenic strains could convert to toxigenic forms. In 1955, Parsons (21), showed that under certain conditions Tox+ strains could emerge after exposure of a Tox− strain to the lysate of a second Tox− strain. In 1956, these observations were confirmed by Groman (10). These studies suggested that Tox− strains might themselves carry all or part of the tox gene and that under certain conditions a fully expressed tox gene could be recovered. In 1983, Groman et al. (9) demonstrated that nontoxigenic strains represent a potential reservoir for the tox gene and for the possible reemergence of toxigenic strains, assuming that these nontoxigenic strains possess functional diphtheria toxin repressor (dtxR) genes. Since there is a significant reservoir of nontoxigenic C. diphtheriae strains circulating in the United Kingdom, knowledge of the carriage rate of dtxR by these strains is essential for the understanding of the epidemiology of diphtheria and assessment of the risk of reemergence of fully toxigenic C. diphtheriae strains from indigenous strains. Therefore, studies were undertaken to determine whether the predominant strain of nontoxigenic C. diphtheriae circulating in the United Kingdom possessed fully functional dtxR genes. Twenty-six nontoxigenic strains, from different geographical regions in the United Kingdom, which belonged to the predominant ribotype, together with four nontoxigenic strains from other countries (Australia, Uzbekistan, Thailand, and Russia) and the genome-sequenced strain were analyzed by PCR amplification and sequencing of the dtxR gene. To determine whether intact dtxR genes were present and to examine their functionality, the strains were tested for the production of siderophore in high-iron and iron-limiting conditions.
MATERIALS AND METHODS
Bacterial strains.
For dtxR gene analysis, a total of 26 nontoxigenic C. diphtheriae strains of the predominant ribotype, isolated in the United Kingdom in 1995, were chosen. The isolates represented all geographical regions in the United Kingdom in which the predominant strain was circulating (Table 1). The dtxR gene of four nontoxigenic strains isolated in other countries (Australia, Uzbekistan, Thailand, and Russia [the Russian strain was a nontoxigenic tox-gene-bearing (NTTB) isolate of C. diphtheriae]), and the dtxR gene of the genome-sequenced strain C. diphtheriae NCTC 13129 (GenBank accession no. BX248353) were also analyzed for comparison. The siderophore assay was performed only on the isolates from the United Kingdom. The toxigenic C. diphtheriae strain Park-Williams no. 8 (PW8) and the nontoxigenic strains C7(β) and C7(β)hm723 were used as controls in the siderophore assay.
TABLE 1.
Summary of the 26 nontoxigenic C. diphtheriae isolates from the United Kingdom analyzed in this study
| Lab. no. | City and area of sending laboratory in the UK | Site of isolation | Biotype |
|---|---|---|---|
| 95/343 | Southwest London | Throat | Gravis |
| 95/392 | North central London | Throat | Gravis |
| 95/282 | Southwest London | Throat | Gravis |
| 95/311 | North London | Throat | Gravis |
| 95/421 | North central London | Throat | Gravis |
| 95/454 | North central London | Throat | Gravis |
| 95/429 | Bristol, Southwest England | Nose/throat | Gravis |
| 95/211 | Exeter, Southwest England | Throat | Gravis |
| 95/212 | Exeter, Southwest England | Throat | Gravis |
| 95/357 | Leeds, Yorkshire and Humber | Throat | Gravis |
| 95/475 | Sheffield, Yorkshire and Humber | Throat | Gravis |
| 95/354 | Derbyshire, East Midlands | Throat | Gravis |
| 95/412 | Derbyshire, East Midlands | Throat | Gravis |
| 95/12 | Middlesbrough, Northeast England | Throat | Gravis |
| 95/305 | Southport, Northwest England | Throat | Gravis |
| 95/476 | Preston, Northwest England | Throat | Gravis |
| 95/312 | Chester, Northwest England | Throat | Gravis |
| 95/407 | Chester, Northwest England | Throat | Gravis |
| 95/411 | Chester, Northwest England | Throat | Gravis |
| 95/403 | Manchester, Northwest England | Throat | Gravis |
| 95/390 | Cheshire, Northwest England | Throat | Gravis |
| 95/451 | Cheshire, Northwest England | Throat | Gravis |
| 95/22 | Jersey, Channel Islands | Throat | Gravis |
| 95/313 | Swansea, Wales | Throat | Gravis |
| 95/329 | Swansea, Wales | Throat | Gravis |
| 95/409 | Isle of Man | Throat | Gravis |
Amplification of the dtxR gene.
