Abstract
The development of PCR-based genotyping modalities (spoligotyping and mycobacterial interspersed repetitive unit-variable-number tandem repeat [MIRU-VNTR] typing) offers promise for real-time molecular epidemiological studies of tuberculosis (TB). However, the utility of these methods depends on their capacity to appropriately classify isolates. To determine the operating parameters of spoligotyping and MIRU-VNTR typing, we have compared results generated by these newer tests to the standard typing method, IS6110 restriction fragment length polymorphism, in analyses restricted to high-copy-number IS6110 isolates. Sensitivities of the newer tests were estimated as the percentages of isolates with identical IS6110 fingerprints that had identical spoligotypes and MIRU-VNTR types. The specificities of these tests were estimated as the percentages of isolates with unique IS6110 fingerprints that had unique spoligotypes and MIRU-VNTR types. The sensitivity of MIRU-VNTR typing was 52% (95% confidence interval [CI], 31 to 72%), and the sensitivity of spoligotyping was 83% (95% CI, 63 to 95%). The specificity of MIRU-VNTR typing was 56% (95% CI, 51 to 62%), and the specificity of spoligotyping was 40% (95% CI, 35 to 46%). The proportion of isolates estimated to be due to recent transmission was 4% by identical IS6110 patterns, 19% by near-identical IS6110 patterns, 33% by MIRU-VNTR typing, and 53% by spoligotyping. The low calculated specificities of spoligotyping and MIRU-VNTR typing led to misclassification of cases, inflated estimates of TB transmission, and low positive predictive values, suggesting that these techniques have unsuitable operating parameters for population-based molecular epidemiology studies.
Tuberculosis (TB) molecular epidemiology exploits selected bacterial DNA targets to serve as markers for Mycobacterium tuberculosis strains. The most common method of DNA fingerprinting used is IS6110-based restriction fragment length polymorphism (RFLP). In a number of studies over the past decade, this modality has been validated for tracking TB transmission through two sets of observations. First, isolates from epidemiologically linked patients generally share identical or similar patterns (2, 3, 5). Second, matched RFLP patterns, when occurring among patients without known epidemiological links, are generally observed within groups with clear risk factors for TB transmission (1, 10, 13, 24).
An important practical limitation of IS6110 RFLP is that results are usually obtained weeks to months after the initial diagnosis of TB. This limitation stems from the need to grow large numbers of bacteria to extract DNA of sufficient quantity and quality for RFLP analysis. Therefore, while useful for documenting transmission events, IS6110 RFLP often provides data once outbreaks are well established. In contrast, a number of PCR-based typing modalities have been recently developed, including spoligotyping and mycobacterial interspersed repetitive unit-variable-number tandem repeat analysis (MIRU-VNTR) typing, which offer the possibility of obtaining DNA fingerprints from small numbers of bacteria or even directly from clinical specimens (9, 16). The major advantage of rapid typing of isolates is the capacity to immediately provide a strain designation, which in turn may facilitate prompt public health intervention. While the technical feasibility of these methods has been well demonstrated (12, 26), their public health utility remains to be determined.
To date, studies of small outbreaks have shown that results from spoligotyping and MIRU-VNTR typing are often identical among isolates clustered by IS6110-based RFLP (6, 7, 8, 11, 15, 16, 18, 23). However, the capacity of these modalities to provide unique patterns among unrelated isolates (i.e., their specificities) in a population-based study has not yet been evaluated. As part of an ongoing study of TB on the island of Montreal, we have typed all isolates over a 3-year period by using IS6110-based RFLP, spoligotyping, and MIRU-VNTR typing. Using IS6110-based RFLP as the reference typing modality, we have determined the sensitivities of these newer methods for detecting IS6110-based clusters and the specificities of these methods for providing distinct patterns among isolates unmatched by IS6110.
(Part of the information in this study was presented in abstract form [no. 74] at the International Union Against Tuberculosis and Lung Disease North American Region Conference, Austin, Tex., 25 to 28 February 2004.)
MATERIALS AND METHODS
Study setting.
The island of Montreal has a population of 1.8 million people with an average of 180 cases of tuberculosis diagnosed each year (10 cases per 100,000 people). The descriptive and molecular epidemiologies of TB in Montreal have been described elsewhere (14, 22). The cohort consisted of all Montreal TB patients notified between 1 January 1996 and 31 December 1998. This study was granted ethics approval by the Faculty of Medicine Institutional Review Board of McGill University.
