Abstract
A multilocus sequence typing (MLST) scheme that uses the same loci as a previously described system for Campylobacter jejuni was developed for Campylobacter coli. The C. coli-specific primers were validated with 53 isolates from humans, chickens, and pigs, together with 15 Penner serotype reference isolates. The nucleotide sequence of the flaA short variable region (SVR) was determined for each isolate. These sequence data were compared to equivalent information for 17 C. jejuni isolates representing the known genetic diversity of this species. C. coli and C. jejuni share approximately 86.5% identity at the nucleotide sequence level within the MLST loci. There is evidence of genetic exchange of the housekeeping genes between the two species, but at a very low rate; only one sequence type from each species showed evidence of imported DNA. The flaA gene was more variable and has been exchanged many times between the two species, making it an unreliable marker for species identification but useful for distinguishing closely related strains. All but 3 of 21 human C. coli clinical isolates were distinct, according to the combined MLST and SVR sequences. The use of a common MLST scheme allows direct comparisons of the population biology and molecular epidemiology of these two closely related human pathogens.
Campylobacter is the most common bacterial cause of gastroenteritis in industrialized countries (10), with an estimated 2.5 million cases in the United States each year (19). Campylobacter jejuni accounts for approximately 90% of infections, and Campylobacter coli accounts for most of the remainder (11). C. coli alone caused more than 25,000 cases of gastroenteritis in England and Wales during 2000 (26). Multilocus sequence typing (MLST) is a high-resolution bacterial genotyping technique which has been useful for studies of the population structure and molecular epidemiology of C. jejuni (3, 4, 5, 7, 23). MLST data are directly comparable and easily shared via the Internet (http://pubmlst.org/campylobacter/). They are insensitive to genome instability and are able to overcome certain limitations of serotyping, pulsed-field gel electrophoresis (PFGE), and flaA typing (12, 13, 24, 29).
MLST of C. jejuni has identified a number of clonal complexes or lineages (5, 25) which comprise groups of closely related strains. Several of the clonal complexes that are responsible for human disease have also been shown to colonize mammalian or avian species (5, 18). A combination of MLST and sequencing of the short variable region (SVR) of the flaA flagellin gene allow resolution equivalent to that of PFGE for outbreak investigations (18, 23). A similarly rigorous typing scheme is also required for C. coli, since the epidemiologies of C. coli and C. jejuni infections may be different (11). The present study extended the existing MLST and flaA typing schemes for C. jejuni to C. coli by developing new MLST primers specific for C. coli. The use of a common typing scheme for both species allows their genetic variability, relatedness, and population biology to be compared.
MATERIALS AND METHODS
Campylobacter isolates.
C. coli isolates (n = 68) were obtained from a variety of sources and locations by standard microbiological procedures. Isolates (n = 53) were obtained from the stools of human patients with gastroenteritis (n = 21), colonized chickens (n = 24), and colonized pigs (n = 8). The human isolates comprised consecutive C. coli isolates obtained in a clinical microbiology laboratory (John Radcliffe Hospital, Oxford, United Kingdom) between 15 September 2003 and 30 January 2004. The chicken isolates were from anal swab specimens taken from live birds on the Oxford University farm and at Northmoor Trust site (both in Oxfordshire, United Kingdom) between 7 January 2003 and 1 August 2003. The pig isolates were from pig feces collected from the Oxford University farm on 2 February 2004. An additional 15 C. coli reference isolates from the Penner serotyping scheme (21) were included as representatives of the known phenotypic capsular diversity of the species (see Table 2).
TABLE 2.