The dtxR gene was amplified using primers previously published by Nakao et al. (18). Primers DtxR 1F (5′-GGGACTGCAACTTAACAAG AA-3′) and DtxR 4R (5′-GCTAATTTCGCCGCCTTTAGT-3′) were used for amplification of the dtxR gene. Template DNA for the PCR amplifications was extracted as described previously by De Zoysa et al. (5). PCRs were performed with 0.5-ml microcentrifuge tubes (Thermoquest). A 100-μl reaction mix contained the following reagents: 10 μl of 10× PCR buffer (200 mM Tris-HCl [pH 8.4], 500 mM KCl) (Life Technologies), 3 μl of 50 mM MgCl2 (Invitrogen), 2 μl of each of 10 mM dATP, dCTP, dGTP, and dTTP (Invitrogen), 20 pmol of each of the two oligonucleotide primers (DtxR 1F and DtxR 4R) (Bioline), 1 U of Taq polymerase (5 U/μl), 20 ng of template DNA, and nuclease-free water (Promega) to a final volume of 100 μl. A control reaction mix, which contained all the reagents except the DNA template, was prepared each time the PCR amplifications were performed. Amplification was performed in a DNA Engine apparatus (MJ Research) using the following temperature profile: 95°C for 2 min, followed by 35 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min, and ending with a final 10-min extension at 72°C. After completion of the cycling, 10 μl of PCR product mixed with 5 μl of stop mix (10% Ficoll, 0.25% bromophenol blue, 50 mM EDTA) was electrophoresed on a 1% (wt/vol) agarose gel (Ultrapure; Invitrogen) in 1× TBE buffer (0.089 M Tris, 0.089 M boric acid, 0.002 M EDTA [pH 8.0]). A 100-bp ladder (Life Technologies) was run to gauge the size of the amplified PCR product. The gels were stained in ethidium bromide (1.0 μg/ml) and visualized under UV transillumination.
dtxR gene sequencing.
The amplicons were purified using the Wizard PCR Preps DNA purification system (Promega Corp., Madison, Wis.) as specified by the manufacturer. Sequencing was performed on each strand with the primers used in the initial PCR amplification (DtxR 1F and DtxR 4R) and primers DtxR 2F (5′-CTGAGCGTCTGGAACAATCT-3′) and DtxR 3R (5′-ATCCAACACGGATGTCAGCAT-3′). Nucleotide sequences were determined using the Dye Terminator cycle-sequencing kit (Beckman Coulter) and analyzed on a CEQ 2000 DNA analysis system (Beckman Coulter). Raw chromatograph data were edited using Genebuilder sequence analysis software, and multiple alignments were made in CLUSTAL W (35), which is part of the BioEdit software package (11). Sequences obtained in this study were compared with previously published dtxR sequences of C. diphtheriae strains C7(−) (GenBank accession no. M34239), C7hm723 (GenBank accession no. M80337), 1030(−) (GenBank accession no. M80336), and PW8 (GenBank accession no. M80338) (2, 3).
Siderophore assay.
The production of siderophores was determined using the Chrome Azurol S (CAS) agar plate assay as described by Tai et al. (33) with slight modifications. Isolates of C. diphtheriae were grown overnight at 37°C on deferrated tryptose yeast (TY) agar, and a dense cell suspension was prepared by harvesting the cells into 3 ml of modified PGT medium, using a sterile loop to achieve turbidity greater than a McFarland no. 6 standard. A 10-μl volume of bacterial suspension was pipetted onto the surface of agar plates containing 10 ml of deferrated TY agar medium (with no added iron [Fe3+] or with 2 μg of Fe3+ per ml added). The plates were incubated for 48 h at 37°C, and 25 ml of blue CAS top agarose was overlaid on each culture plate. The blue CAS top agarose contained 5 volumes of the CAS dye mixture, 2.5 volumes of modified PGT medium, and 2.5 volumes of melted 1% agarose. The plates were incubated at room temperature for 2 days. Siderophore production was apparent as a peach-colored halo around the spots of bacterial growth, whereas the medium surrounding spots of bacterial growth that did not produce siderophore remained blue. The sizes of the halos provided a semiquantitative measure of siderophore production by each strain.