Diagnosis and public health.
All diagnosed cases of tuberculosis are reported to the local public health authorities. Clinical and demographic information are entered into the provincial reportable disease registry, from which the data for this study were obtained in non-nominal format. About 85 to 90% of cases are culture positive, with culture confirmation, speciation, and antimicrobial susceptibility testing being performed at the provincial public health laboratory (Laboratoire de Santé Publique du Québec). There, an aliquot of each sample is stored at −80°C for future use and was later retrieved for this study.
Typing methods.
DNA extraction, IS6110 RFLP typing, spoligotyping, and MIRU-VNTR typing were performed per standard methods (11, 16, 27, 29). Results of all typing methods were read by three independent readers, and genotypes were assigned by consensus. IS6110 fingerprints were scanned into Gelcompar II (1998 version; Applied Maths) and only isolates with six or more IS6110 copies were included, based on the lower resolution of RFLP patterns for those with five bands or fewer (21). Spoligotypes and MIRU-VNTR types were entered manually into Microsoft Excel.
Fingerprint comparison.
IS6110 patterns were compared by using Molecular Fingerprint Analyzer 2.0 (Stanford University). Two criteria were applied to define matching IS6110 patterns. In the conservative analysis, a pattern was defined as identical if there was at least one other pattern in the database that had that same number of bands with the same molecular weights. If this condition was not met, the pattern was considered unique. In the less restrictive analysis, patterns were defined as similar if there was a single-band difference with at least one other pattern in the database and were defined as dissimilar if they differed by more than one band from all other patterns in the database. For spoligotypes and MIRU-VNTR patterns, we also performed two forms of analysis for matching. First, patterns were compared for identical matching of these modalities, defined as “identical” and “unique.” Next, we looked for whether spoligotypes or MIRU-VNTR patterns differed by only one spoligotype direct variable region or by one MIRU tandem repeat, by using a novel program that we created to facilitate spoligotype and MIRU-VNTR typing comparisons (available at http://www.med.mcgill.ca/epidemiology/Joseph/software.html). For any modality, when two or more isolates had identical or similar genotypes, they were considered to be clustered by that method. When MIRU-VNTR typing and spoligotyping was used in combination, both spoligotypes and MIRU-VNTR type patterns had to be identical for isolates to be considered clustered.
Sensitivity and specificity.
We use the terms sensitivity and specificity according to conventional epidemiological definitions, with results of IS6110-based RFLP serving as our referent. As sensitivity is defined as the likelihood that the diagnosis of interest will be detected by the test, we employed sensitivity to describe the proportion of IS6110-identical cases classified as identical by the newer typing methods. This definition was further restricted to require that identical matches involve the same isolates, as we were not only interested in whether the isolate was matched or unmatched, but also whether it matched an isolate with the same IS6110 pattern. As specificity refers to the likelihood that an individual without the diagnosis will test negative, we estimated specificity as the proportion of IS6110-unique cases classified as unique by the newer typing methods.
Predictive values.
Positive predictive value (PPV) and negative predictive value (NPV) were calculated from the two-by-two tables generated in the study and thus reflected the prevalence of IS6110-defined clustering found in this population. PPV was calculated as the percentage of individuals with a matched MIRU-VNTR typing or spoligotyping result that were clustered by IS6110; conversely, NPV was calculated as the percentage of individuals unmatched by MIRU-VNTR and spoligotyping for whom IS6110 results were unique.
Data analysis and statistics.
The sensitivities and specificities of spoligotyping and MIRU-VNTR typing were calculated when they were used alone or in combination, the latter requiring that isolates be identical by both spoligotyping and MIRU-VNTR typing. For each typing method, the percentage of clustering was determined and the percentage of transmission was inferred by using the n − 1 method, where the percentage of transmission = (100 × [number of clustered isolates − number of clusters]/total isolates) (24). SAS version 8.2 (Cary, N.C.) was used to calculate basic demographic and clinical characteristics and to perform bivariate analysis. We evaluated risk factors for clustering based on each of the following typing methods: (i) IS6110-identical matches, (ii) IS6110-similar matches, (iii) spoligotype-identical matches, (iv) MIRU-VNTR-identical matches, and (v) matches identical by a combination of spoligotype and MIRU-VNTR typing.
RESULTS
Data flow.