MLST and flaA SVR data
| Isolate | Isolation date (yr.mo.day) | Isolation source | Countrya | STb | Allele nucleotide sequence no.c
|
SVR
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aspA | glnA | gltA | glyA | pgm | tkt | uncA | Allele no. | Peptide no. | |||||
| 2 | 2003.09.15 | Human stool | UK | 868 | 81 | 104 | 81 | 113 | 143 | 119 | 67 | 294 | 98 |
| 12 | 2003.09.18 | Human stool | UK (Greece)d | 825 | 33 | 39 | 30 | 82 | 113 | 47 | 17 | 336 | 106 |
| 18 | 2003.09.22 | Human stool | UK (Malawi)d | 832 | 33 | 39 | 30 | 79 | 113 | 43 | 17 | 243 | 1 |
| 23 | 2003.09.26 | Human stool | UK | 826 | 33 | 39 | 30 | 114 | 104 | 35 | 17 | 256 | 97 |
| 26 | 2003.09.29 | Human stool | UK | 827 | 33 | 39 | 30 | 82 | 104 | 56 | 17 | 255 | 1 |
| 27 | 2003.09.29 | Human stool | UK (Pakistan)d | 872 | 33 | 39 | 30 | 82 | 113 | 44 | 17 | 30 | 11 |
| 29 | 2003.09.29 | Human stool | UK | 825 | 33 | 39 | 30 | 82 | 113 | 47 | 17 | 253 | 1 |
| 40 | 2003.10.06 | Human stool | UK (Mexico)d | 825 | 33 | 39 | 30 | 82 | 113 | 47 | 17 | 292 | 1 |
| 44 | 2003.10.08 | Human stool | UK (Israel)d | 828 | 33 | 39 | 30 | 82 | 104 | 43 | 17 | 271 | 10 |
| 47 | 2003.10.09 | Human stool | UK | 829 | 33 | 39 | 30 | 82 | 113 | 43 | 17 | 16 | 12 |
| 48 | 2003.10.09 | Human stool | UK | 827 | 33 | 39 | 30 | 82 | 104 | 56 | 17 | 255 | 1 |
| 50 | 2003.10.10 | Human stool | UK | 830 | 33 | 39 | 30 | 79 | 104 | 47 | 17 | 272 | 1 |
| 65 | 2003.10.20 | Human stool | UK | 831 | 33 | 39 | 66 | 82 | 104 | 44 | 41 | 17 | 11 |
| 71 | 2003.10.24 | Human stool | UK | 832 | 33 | 39 | 30 | 79 | 113 | 43 | 17 | 66 | 1 |
| 90 | 2003.11.05 | Human stool | UK | 827 | 33 | 39 | 30 | 82 | 104 | 56 | 17 | 255 | 1 |
| 125 | 2003.11.24 | Human stool | UK | 860 | 33 | 39 | 30 | 79 | 113 | 47 | 17 | 311 | 1 |
| 138 | 2003.12.01 | Human stool | UK | 825 | 33 | 39 | 30 | 82 | 113 | 47 | 17 | 16 | 12 |
| 142 | 2003.12.03 | Human stool | UK | 866 | 33 | 110 | 30 | 116 | 104 | 47 | 41 | 318 | 8 |
| 143 | 2003.12.04 | Human stool | UK | 830 | 33 | 39 | 30 | 79 | 104 | 47 | 17 | 17 | 11 |
| 146 | 2003.12.09 | Human stool | UK | 867 | 33 | 38 | 30 | 115 | 104 | 85 | 17 | 17 | 11 |
| 219 | 2003.01.30 | Human stool | UK | 902 | 33 | 39 | 30 | 79 | 104 | 43 | 17 | 66 | 1 |
| 1c We | 2003.01.07 | Chicken anal swab | UK | 854 | 33 | 38 | 30 | 82 | 104 | 43 | 17 | 319 | 15 |
| 2c W | 2003.01.07 | Chicken anal swab | UK | 854 | 33 | 38 | 30 | 82 | 104 | 43 | 17 | 319 | 15 |
| 3c W | 2003.01.14 | Chicken anal swab | UK | 854 | 33 | 38 | 30 | 82 | 104 | 43 | 17 | 319 | 15 |
| 4c W | 2003.01.21 | Chicken anal swab | UK | 854 | 33 | 38 | 30 | 82 | 104 | 43 | 17 | 319 | 15 |
| 5c Nf | 2003.03.14 | Chicken anal swab | UK | 854 | 33 | 38 | 30 | 82 | 104 | 43 | 17 | 319 | 15 |
| 6c W | 2003.02.21 | Chicken anal swab | UK | 854 | 33 | 38 | 30 | 82 | 104 | 43 | 17 | 319 | 15 |