Nucleotide sequence accession numbers.
Sequences of DtxR variants 5 through 7 have been deposited in GenBank under accession numbers AY741368 through AY741370, respectively.
RESULTS
Amplification and sequence analysis of the dtxR gene.
PCR amplification showed that all 26 C. diphtheriae isolates from the United Kingdom and the 4 nontoxigenic isolates from Australia, Uzbekistan, Thailand, and Russia possessed the dtxR gene.
The DNA sequence data obtained spanned positions −89 to 689 of the dtxR gene. Among the strains analysed from the United Kingdom and overseas, a total of 32 point mutations were detected within the 826-bp amplified fragment. Of these, 29 were located in the open reading frame (ORF) of the dtxR gene; the other 3 were in the 5′ noncoding region. In the amino-terminal half, three base substitutions were detected, and none resulted in an amino acid substitution (i.e., they were synonymous). In the carboxy-terminal half, 26 base substitutions were detected and 7 resulted in amino acid substitutions (i.e., they were nonsynonymous) (A147V, D199V, H201P, H201R, L214I, I221T, and I221M) (Fig. 1). The sequences were compared with previously published dtxR sequences, and an alignment of all the amino acid sequences is shown in Fig. 1. Numbers (1 through 7) were assigned to differentiate the different DtxR variants (Fig. 1). Each variant represents a different nucleotide sequence. Variants 1 through 4 have been published previously (refer to the bacterial strains for GenBank accession numbers). Five DtxR protein variants (variants 1, 4, 5, 6, and 7) were seen among the nontoxigenic strains from the United Kingdom and overseas. The previously published sequence of strain C7(−) (designated variant 1 in this study) does not have any amino acid changes, and this variant was found in 24 wild-type strains analysed (22 strains from the United Kingdom, the Russian NTTB strain, and the strain from Uzbekistan). The previously published sequences of strains C7hm723 and 1030 (designated variants 2 and 3, respectively, in this study) were not seen in any of the wild-type isolates analysed. DtxR variants 5, 6, and 7 were novel sequences and have been deposited in the GenBank sequence database. Variant 6 has one amino acid substitution (isoleucine to methionine at residue 221), and it was found in two isolates from the United Kingdom; variant 7 also has one amino acid substitution at residue 201 (histidine to arginine), and a single isolate from the United Kingdom produced this variant; variant 5 has three amino acid substitutions (aspartic acid to valine at residue 199, histidine to proline at residue 201, and isoleucine to threonine at residue 221), and a single isolate from the United Kingdom produced variant 5; variant 4 has two amino acid substitutions (alanine to valine at residue 147, leucine to isoleucine at residue 214), and it was found in the genome-sequenced strain NCTC 13129 and the nontoxigenic isolate from Thailand.
FIG. 1.
Deduced amino acid sequences of the dtxR alleles. Amino acids are shown only where changes were identified; amino acids identical to the consensus sequence are indicated by dots. The numbers on the left at the beginning of the sequences represent the different variants of the DtxR protein. (a) 95/392, 95/421, 95/12, 95/357, 95/313, 95/329; (b) 95/211, 95/212; (c) 95/343, 95/282, 95/311, 95/454, 95/476, 95/312, 95/411, 95/475, 95/395, 95/390, 95/451, 95/409, 95/22, 95/403, 92/100 (strain from Australia).
Siderophore production.