Five hundred twenty-seven TB cases were reported between 1 January 1996 and 13 December 1998; 453 isolates were sent for molecular typing. The majority of cases without genotyping were clinically diagnosed, culture-negative TB (n = 55). Upon IS6110 RFLP typing, 84 isolates had five bands or less and 22 isolates could not be typed, leaving 347 isolates with high-copy-number, interpretable IS6110 fingerprints. Of the 347 isolates, 326 (94%) were successfully typed by MIRU-VNTR, and 323 (93%) were successfully spoligotyped. There were no evident differences between patients included in the study and those excluded for reasons noted above, save for a greater proportion of foreign-born individuals in the group with low copy numbers of IS6110 (Table 1).
TABLE 1.
Patient characteristicsa
| Characteristic | No. of:
|
|||||
|---|---|---|---|---|---|---|
| Reported cases | Cultures testing negative | Isolates included in analysis | Isolates excluded from analysis | Isolates with a high copy no. | Isolates with a low copy no. | |
| No. of patients | 527 | 55 | 431 | 41 | 347 | 84 |
| Median age (yr) (IQR) | 39 (28-63) | 33 (18-62) | 40 (29-63) | 39 (31-64) | 40 (29-64) | 39 (27-61.5) |
| No. female (%) | 241 (46) | 31 (56) | 195 (45) | 15 (37) | 155 (45) | 40 (48) |
| No. Canadian born (%) | 95 (19) | 13 (24) | 77 (18) | 7 (17) | 68 (20) | 9 (11) |
| Median time (yr) since migration (IQR) | 5 (1-13) | 3 (1-13) | 5 (2-13) | 4 (1-14) | 5 (1-14) | 6.5 (3-13) |
| No. with pulmonary disease (%) | 333 (63) | 36 (66) | 270 (63) | 27 (66) | 221 (64) | 49 (58) |
| No. susceptible to all drugs (%) | 400 (85) | NA | 370 (86) | 30 (77) | 297 (86) | 73 (87) |
Demographic and clinical characteristics of patients included in and excluded from the study. IQR, interquartile range; NA, not applicable.
Test characteristics.
By using identical matches by IS6110 as the referent, the sensitivities, specificities, PPVs, and NPVs of spoligotyping, MIRU-VNTR, and the combination of the two tests were determined (Table 2). Of note, sensitivity estimates were based on relatively small denominators, hence their broad confidence intervals (CIs). In contrast, specificity estimates were based on 298 isolates with unique IS6110 RFLP profiles, assuring precision for point estimates. Of note, 178 of these 298 were matched by spoligotyping, yielding a specificity of just 40%. MIRU-VNTR typing fared only slightly better; 131 isolates were matched by MIRU-VNTR, resulting in a calculated specificity of 56%. From the calculated PPVs, it can be seen that when two individuals shared the same spoligotype or MIRU-VNTR, there was only a 1-in-10 chance that their IS6110 RFLPs were identical.
TABLE 2.
Operating characteristicsa
| Parameter evaluated | Operating characteristics of typing method(s)b
|
||
|---|---|---|---|
| MIRU-VNTR typing | Spoligotyping | Combined MIRU-VNTR-spoligotyping | |
| Sensitivity | 52 (31-72) [13/25] | 83 (63-95) [20/24] | 50 (29-71) [12/24] |
| Specificity | 56 (51-62) [169/300] | 40 (35-46) [120/298] | 70 (65-76) [204/290] |
| PPV | 9 (4-14) [13/144] | 10 (6-14) [20/198] | 12 (6-19) [12/98] |
| NPV | 93 (90-97) [169/181] | 97 (94-99.8) [120/124] | 94 (91-98) [204/216] |
Test characteristics of spoligotyping, MIRU-VNTR typing, and combined MIRU-VNTR-spoligotyping. Identical IS6110RFLP was used as the reference standard. When MIRU-VNTR and spoligotyping were used in combination, spoligotypes and MIRU-VNTRs both had to be identical for isolates to be considered clustered.
Values are percentages. Values in parentheses are 95% CIs. Values in brackets differ for each parameter and are as follows. Sensitivity is the number of isolates identical by the indicated typing method out of all isolates identical by IS6110 RFLP typing. Specificity is the number of isolates considered unique by the indicated typing method, out of all isolates unique by IS6110 RFLP typing. PPV is the number of isolates identical by IS6110 RFLP out of all isolates considered identical by the indicated typing method. NPV is the number of isolates unique by IS6110 RFLP out of all isolates considered unique by the indicated typing method.