| 7c W | 2003.02.28 | Chicken anal swab | UK | 855 | 33 | 39 | 30 | 79 | 104 | 35 | 17 | 66 | 1 |
| 8c W | 2003.04.04 | Chicken anal swab | UK | 855 | 33 | 39 | 30 | 79 | 104 | 35 | 17 | 66 | 1 |
| 9c N | 2003.04.11 | Chicken anal swab | UK | 855 | 33 | 39 | 30 | 79 | 104 | 35 | 17 | 66 | 1 |
| 10c N | 2003.04.25 | Chicken anal swab | UK | 855 | 33 | 39 | 30 | 79 | 104 | 35 | 17 | 66 | 1 |
| 11c W | 2003.05.02 | Chicken anal swab | UK | 855 | 33 | 39 | 30 | 79 | 104 | 35 | 17 | 66 | 1 |
| 12c W | 2003.05.09 | Chicken anal swab | UK | 855 | 33 | 39 | 30 | 79 | 104 | 35 | 17 | 66 | 1 |
| 13c W | 2003.05.16 | Chicken anal swab | UK | 855 | 33 | 39 | 30 | 79 | 104 | 35 | 17 | 66 | 1 |
| 14c N | 2003.05.23 | Chicken anal swab | UK | 855 | 33 | 39 | 30 | 79 | 104 | 35 | 17 | 66 | 1 |
| 15c N | 2003.05.30 | Chicken anal swab | UK | 855 | 33 | 39 | 30 | 79 | 104 | 35 | 17 | 66 | 1 |
| 16c N | 2003.06.06 | Chicken anal swab | UK | 855 | 33 | 39 | 30 | 79 | 104 | 35 | 17 | 66 | 1 |
| 17c W | 2003.06.13 | Chicken anal swab | UK | 855 | 33 | 39 | 30 | 79 | 104 | 35 | 17 | 66 | 1 |
| 18c W | 2003.06.20 | Chicken anal swab | UK | 855 | 33 | 39 | 30 | 79 | 104 | 35 | 17 | 66 | 1 |
| 19c W | 2003.06.21 | Chicken anal swab | UK | 854 | 33 | 38 | 30 | 82 | 104 | 43 | 17 | 319 | 15 |
| 20c N | 2003.07.03 | Chicken anal swab | UK | 855 | 33 | 39 | 30 | 79 | 104 | 35 | 17 | 66 | 1 |
| 21c N | 2003.07.11 | Chicken anal swab | UK | 855 | 33 | 39 | 30 | 79 | 104 | 35 | 17 | 191 | 33 |
| 22c N | 2003.07.18 | Chicken anal swab | UK | 855 | 33 | 39 | 30 | 79 | 104 | 35 | 17 | 66 | 1 |
| 23c W | 2003.07.25 | Chicken anal swab | UK | 871 | 33 | 108 | 30 | 79 | 104 | 85 | 17 | 30 | 11 |
| 24c W | 2008.08.01 | Chicken anal swab | UK | 855 | 33 | 39 | 30 | 79 | 104 | 35 | 17 | 191 | 11 |
| P1 W | 2004.02.02 | Pig feces | UK | 886 | 53 | 38 | 83 | 82 | 104 | 43 | 17 | 325 | 15 |
| P3 W | 2004.02.02 | Pig feces | UK | 887 | 33 | 38 | 30 | 82 | 104 | 85 | 68 | 291 | 1 |
| P4 W | 2004.02.02 | Pig feces | UK | 886 | 53 | 38 | 83 | 82 | 104 | 43 | 17 | 325 | 15 |
| P5 W | 2004.02.02 | Pig feces | UK | 886 | 53 | 38 | 83 | 82 | 104 | 43 | 17 | 325 | 15 |
| P6 W | 2004.02.02 | Pig feces | UK | 888 | 32 | 39 | 30 | 82 | 104 | 44 | 36 | 27 | 25 |
| P7 W | 2004.02.02 | Pig feces | UK | 886 | 53 | 38 | 83 | 82 | 104 | 43 | 17 | 325 | 15 |
| P9 W | 2004.02.02 | Pig feces | UK | 888 | 32 | 39 | 30 | 82 | 104 | 44 | 36 | 27 | 25 |
| P10 W | 2004.02.02 | Pig feces | UK | 887 | 33 | 38 | 30 | 82 | 104 | 85 | 68 | 291 | 1 |
| Penner 14 | Human stool (12526)g | Belgium | 889 | 33 | 39 | 30 | 82 | 113 | 47 | 41 | 9 | 10 | |
| Penner 25 | Sheep feces (12529) | Canada | 890 | 33 | 38 | 30 | 82 | 104 | 35 | 36 | 13 | 21 | |
| Penner 28 | Marmoset feces (12531) | NK | 891 | 33 | 39 | 30 | 118 | 104 | 64 | 17 | 16 | 12 | |