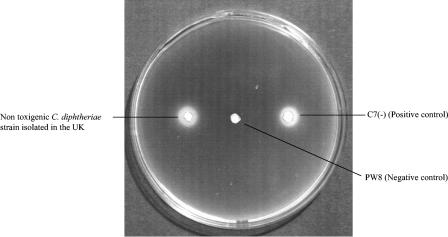
To determine whether the dtxR genes were functioning, siderophore production by the 26 nontoxigenic strains isolated in the United Kingdom was tested using the CAS agar plate assay. Isolates were grown on medium without any added iron and in medium containing 2.0 μg of Fe3+ per ml. The production of siderophore was apparent as a peach-colored halo around the spots of bacterial growth (Fig. 2). All isolates analyzed produced the siderophore, and the size of the halo produced by each strain was determined to give a semiquantitative measure of siderophore production by each strain. All strains produced halos larger than 5 mm in diameter. As expected, the mutant C7hm723 strain produced detectable siderophore in medium which did not contain any iron and in medium which contained 2.0 μg of Fe3+ per ml.
FIG. 2.
Production of siderophore. An agar plate containing deferrated TY agar medium was inoculated with a strain of nontoxigenic C. diphtheriae isolated in the United Kingdom with positive and negative control strains and was overlaid with CAS top agarose. Halos indicate siderophore production.
DISCUSSION
A total of 26 nontoxigenic strains isolated in the United Kingdom, together with 4 strains isolated from other countries (Australia, Thailand, Uzbekistan, and Russia) and strain NCTC 13129 (used as a control) were analyzed by PCR for the presence of the diphtheria toxin regulatory element (DtxR). The amplified products were studied by DNA direct sequencing and to determine whether the dtxR genes were functioning, the strains were tested for siderophore production. Amplification by PCR showed that all 26 nontoxigenic strains isolated in the United Kingdom in 1995 (belonging to the predominant ribotype) possessed the dtxR gene. Among the strains analyzed from the United Kingdom and overseas, sequencing revealed dtxR alleles that encode five variants of the DtxR protein. Four DtxR variants were found in the 26 nontoxigenic strains from the United Kingdom; 22 nontoxigenic isolates from the United Kingdom had entire dtxR sequences identical to that of PW8 strain. The dtxR allele from the genome-sequenced strain (NCTC 13129, a highly virulent toxigenic Gravis strain responsible for the Eastern European epidemic) was identical to that from 96/261, which is a nontoxigenic Gravis strain from Thailand. Sequencing detected considerable heterogeneity within the ORF of the dtxR gene. However, none of the amino acid substitutions appear to have inactivated the dtxR gene, since siderophore production was repressed in medium containing a high concentration of iron and derepressed in medium containing a low concentration of iron. Also, it was observed that the amount of siderophore produced by the toxigenic strain was no different from the amount produced by the nontoxigenic strains. Therefore, toxigenic strains do not necessarily produce more siderophore than nontoxigenic strains. However, whether variants of dtxR genes could be associated with increased or decreased toxin production is still unknown and should be tested.
In 1997, Nakao et al. (17) analyzed the heterogeneity of the dtxR gene from 72 C. diphtheriae isolates from Russia and Ukraine. These workers found 35 point mutations within the ORF; of these, 26 were silent mutations; 9 resulted in amino acid substitutions in the carboxyl-terminal half of the DtxR. A total of five variants of the DtxR protein were revealed in the strains from Russia and Ukraine. Some of the DtxR variants seen in this study were also seen by Nakao et al. (17) in Russian and Ukranian strains they analysed. DtxR variants 1, 4, and 5 were seen in the 72 isolates analyzed by Nakao et al. (17). As reported previously, mutant strain C7hm723, which produces siderophore under high- and low-iron conditions, contains a single nucleotide substitution resulting in substitution of histidine for arginine at residue 47 of DtxR; this single amino acid substitution is sufficient to almost abolish the regulatory activity of DtxR from strain C7hm723 (2, 30). The dtxR allele from strain PW8 is identical to that from strain C7(−), but dtxR from C. diphtheriae 1030 biotype Belfanti is only 91.7% identical to the allele from C7(−). Strain 1030 has 56 base substitutions and encodes a DtxR variant that differs from C7(−) DtxR by 6 amino acid substitutions in the carboxyl-terminal region (I165V, V174A, S191T, S205R, L214Y, and L218T) (2). The amino-terminal half includes the DNA-binding motif and the metal-binding motif (37). Boyd et al. (2) reported in 1992 that all six amino acid substitutions in C. diphtheriae 1030 are silent mutations and do not affect DtxR activity.