Analysis incorporating IS6110 pattern evolution.
Because poor specificity of a novel test can occur when an overly stringent test is used as the referent, we recalculated the operating parameters using matching by similar IS6110 patterns. The sensitivities of the newer tests generally decreased while the specificities increased, although changes were modest and not statistically significant. For spoligotyping, the estimated specificity increased to 49% (95% CI, 42 to 56%) but the sensitivity dropped to 63% (95% CI, 53 to 73%). For MIRU-VNTR, specificity increased to 65% (95% CI, 59 to 72%) but the sensitivity was only 35% (95% CI, 25 to 45%). For the combined MIRU-VNTR-spoligotype match, the specificity was 80% (95% CI, 75 to 85%) but the sensitivity was only 33% (95% CI, 24 to 44%). In the latter scenario, the PPV was 42% (95% CI, 31 to 53%).
Analysis incorporating spoligotype and MIRU-VNTR pattern evolution.
Because the novel tests may also be susceptible to genotype evolution, we determined the sensitivities and specificities of these markers allowing single differences in patterns. For spoligotyping, when one spacer difference was allowed between matching patterns, the sensitivity of spoligotyping increased to 100% (95% CI, 88 to 100%) but the specificity decreased to 24% (95% CI, 19 to 29%). In the case of MIRU-VNTR, allowing a difference in one MIRU locus increased the sensitivity of MIRU-VNTR to 76% (95% CI, 55 to 91%) but the specificity decreased to 25% (95% CI, 21 to 31%).
Epidemiological inferences.
Based on the proportion of clustering by each modality, the estimated percentage of TB due to ongoing transmission was calculated, with the lowest estimate observed with IS6110 identity and the highest estimate with spoligotyping (Table 3). Using clusters so derived, we determined the risk factors for clustering (Table 4). By using IS6110 identity or similar IS6110 fingerprints, individuals born in Haiti had elevated odds ratios (ORs) for clustering (4.51 and 4.00, respectively). By MIRU-VNTR alone, being born in Haiti born was no longer unambiguously identifiable as a strong risk factor (OR, 1.59; 95% CI, 0.91 to 2.76); instead, being born in Canada emerged as a risk factor (OR, 2.50; 95% CI, 1.41 to 4.43). By employing spoligotyping, neither Canadian nor Haitian birth appeared to be a risk factor, but with combined MIRU-VNTR-spoligotyping, Canadian birth was a risk factor (OR, 1.84; 95% CI, 1.04 to 3.25).
TABLE 3.
Clusteringa
| Characteristic measured | Results for typing method indicated
|
||||
|---|---|---|---|---|---|
| IS6110 identical | IS6110 similar | MIRU-VNTR identical | Spoligotype identical | MIRU-VNTR and spoligotype identical | |
| No. of isolates | 347 | 347 | 326 | 323 | 315 |
| No. of clusters | 12 | 38 | 40 | 34 | 29 |
| No. of matched isolates (%) | 27 (8) | 104 (30) | 148 (45) | 205 (64) | 101 (32) |
| Median no. of isolates/cluster (range) | 2 (2-3) | 2 (2-8) | 2.5 (2-11) | 3.5 (2-32) | 2 (2-9) |
| % Transmission | 4 | 19 | 33 | 53 | 23 |
Clustering by each typing method. To be clustered by the combination of MIRU-VNTR and spoligotyping, isolates had to be identical by both methods.
TABLE 4.
Risk factors for clusteringa
| Risk factor | OR (95% CI) for typing method indicated
|
||||
|---|---|---|---|---|---|
| IS6110 identical | IS6110 similar | MIRU-VNTR identical | Spoligotype identical | MIRU-VNTR and spoligotype identical | |
| Age <50 yr | 1.76 (0.72-4.29) | 0.98 (0.61-1.58) | 0.95 (0.60-1.49) | 1.52 (0.96-2.42) | 1.3 (0.83-2.19) |
| Female | 1.61 (0.73-3.54) | 1.29 (0.81-2.04) | 0.89 (0.56-1.38) | 1.15 (0.73-1.82) | 1.01 (0.63-1.60) |
| Born in Canada | 1.17 (0.45-3.02) | 1.38 (0.79-2.42) | 2.50 (1.41-4.43) | 0.88 (0.50-1.55) | 1.84 (1.04-3.25) |
| Born in Haiti | 4.51 (2.01-10.13) | 4.00 (2.30-6.98) | 1.59 (0.91-2.76) | 1.31 (0.72-2.39) | 1.47 (0.83-2.62) |
ORs (95% CIs) for putative risk factors of clustering in Montreal. To be clustered by the combination of MIRU-VNTR and spoligotyping, isolates had to be identical by both methods. ORs were calculated by using unclustered isolates as the reference group.