| Penner 30 | Human (12532) | Canada | 892 | 33 | 38 | 30 | 115 | 113 | 43 | 17 | 17 | 11 | |
| Penner 34 | Pig (12533) | NK | 893 | 33 | 39 | 30 | 78 | 104 | 35 | 36 | 20 | 25 | |
| Penner 39 | Turkey (12534) | USA | 894 | 33 | 39 | 65 | 82 | 113 | 47 | 13 | 23 | 1 | |
| Penner 46 | Human (12569) | Canada | 895 | 82 | 38 | 30 | 82 | 104 | 35 | 36 | 17 | 11 | |
| Penner 47 | NK (12535) | NK | 896 | 33 | 38 | 30 | 78 | 104 | 122 | 17 | 17 | 11 | |
| Penner 48 | NK (12536) | NK | 897 | 53 | 39 | 44 | 82 | 118 | 44 | 36 | 27 | 25 | |
| Penner 49 | Human (12570) | USA | 898 | 32 | 42 | 30 | 82 | 104 | 43 | 17 | 28 | 10 | |
| Penner 51 | NK (12550) | NK | 832 | 33 | 39 | 30 | 79 | 113 | 43 | 17 | 29 | 28 | |
| Penner 54 | NK (12551) | NK | 903 | 33 | 39 | 32 | 79 | 104 | 47 | 17 | 30 | 11 | |
| Penner 56 | Human (12567) | Israel | 899 | 33 | 39 | 30 | 82 | 113 | 35 | 17 | 16 | 12 | |
| Penner 59 | Pig (12568) | NK | 900 | 32 | 38 | 30 | 82 | 152 | 35 | 17 | 13 | 21 | |
| Penner 61 | NK (12570) | NK | 901 | 33 | 39 | 30 | 79 | 104 | 43 | 41 | 33 | 1 | |
Abbreviations: UK, United Kingdom; USA, United States; NK, not known.
ST numbers are unique numbers assigned to the numbers making up the aspA glnA gltA glyA pgm tkt uncA allelic profile.
Allele nucleotide sequence number in the MLST database (http://pubmlst.org/campylobacter/).
Illness occurred in the United Kingdom after recent travel to the country indicated.
W, Oxford University Farm, Wytham, Oxford, United Kingdom.
N, Northmoor Trust site, Oxfordshire, United Kingdom.
The numbers in parentheses indicate the National Collection of Type Culture accession numbers for the C. coli reference isolates of the Penner serotyping scheme.
Seventeen C. jejuni isolates representing the known genetic diversity of this bacterium were included to allow comparisons with C. coli. The 17 C. jejuni isolates corresponded to the central genotypes of the 17 clonal complexes described to date (5): 13 of these isolates have been submitted to the United Kingdom National Collection of Type Cultures (28). The following other four isolates have the indicated isolate numbers in the MLST database (http://pubmlst.org/campylobacter/): sequence type (ST) 179 (ST-179), 78972; ST-403, 401313; ST-353, 7086; and ST-433, 2632.
Preparation of chromosomal DNA.
A thick suspension of Campylobacter cells was made in 125 μl of molecular biology-grade water (Sigma Aldrich Company Ltd., Dorset, United Kingdom) in a 0.2-ml PCR tube. The suspension was vortexed briefly, immediately transferred to a thermocycler, and held at 100°C for 10 min. This was followed by centrifugation in an Eppendorf microcentrifuge at 13,000 rpm for 10 min. The supernatant was removed and stored at −20°C until it was required for PCR amplification.
Species identification.