In 1993, Tao and Murphy (34) analyzed the function of DtxR by introducing mutations into the cloned dtxR gene and analyzing their effects on the expression of DtxR-regulated reporter genes in Escherichia coli. These workers established that all possible amino acid substitutions for C102 except aspartate abolished DtxR activity, while the C102D variant was significantly less active than wild-type DtxR. In 1994, Wang et al. (37) identified 20 mutations that affected single codons after random bisulfite mutagenesis of dtxR followed by phenotypic screening and nucleotide sequencing. Of these 20 mutations, 18 resulted in single amino acid substitutions in DtxR and 2 were chain terminating. Two DtxR variants with amino acid substitutions in domain 1 had the wild-type phenotype, and all the other DtxR variants exhibited decreased repressor activity varying from slight to considerable (37). Most of the substitutions in DtxR variants with decreased activity affected residues in or immediately adjacent to the DNA-binding motif, the dimer interface, or metal-binding site 2 (24, 37). In addition, the use of single alanine substitutions for each of the amino acids that serve as ligands for metal binding at site 1 (H79, E83, and H98) and site 2 (C102, E105, and H106) demonstrated that substitution at site 1 had little or no effect on repressor activity, and substitution at site 2 abolished repressor activity (6, 34). Furthermore, single alanine substitutions for residues D6, E9, M10, R13, and E17 in domain 1, which interact directly or indirectly with amino acids in domain 2 which take part in metal binding, resulted in decreased repressor activity. These workers concluded that metal-binding site 2 is the main binding site that functions directly in activation of DtxR and that site 1 plays a minor role in DtxR activation (6).
However, in 1999, Goranson-Siekierke et al. (8) reinvestigated single alanine substitutions for metal-binding residues at sites 1 and 2 and extended the investigation to include substitutions for anion-binding residues R80, S126, and N130 as well residue E20 in domain 1, which interacts directly with R80 in domain 2 via two hydrogen bonds. Their results confirmed that substitutions for metal-binding residues at site 1 did not decrease DtxR activity as much as substitutions at site 2 did. In contrast, replacing R80, S126, N130, or E20 with alanine decreased DtxR activity almost as much as the site 2 substitutions did. Therefore, their findings show that residues involved in anion binding at site 1 and the R80-E20 interaction are essential for repressor activity and support the conclusion that both the anion-cation-binding site 1 and cation-binding site 2 are important for DtxR function.
In conclusion, the wild-type strains analyzed in our study did not show any of the above amino acid substitutions, and those that were found are unlikely to affect dtxR function. When the strains were tested for siderophore production, all were positive. The currently circulating nontoxigenic C. diphtheriae strains therefore represent a potential source for the emergence of toxigenic C. diphtheriae in the United Kingdom.