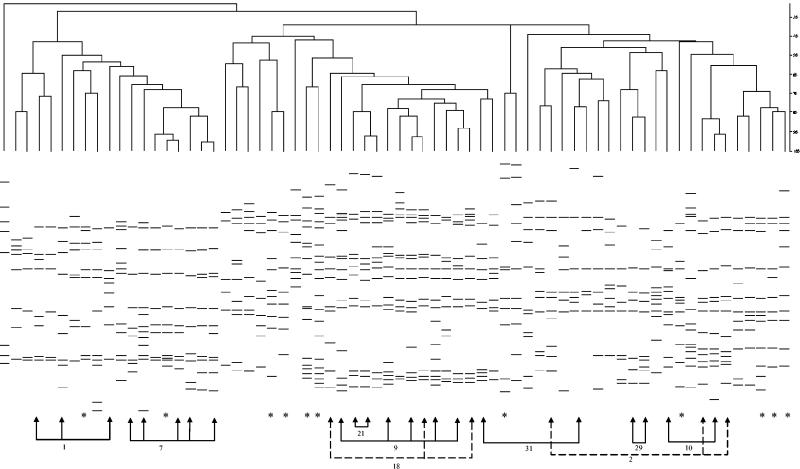
To explore MIRU-VNTR clustering in the Canadian born, we looked more closely at the isolates from Canadian-born individuals (Fig. 1). As we have previously documented an endemic strain of M. tuberculosis in Quebec (17), we wondered about the possibility of an endemic strain. Of note, while MIRU-VNTR clustered isolates with some IS6110 RFLP pattern similarity, the differences were usually much greater than are accepted as evidence of transmission and were often quite striking. Moreover, Canadian-born individuals represented only about 20% of all cases in this study; therefore, an endemic strain could have only minimal impact on overall estimates.
FIG. 1.
IS6110 RFLP patterns of isolates from Canadian-born individuals. Arrows indicate isolates with identical MIRU-VNTR types; the MIRU-VNTR cluster number is shown. Asterisks indicate isolates whose matching MIRU-VNTR result is from an individual born outside of Canada.
DISCUSSION
Genotypic analysis using IS6110-based RFLP has considerably advanced our understanding of TB epidemiology. With the advent of PCR-based genotyping techniques comes the potential for real-time molecular epidemiological investigation, whereby isolates can be rapidly typed within the same time frame as speciation and antibiogram determination. Our results, unfortunately, suggest significant limitations in the operating characteristics of these newer techniques, which likely compromise the epidemiological inferences so derived.
With respect to sensitivities, the denominators were small, so the estimates obtained had low precision. We found that spoligotyping missed two IS6110-identical pairs and that MIRU-VNTR typing failed to detect about half of the IS6110-identical matches. These results concur with observations about spoligotyping (6, 7) but appear to contrast with earlier reports about MIRU-VNTR, which suggested that patterns are generally identical within epidemiologically linked clusters (8). On further examination, we noted that for the discordant pairs, most IS6110-defined clusters had only one MIRU-VNTR locus difference between them, and spoligotypes were either identical or differed by one spacer. Therefore, it appears that we merely observed subtle changes in genotype in the same clone over time.
Of much greater concern were the extremely poor calculated specificities for both spoligotyping and MIRU-VNTR typing. Consequently, for every 10 clusters suggested by spoligotyping or MIRU-VNTR typing, only 1 was confirmed by IS6110. In Montreal, this nonspecificity generated estimates of ongoing transmission that were much higher than those obtained by using IS6110, and calculations aimed at determining risk factors for clustering failed to identify the only established risk group in our population (14, 22). While the rate of TB transmission in Montreal is low, the pretest probability of transmission links between two individuals in Montreal is still likely greater than two individuals across a country; therefore, the proportion of false-positive matches generated by these modalities could only be more problematic in national and international studies. Although combining these two modalities resulted in an estimate of ongoing transmission more in keeping with expectations, the risk factor analysis still failed to identify the known risk group, suggesting that combining these modalities does not overcome the problem of significant misclassification.