All isolates were identified as or confirmed to be C. coli or C. jejuni by use of the primers from a previously described multiplex PCR assay (27). A 323-bp amplicon was generated for C. jejuni and a 126-bp amplicon was generated for C. coli by using a mix of oligonucleotide primers which hybridize to the C. jejuni hipO gene (primers CJF and CJR) or the C. coli glyA gene (primers CCF and CCR). Each 10-μl PCR mixture contained 8.15 μl of molecular biology-grade water (Sigma Aldrich Company Ltd.), 1 μl of 10× reaction buffer (Qiagen Ltd., Crawley, United Kingdom), 0.2 μl of 10 mM deoxynucleoside triphosphates (Invitrogen Ltd., Paisley, United Kingdom), 0.2 μl of 10 μM primer mixture, 0.05 μl of HotStar Taq DNA polymerase (Qiagen Ltd.), and 0.4 μl of chromosomal DNA. The reaction conditions were modified slightly from those published previously (27); they were 95°C for 15 min, followed by 30 cycles of 94°C for 20 s, 55°C for 20 s, and 72°C for 30 s and then a single cycle of 72°C for 5 min and storage at 4°C. The entire 10-μl reaction mixture was analyzed by 2% agarose gel electrophoresis, and the Campylobacter species was identified on the basis of the amplicon size.
C. coli MLST primer design.
C. coli sequences that corresponded to the seven loci used in a previously described C. jejuni MLST scheme (3), aspA, glnA, gltA, glyA, pgm, tkt, and uncA, were required. The primers for the C. jejuni MLST (3) were used in various combinations to amplify these sequences from C. coli. Less stringent amplification conditions with a reduced annealing temperature of 48°C were used to enhance the binding of the C. jejuni primers to C. coli DNA. The primers that provided amplification products were primers aspA A9 and aspA A10, glnA A1 and A2, gltA A1 and gltA A2, glyA A1 and glyA A2, pgm A3 and pgm A4, tkt A5 and tkt A4, and uncA A7 and uncA A2 (the sequences of all primers except tkt A5 are available at http://pubmlst.org/campylobacter/; the sequence of tkt A5 is 5′-TTTAAGTGCTGATATGGTGC-3′). This approach allowed PCR amplicons representing each locus to be obtained from some of 11 C. coli isolates tested. The amplification products were purified by precipitation with 20% polyethylene glycol-2.5 M NaCl (8). The C. coli amplicons were sequenced directly by using the same primers specific for C. jejuni. Nucleotide sequence extension reactions were carried out with the BigDye Ready Reaction Mix (version 3; Applied Biosystems, Foster City, Calif.), in accordance with the instructions of the manufacturer. The reaction products were separated with an ABI 3730 automated DNA sequencer (PE Biosystems).
New C. coli-specific primers for MLST of this organism were designed from sequences within these sequences (Table 1). At least 28 nucleotides separated the MLST trimming site and the 3′ end of the primer. The MLST allele trimming sites for C. coli were chosen to be identical to those for C. jejuni (3). Due to the relatively short lengths of the C. coli sequences available for primer design, C. coli MLST was performed with the same primer pairs used for both amplification and sequencing.
TABLE 1.
Nucleotide sequences of oligonucleotide primers used to perform C. coli MLST
| Locus | Forward primer (sequence) | Reverse primer (sequence) |
|---|---|---|
| aspA | Aspcoli S1(5′-CAACTTCAAGATGCAGTACC-3′) | Aspcoli S2 (5′-ATCTGCTAAAGTATGCATTGC-3′) |
| glnA | Glncoli S1 (5′-TTCATGGATGGCAACCTATTG-3′) | Glncoli S2 (5′-GCTTTGGCATAAAAGTTGCAG-3′) |
| gltA | Gltcoli S1 (5′-GATGTAGTGCATCTTTTACTC-3′) | Gltcoli S2 (5′-AAGCGCTCCAATACCTGCTG-3′) |
| glyA | Glycoli S1 (5′-TCAAGGCGTTTATGCTGCAC-3′) | Glycoli S2 (5′-CCATCACTTACAAGCTTATAC-3′) |
| pgm | Pgmcoli S1 (5′-TTATAAGGTAGCTCCGACTG-3′) | Pgmcoli S2 (5′-GTTCCGAATAGCGAAATAACAC-3′) |
| tkt | Tktcoli S1 (5′-AGGCTTGTGTTTTCAGGCGG-3′) | Tktcoli S2 (5′-TGACTTCCTTCAAGCTCTCC-3′) |
| uncA | Unccoli S1 (5′-AAGCACAGTGGCTCAAGTTG-3′) | Unccoli S2 (5′-CTACTTGCCTCATCCAATCAC-3′) |