REFERENCES
- 1.Bagg, A., and J. B. Neilands. 1987. Molecular mechanism of regulation of siderophore-mediated iron assimilation. Microbiol. Rev. 51:509-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyd, J. M., K. C. Hall, and J. R. Murphy. 1992. DNA sequences and characterization of dtxR alleles from Corynebacterium diphtheriae PW8(−), 1030(−), and C7hm723(−). J. Bacteriol. 174:1268-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd, J. M., M. N. Oza, and J. R. Murphy. 1990. Molecular cloning and DNA sequence analysis of a diphtheria tox iron-dependent regulatory element (dtxR) from Corynebacterium diphtheriae. Proc. Natl. Acad. Sci. USA 87:5968-5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bullen, J. J., H. J. Rogers, and E. Griffiths. 1978. Role of iron in bacterial infection. Curr. Top. Microbiol. Immunol. 80:1-35. [DOI] [PubMed] [Google Scholar]
- 5.De Zoysa, A., A. Efstratiou, R. C. George, M. Jahkola, J. Vuopio-Varkila, S. Deshevoi, G. Tseneva, and Y. Rikushin. 1995. Molecular epidemiology of Corynebacterium diphtheriae from northwestern Russia and surrounding countries studied by using ribotyping and pulsed-field gel electrophoresis. J. Clin. Microbiol. 33:1080-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding, X., H. Zeng, N. Schiering, D. Ringe, and J. R. Murphy. 1996. Identification of the primary metal ion-activation sites of the diphtheria tox repressor by x-ray crystallography and site-directed mutational analysis. Nat. Struct. Biol. 3:382-387. [DOI] [PubMed] [Google Scholar]
- 7.Freeman, V. J. 1951. Studies on the virulence of bacteriophage-infected strains of Corynebacterium diphtheriae. J. Bacteriol. 61:675-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goranson-Siekierke, J., E. Pohl, W. G. J. Hol, and R. K. Holmes. 1999. Anion-coordinating residues at binding site 1 are essential for the biological activity of the diphtheria toxin repressor. Infect. Immun. 67:1806-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groman, N. B., N. Cianciotto, M. Bjorn, and M. Rabin. 1983. Detection and expression of DNA homologous to the tox gene in non-toxigenic isolates of Corynebacterium diphtheriae. Infect. Immun. 42:48-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Groman, N. B. 1956. Conversion in Corynebacterium diphtheriae with phages originating from non-toxigenic strains. Virology 2:843-844. [DOI] [PubMed] [Google Scholar]
- 11.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 12.Hantke, K. 1984. Cloning of the repressor protein gene of iron regulated systems in Escherichia coli K-12. Mol. Gen. Genet. 197:337-341. [DOI] [PubMed] [Google Scholar]
- 13.Holmes, R. K. 1975. Genetic aspects of toxinogenesis in bacteria, p. 296-301. In D. Schlessinger (ed.), Microbiology—1975. American Society for Microbiology, Washington, D.C.
- 14.Jakubovics, N. S., A. W. Smith, and H. F. Jenkinson. 2000. Expression of the virulence-related Sca (Mn2+) permease in Streptococcus gordonii is regulated by a diphtheria toxin metallorepressor-like protein, ScaR. Mol. Microbiol. 38:140-153. [DOI] [PubMed] [Google Scholar]
- 15.Lee, J. H., T. Wang, K. Ault, J. Liu, M. P. Schmitt, and R. K. Holmes. 1997. Identification and characterization of three new promoter/operators from Corynebacterium diphtheriae that are regulated by the diphtheria toxin repressor (DtxR) and iron. Infect. Immun. 65:4273-4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy, J. R., P. Bacha, and R. K. Holmes. 1979. Regulation of diphtheria toxin production, p. 181-186. In D. Schlessinger (ed.), Microbiology—1979. American Society for Microbiology, Washington, D.C.
- 17.Nakao, H., I. K. Mazurova, T. Glushkevich, and T. Popovic. 1997. Analysis of heterogeneity of Corynebacterium diphtheriae toxin gene, tox, and its regulatory element, dtxR, by direct sequencing. Res. Microbiol. 148:45-54. [DOI] [PubMed] [Google Scholar]
- 18.Nakao, H., J. M. Pruckler, I. K. Mazurova, O. V. Narvskaia, T. Glushkevich, V. F. Marijevski, A. N. Kravetz, B. S. Fields, I. K. Wachsmuth, and T. Popovic. 1996. Heterogeneity of diphtheria toxin gene, tox, and its regulatory element, dtxR, in Corynebacterium diphtheriae strains causing epidemic diphtheria in Russia and the Ukraine. J. Clin. Microbiol. 34:1711-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neilands, J. B. 1982. Microbial envelope proteins related to iron. Annu. Rev. Microbiol. 36:285-309. [DOI] [PubMed] [Google Scholar]