To explore reasons for the poor calculated specificities, we wondered about nonspecific patterns, akin to strains with low IS6110 copy numbers that produce RFLP patterns with low discrimination. Discarding the four most common spoligotypes (responsible for clusters of 32, 26, 16, and 14 isolates, respectively) increased the specificity only to 55%, and we did not have any large clusters defined by MIRU-VNTR (maximum number of isolates = 11). We noted that Canadian birth appeared to become a risk factor for clustering by MIRU-VNTR, but when we restricted our analysis to foreign-born individuals, the specificity of MIRU-VNTR was still only 62%. Together, these observations suggest that the low calculated specificities of these tests are a function of the techniques and not an artifact of our local TB epidemiology.
We hypothesize at least two reasons for the low calculated specificity: slower molecular clocks and/or genetic convergence. Previous studies looking at the numbers of different patterns obtained in cross sections of isolates have also reported the least diversity with spoligotyping, although MIRU-VNTR typing and IS6110-based RFLP have been seen as roughly equivalent (6, 7, 8, 16, 25). In a detailed analysis of pattern evolution within strain families in South Africa, Warren and colleagues were able to conclusively demonstrate convergence of spoligotypes (30). Together, these studies confirm that spoligotype patterns suffer from slower evolution and convergence. In an analysis of a genomically defined endemic strain in Quebec, Canada, we found the least variability with spoligotyping, but we also noted that variability of IS6110 RFLP results was considerably greater than that of MIRU-VNTR results (19). Confirming previous observations, we noted matching spoligotypes between isolates that had different deletion profiles and belonged to different principal genetic groups, but we also observed matching MIRU-VNTR patterns across these different groups (Nguyen et al., unpublished data). The latter observation suggests that MIRU-VNTR may also be prone to pattern convergence. While further studies are needed to clarify precisely why these tests manifest low specificities, the epidemiological consequences nonetheless appear considerable.
An important limitation of this study is the fact that IS6110 RFLP typing may not be a perfect “gold standard,” which potentially adversely affects the estimates obtained for the newer modalities. As shown in Fig. 1, a number of MIRU-VNTR clusters had similar but nonidentical IS6110 patterns, indicating either endemic strains or overly restrictive matching criteria for IS6110. While subtle changes in RFLP patterns have been observed with serial isolates (4, 20), with case contacts (3), and in population-based studies (1, 5, 24), we observed spoligotype and MIRU-VNTR matches with isolates with clearly different IS6110 RFLP patterns; therefore, this concern is unlikely to fully explain the nonspecificity of these modalities.
A second limitation is that to compare newer typing methods to IS6110, we have restricted our study to isolates with six or more IS6110 copies. This restriction follows from observations that IS6110 RFLP typing discriminates poorly among low-copy-number IS6110 isolates (21, 28). Consequently, we could not formally study the operating parameters of MIRU-VNTR typing and spoligotyping in these low-copy-number isolates, where they are often used as secondary typing modalities. As an approximation, the estimated proportion of unique isolates by these modalities was examined as a function of IS6110 copy number in the 84 low-copy-number isolates of our data set (data not shown). MIRU-VNTR resulted in a higher proportion of unique isolates than spoligotyping, regardless of IS6110 copy number. Together with the observation of a higher specificity by MIRU-VNTR in high-copy-number isolates, our results suggest that for classifying low-copy-number isolates, MIRU-VNTR typing appears more specific than spoligotyping.
Sensitivity and specificity are measures of how well a new test performs in securing a diagnosis. In the case of tuberculosis molecular epidemiology, an insensitive technique will overlook transmission events, while a nonspecific method will suggest transmission when none has occurred. The consequences of low sensitivity and specificity may be context dependent, but our data strongly suggest that both PCR-based modalities will result in greater estimates of ongoing transmission and a lesser capacity to detect high-risk groups. While potentially useful for outbreak investigations, to confirm laboratory cross-contamination, and as secondary modalities in low-copy-number strains, these data suggest that MIRU-VNTR typing and spoligotyping have unacceptable operating parameters for population-level epidemiological studies.
Acknowledgments
We thank Patrick Belisle, Dao Nguyen, Michael Purdy, and Jennifer Westley for technical assistance.
This work was supported by grants to K.S. from the Canadian Institutes for Health Research and M.A.B. from the Association Pulmonaire du Quebec and the Sequella Global Tuberculosis Foundation. D.M. is a Chercheur-Boursier of the FRSQ, L.J. is a Senior Investigator of CIHR, K.S. is a Chercheur-Boursier Clinicien of the FRSQ, and M.A.B. is a New Investigator of CIHR.