C. coli sequence typing.
MLST was performed as follows. Seven PCR amplicons were obtained for each isolate by using the primers shown in Table 1. Each 50-μl PCR mixture contained 39.75 μl of molecular biology-grade water (Sigma Aldrich Company Ltd.), 5 μl of 10× PCR buffer (Qiagen Ltd.), 1 μl of 10 μM each forward and reverse primers, 1 μl of a 10 mM deoxynucleoside triphosphate mixture (Invitrogen Ltd.), 0.25 μl of HotStar Taq DNA polymerase (Qiagen Ltd.), and 2 μl (approximately 10 ng) of C. coli chromosomal DNA. The amplification conditions were 95°C for 15 min, followed by 35 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min, with a final extension at 72°C for 5 min and storage at 4°C. Nucleotide sequencing was performed with the same primers (diluted 1:15 in water) and 30 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 2 min. Data for the newly described C. coli alleles and STs were deposited in the Campylobacter MLST database (http://pubmlst.org/campylobacter/).
A 321-bp sequence containing the flaA SVR was also obtained for each isolate, as described previously (5, 20). SVR nucleotide allele numbers and peptide numbers were assigned by using the database at http://phoenix.medawar.ox.ac.uk/flaA/, and newly described sequences were deposited in that database.
Data analysis.
Phylogenetic analysis was performed with MEGA software (version 2.1), available at http://www.megasoftware.net (16), by using the concatenated MLST gene sequence fragments for each isolate. Analysis of fixed differences, shared mutations, and estimates of the gene flow between populations (FST) was performed with the DnaSP software package (version 4.00), available at http://www.ub.edu/dnasp/ (22). An FST value of 1 indicates that two populations are genetically distinct, and a value of 0 indicates that two populations are indistinguishable.
The linkage model of the STRUCTURE program (9) was used to find evidence of gene exchange between C. jejuni and C. coli by identifying regions of the C. coli MLST sequences containing polymorphisms which were characteristic of C. jejuni, and vice versa. Each of the polymorphic nucleotides in the seven MLST gene fragments was treated as a separate locus. Map distances were assumed to be proportional to the number of base pairs between sites, but with sites on different gene fragments being treated as unlinked. The STRUCTURE program was run for a burn in of 10,000 iterations and 20,000 subsequent iterations, with K equal to 2 populations assumed.
RESULTS
Sequence typing of C. coli isolates.
A total of 34 STs and 26 SVR nucleotide sequences in 42 combinations were identified among the 68 C. coli isolates (Table 2). An individual ST occurred in association with multiple different flaA SVR alleles; for example, ST-825 occurred in association with SVR alleles 336, 253, 292, and 16. Conversely, a single SVR allele sequence occurred with multiple different STs; for example, SVR allele 16 occurred in association with ST-825, ST-829, ST-891, and ST-899. Therefore, the combination of STs and SVR allele sequences provided a high level of discrimination, but the ST was an unreliable indicator of the SVR allele, and vice versa.
Among the 21 contemporary human C. coli isolates from patients living in Oxfordshire, United Kingdom, 14 STs were found in association with 15 SVR alleles in 19 combinations (Table 2). Only three of the human C. coli isolates were indistinguishable by this approach, having ST-827 and SVR allele 255. They were not clustered temporally or geographically within this data set. A total of 24 chicken isolates from two farms were studied, and three STs with four SVR nucleotide sequences were identified (Table 2). Two STs predominated, with ST-855 appearing to replace ST-854 over the study period. Three STs were identified among the isolates from pigs on one of the same farms.
Fifteen C. coli reference isolates of the Penner serotyping scheme were included as representatives of the capsular diversity of the species and because they were isolated more than 20 years ago in a variety of countries (Table 2). They also introduced three additional isolation sources (sheep, marmoset, and turkey feces). Each of these 15 reference isolates exhibited a unique ST, but they contained only 11 SVR alleles (Table 2).
Comparison of diversity within the C. coli and C. jejuni housekeeping genes.