- 20.Neilands, J. B. 1995. Siderophores: structure and function of microbial iron transport compounds. J. Biol. Chem. 270:26723-26726. [DOI] [PubMed] [Google Scholar]
- 21.Parsons, E. I. 1955. Induction of toxigenicity in non-toxigenic strains of C. diphtheriae with bacteriophage derived from non-toxigenic strains. Proc. Soc. Exp. Biol. Med. 90:91-93. [DOI] [PubMed] [Google Scholar]
- 22.Patzer, S. I., and K. Hantke. 2002. Dual repression by Fe2+-Fur and Mn2+ MntR of the mntH gene, encoding an NRAMP-like Mn2+ transporter in Escherichia coli. J. Bacteriol. 183:4806-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qian, Y., J. H. Lee, and R. K. Holmes. 2002. Identification of a DtxR regulated operon that is essential for siderophore-dependent iron uptake in Corynebacterium diphtheriae. J. Bacteriol. 184:4846-4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiu, X., C. L. Verlinde, S. Zhang, M. P. Schmitt, R. K. Holmes, and W. G. Hol. 1995. Three-dimensional structure of the diphtheria toxin repressor in complex with divalent cation co-repressors. Structure 3:87-100. [DOI] [PubMed] [Google Scholar]
- 25.Que, Q., and J. D. Helmann. 2000. Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol. Microbiol. 35:1454-1468. [DOI] [PubMed] [Google Scholar]
- 26.Reacher, M., M. Ramsay, J. White, A. De Zoysa, A. Efstratiou, G. Mann, A. Mackay, and R. C. George. 2000. Non-toxigenic Corynebacterium diphtheriae: an emerging pathogen in England and Wales? Emerg. Infect. Dis. 6:640-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmitt, M. P. 1997. Utilisation of host iron sources by Corynebacterium diphtheriae: identification of a gene whose product is homologous to eukaryotic heme oxygenases and is required for acquisition of iron from heme and hemoglobin. J. Bacteriol. 179:838-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmitt, M. P. 1997. Transcription of the Corynebacterium diphtheriae hmuO gene is regulated by iron and heme. Infect. Immun. 65:4634-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmitt, M. P., and R. K. Holmes. 1991. Iron-dependent regulation of diphtheria toxin and siderophore expression by the cloned Corynebacterium diphtheriae repressor gene dtxR in C. diphtheriae C7 strains. Infect. Immun. 59:1899-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmitt, M. P., and R. K. Holmes. 1991. Characterisation of a defective diphtheria toxin repressor (dtxR) allele and analysis of dtxR transcription in wild type and mutant strains of Corynebacterium diphtheriae. Infect. Immun. 59:3903-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitt, M. P., and R. K. Holmes. 1994. Cloning, sequence and footprint analysis of two promoter/operators from Corynebacterium diphtheriae that are regulated by the diphtheria toxin repressor and iron. J. Bacteriol. 176:1141-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmitt, M. P., M. Predich, L. Doukhan, I. Smith, and R. Holmes. 1995. Characterization of an iron-dependent regulatory protein (IdeR) of Mycobacterium tuberculosis as a functional homolog of the diphtheria toxin repressor (DtxR) from Corynebacterium diphtheriae. Infect. Immun. 63:4284-4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tai, S. P., A. E. Krafft, P. Nootheti, and R. K. Holmes. 1990. Coordinate regulation of siderophore and diphtheria toxin production by iron in Corynebacterium diphtheriae. Microb. Pathog. 9:267-273. [DOI] [PubMed] [Google Scholar]
- 34.Tao, X., and J. R. Murphy. 1993. Cysteine-102 is positioned in the metal binding activation site of the Corynebacterium diphtheriae regulatory element DtxR. Proc. Natl. Acad. Sci. USA 90:8524-8528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uchida, T., D. M. Gill, and A. M. Pappenheimer, Jr. 1971. Mutation in the structural gene for diphtheria toxin carried by temperate phage β. Nat. New Biol. 233:8-11. [DOI] [PubMed] [Google Scholar]
- 37.Wang, Z., M. P. Schmitt, and R. K. Holmes. 1994. Characterization of mutations that inactivate the diphtheria toxin repressor gene (dtxR). Infect. Immun. 62:1600-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weinberg, E. D. 1978. Iron and infection. Microbiol. Rev. 42:45-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilks, A., and M. P. Schmitt. 1998. Expression and characterisation of a heme oxygenase (HmuO) from Corynebacterium diphtheriae. J. Biol. Chem. 273:837-841. [DOI] [PubMed] [Google Scholar]