REFERENCES
- 1.Alland, D., G. E. Kalkut, A. R. Moss, R. A. McAdam, J. A. Hahn, W. Bosworth, E. Drucker, and B. R. Bloom. 1994. Transmission of tuberculosis in New York City. An analysis by DNA fingerprinting and conventional epidemiologic methods. N. Engl. J. Med. 330:1710-1716. [DOI] [PubMed] [Google Scholar]
- 2.Behr, M. A., P. C. Hopewell, E. A. Paz, L. M. Kawamura, G. F. Schecter, and P. M. Small. 1998. Predictive value of contact investigation for identifying recent transmission of Mycobacterium tuberculosis. Am. J. Respir. Crit. Care Med. 158:465-469. [DOI] [PubMed] [Google Scholar]
- 3.Daley, C. L., P. M. Small, G. F. Schecter, G. K. Schoolnik, R. A. McAdam, W. R. Jacobs, Jr., and P. C. Hopewell. 1992. An outbreak of tuberculosis with accelerated progression among persons infected with the human immunodeficiency virus. An analysis using restriction-fragment-length polymorphisms. N. Engl. J. Med. 326:231-235. [DOI] [PubMed] [Google Scholar]
- 4.de Boer, A. S., M. W. Borgdorff, P. E. de Haas, N. J. Nagelkerke, J. D. van Embden, and D. van Soolingen. 1999. Analysis of rate of change of IS6110 RFLP patterns of Mycobacterium tuberculosis based on serial patient isolates. J. Infect. Dis. 180:1238-1244. [DOI] [PubMed] [Google Scholar]
- 5.Genewein, A., A. Telenti, C. Bernasconi, C. Mordasini, S. Weiss, A. M. Maurer, H. L. Rieder, K. Schopfer, and T. Bodmer. 1993. Molecular approach to identifying route of transmission of tuberculosis in the community. Lancet 342:841-844. [DOI] [PubMed] [Google Scholar]
- 6.Goguet de la Salmonière, Y. O., H. M. Li, G. Torrea, A. Bunschoten, J. van Embden, and B. Gicquel. 1997. Evaluation of spoligotyping in a study of the transmission of Mycobacterium tuberculosis. J. Clin. Microbiol. 35:2210-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goyal, M., N. A. Saunders, J. D. van Embden, D. B. Young, and R. J. Shaw. 1997. Differentiation of Mycobacterium tuberculosis isolates by spoligotyping and IS6110 restriction fragment length polymorphism. J. Clin. Microbiol. 35:647-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hawkey, P. M., E. G. Smith, J. T. Evans, P. Monk, G. Bryan, H. H. Mohamed, M. Bardhan, and R. N. Pugh. 2003. Mycobacterial interspersed repetitive unit typing of Mycobacterium tuberculosis compared to IS6110-based restriction fragment length polymorphism analysis for investigation of apparently clustered cases of tuberculosis. J. Clin. Microbiol. 41:3514-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayward, A. C., and J. M. Watson. 1998. Typing of mycobacteria using spoligotyping. Thorax 53:329-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jasmer, R. M., J. A. Hahn, P. M. Small, C. L. Daley, M. A. Behr, A. R. Moss, J. M. Creasman, G. F. Schecter, E. A. Paz, and P. C. Hopewell. 1999. A molecular epidemiologic analysis of tuberculosis trends in San Francisco, 1991-1997. Ann. Intern. Med. 130:971-978. [DOI] [PubMed] [Google Scholar]
- 11.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kremer, K., D. van Soolingen, R. Frothingham, W. H. Haas, P. W. Hermans, C. Martín, P. Palittapongarnpim, B. B. Plikaytis, L. W. Riley, M. A. Yakrus, J. M. Musser, and J. D. van Embden. 1999. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J. Clin. Microbiol. 37:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubín, M., M. Havelková, I. Hynèicová, Z. Švecová, J. Kaustová, K. Kremer, and D. van Soolingen. 1999. A multidrug-resistant tuberculosis microepidemic caused by genetically closely related Mycobacterium tuberculosis strains. J. Clin. Microbiol. 37:2715-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulaga, S., M. Behr, K. Musana, J. Brinkman, D. Menzies, P. Brassard, D. Kunimoto, T. N. Tannenbaum, L. Thibert, L. Joseph, J. F. Boivin, and K. Schwartzman. 2002. Molecular epidemiology of tuberculosis in Montreal. Can. Med. Assoc. J. 167:353-354. [PMC free article] [PubMed] [Google Scholar]