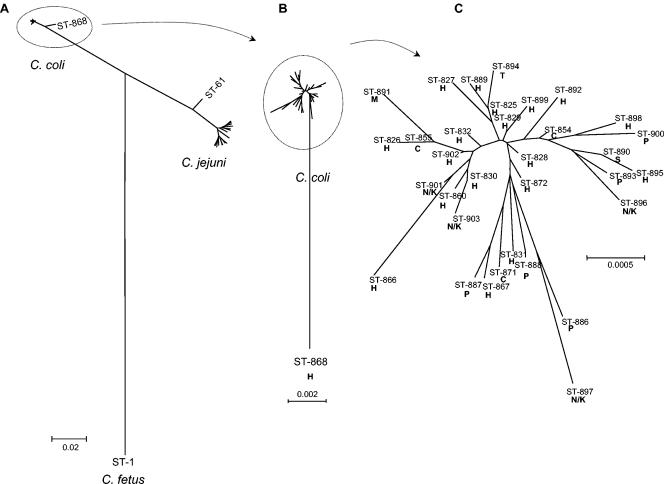
The relationship between the housekeeping genes of C. coli and C. jejuni was examined by using radial neighbor-joining trees constructed from 3,309-bp concatenated MLST allele sequences (Fig. 1). The C. jejuni sequences included were those of the central genotypes of the 17 C. jejuni clonal complexes described to date (5). Of the 68 C. coli isolates, 67 had STs which clustered together (with 34 of 3,309 [1.03%] variable sites) (Fig. 1A). All 15 of the Penner serotype reference isolates occurred within this relatively homogeneous group. There was a single divergent C. coli ST, ST-868, which differed at all seven loci and by about 2% from the remainder of the C. coli STs (Fig. 1B). The level of genetic heterogeneity within the 17 central genotypes of the C. jejuni clonal complexes was greater than that within the C. coli isolates included in the present study (Fig. 1A). The distances between the extremes of C. jejuni diversity were comparable to the distance between outlier C. coli ST-868 and the rest of the STs this species. A second outlier, ST-61, was identified within C. jejuni (Fig. 1A). When a single Campylobacter fetus isolate (unpublished data) was used as the outgroup, C. coli and C. jejuni are inferred to have diverged by approximately the same amount from their inferred common ancestor, implying that the molecular clocks for the two species run at similar rates.
FIG. 1.
Radial neighbor-joining trees constructed with concatenated MLST allele sequences to indicate the relationships between and within C. coli and C. jejuni. (A) Tree constructed with the nucleotide sequences of 34 C. coli STs and the STs of the 17 central genotypes of the C. jejuni clonal complexes described to date (5); (B) tree constructed with the nucleotide sequences of C. coli STs alone; (C) tree constructed with the nucleotide sequences of C. coli STs (excluding divergent ST-868) and their isolation sources, indicated in boldface by H, human; C, chicken; T, turkey; P, pig; S, sheep; and N/K, Penner serotyping scheme reference isolate for which the source is not known.
The levels of identity between C. coli and C. jejuni within the MLST loci were approximately 86.5% at the nucleotide sequence level and 95.0% at the amino acid sequence level. FST between the two species was 0.93170, which is close to the highest possible value (FST = 1.0), which implies that the amount of genetic exchange between them is limited. There were 300 fixed nucleotide differences (363 if ST-61 is excluded) and only 9 shared mutations (Table 3).
TABLE 3.
Nucleotide sequence diversity and gene flow between C. coli and C. jejuni within the flaA SVR and the concatenated sequences of the seven housekeeping genes used in MLST
| Sequence and species | No. of:
|
FSTa | |||
|---|---|---|---|---|---|
| Unique sequences | Polymorphic sites | Fixed differencesa | Shared mutationsa | ||
| flaA SVRb | |||||
| C. coli | 27 | 119 | 0 | 99 | 0.04876 |
| C. jejuni | 17 | 4 | |||
| MLST locic | |||||
| C. coli | 34 | 77 | 300 (363)d | 9 (8) | 0.93170 (0.94049) |
| C. jejuni | 17 (16) | 207 (147) | |||
Data are for C. coli compared with C. jejuni.
The flaA SVR has 321 nucleotides.
MLST loci have a total of 3,309 nucleotides.
Values in parentheses indicate the results obtained when C. jejuni ST-61 (which contains the C. coli uncA17 allele) was excluded from the analysis.