- 15.Kwara, A., R. Schiro, L. S. Cowan, N. E. Hyslop, M. F. Wiser, H. S. Roahen, P. Kissinger, L. Diem, and J. T. Crawford. 2003. Evaluation of the epidemiologic utility of secondary typing methods for differentiation of Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 41:2683-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mazars, E., S. Lesjean, A. L. Banuls, M. Gilbert, V. Vincent, B. Gicquel, M. Tibayrenc, C. Locht, and P. Supply. 2001. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc. Natl. Acad. Sci. USA 98:1901-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen, D., P. Brassard, J. Westley, L. Thibert, M. Proulx, K. Henry, K. Schwartzman, D. Menzies, and M. A. Behr. 2003. Widespread pyrazinamide-resistant Mycobacterium tuberculosis family in a low-incidence setting. J. Clin. Microbiol. 41:2878-2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen, D., J.-F. Proulx, J. Westley, L. Thibert, S. Dery, and M. A. Behr. 2003. Tuberculosis in the Inuit community of Quebec, Canada. Am. J. Respir. Crit. Care Med. 168:1353-1357. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen, D., P. Brassard, D. Menzies, L. Thibert, R. Warren, S. Mostowy, and M. Behr. 2004. Genomic characterization of an endemic Mycobacterium tuberculosis strain: evolutionary and epidemiologic implications. J. Clin. Microbiol. 42:2573-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niemann, S., E. Richter, and S. Rüsch-Gerdes. 1999. Stability of Mycobacterium tuberculosis IS6110 restriction fragment length polymorphism patterns and spoligotypes determined by analyzing serial isolates from patients with drug-resistant tuberculosis. J. Clin. Microbiol. 37:409-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhee, J. T., M. M. Tanaka, M. A. Behr, C. B. Agasino, E. A. Paz, P. C. Hopewell, and P. M. Small. 2000. Use of multiple markers in population-based molecular epidemiologic studies of tuberculosis. Int. J. Tuberc. Lung Dis. 4:1111-1119. [PubMed] [Google Scholar]
- 22.Rivest, P., T. Tannenbaum, and L. Bedard. 1998. Epidemiology of tuberculosis in Montreal. Can. Med. Assoc. J. 158:605-609. [PMC free article] [PubMed] [Google Scholar]
- 23.Savine, E., R. M. Warren, G. D. van der Spuy, N. Beyers, P. D. van Helden, C. Locht, and P. Supply. 2002. Stability of variable-number tandem repeats of mycobacterial interspersed repetitive units from 12 loci in serial isolates of Mycobacterium tuberculosis. J. Clin. Microbiol. 40:4561-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Small, P. M., P. C. Hopewell, S. P. Singh, A. Paz, J. Parsonnet, D. C. Ruston, G. F. Schecter, C. L. Daley, and G. K. Schoolnik. 1994. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N. Engl. J. Med. 330:1703-1709. [DOI] [PubMed] [Google Scholar]
- 25.Sola, C., I. Filliol, E. Legrand, S. Lesjean, C. Locht, P. Supply, and N. Rastogi. 2003. Genotyping of the Mycobacterium tuberculosis complex using MIRUs: association with VNTR and spoligotyping for molecular epidemiology and evolutionary genetics. Infect. Genet. Evol. 3:125-133. [DOI] [PubMed] [Google Scholar]
- 26.Supply, P., S. Lesjean, E. Savine, K. Kremer, D. van Soolingen, and C. Locht. 2001. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39:3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, and T. M. Shinnick. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Soolingen, D., P. E. de Haas, P. W. Hermans, P. M. Groenen, and J. D. van Embden. 1993. Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J. Clin. Microbiol. 31:1987-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Soolingen, D., P. W. Hermans, P. E. de Haas, D. R. Soll, and J. D. van Embden. 1991. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J. Clin. Microbiol. 29:2578-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warren, R. M., E. M. Streicher, S. L. Sampson, G. D. van der Spuy, M. Richardson, D. Nguyen, M. A. Behr, T. C. Victor, and P. D. van Helden. 2002. Microevolution of the direct repeat region of Mycobacterium tuberculosis: implications for interpretation of spoligotyping data. J. Clin. Microbiol. 40:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]