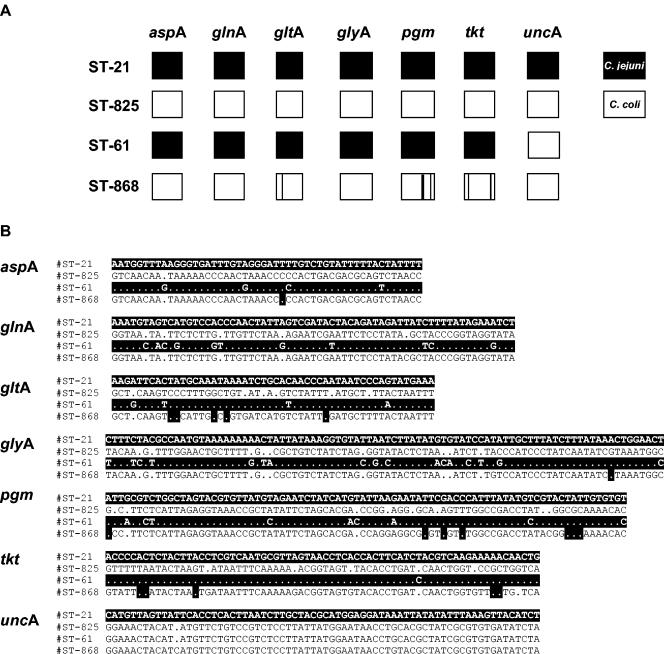
The STRUCTURE program was used to investigate the pattern of genetic exchange between the two species in more detail by using the polymorphic nucleotides in each of the seven gene fragments All but two of the strains were inferred to have inherited all of their nucleotides from a single species (Fig. 2). The two exceptions were ST-868 (C. coli) and ST-61 (C. jejuni). ST-61 contained the complete allele unc-17, which is characteristic of C. coli (Fig. 2). In contrast, the STRUCTURE program inferred that ST-868 had imported five very short gene fragments from C. coli.
FIG. 2.
Analysis of the C. jejuni and C. coli housekeeping genes used to detect nucleotide polymorphisms in the MLST loci of each species characteristic of the other. Black boxes and white text on black highlighting, sequences characteristic of C. jejuni; white boxes and black text with white highlighting, sequences characteristic of C. coli. (A) Diagrammatic output of the STRUCTURE program. The seven MLST loci are represented by seven squares, and four STs are shown. ST-21 and ST-825 are examples of C. jejuni and C. coli, respectively. C. jejuni ST-61 contains an uncA allele characteristic of C. coli. Polymorphisms in C. coli ST-868 which are characteristic of C. jejuni are indicated by vertical black lines. (B) Alignment of the variable nucleotide sites within the MLST loci represented in panel A.
Genetic diversity among C. coli and C. jejuni flaA SVR sequences.
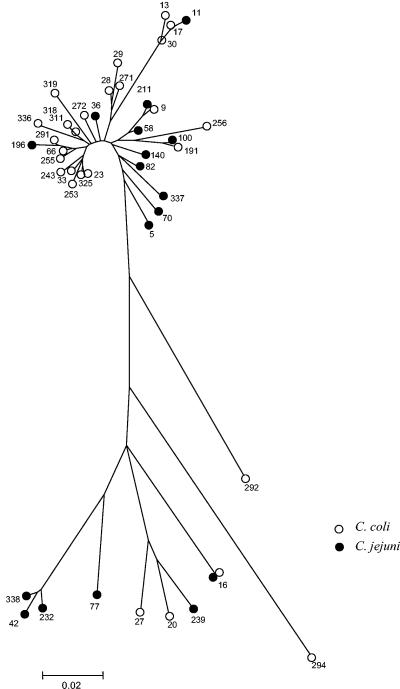
A 321-nucleotide sequence spanning the flaA SVR was obtained for both the C. coli (n = 68) and the C. jejuni (n = 17) isolates. A total of 27 unique sequences were identified in C. coli, and 17 were identified in C. jejuni. The numbers of variable sites within each species was similar, with 119 for C. coli and 94 for C. jejuni. A radial neighbor-joining tree was constructed, as described above for the MLST loci. In contrast to their housekeeping genes, no clustering of allele sequences by microbiological species was detected (Fig. 3). Allele 16 was found in both C. coli and C. jejuni isolates.
FIG. 3.
Radial neighbor-joining tree constructed to indicate (i) the genetic diversity detected among the flaA SVR sequences of C. coli and C. jejuni and (ii) the lack of segregation by species. The C. jejuni sequences were from isolates representing the central genotypes of 17 clonal complexes described to date. The numbers indicate the flaA SVR allele numbers within the SVR database, and the letters C and J represent C. coli and C. jejuni, respectively. Allele 16 was found in both C. jejuni and C. coli isolates.
A high level of gene flow between C. coli and C. jejuni involving this locus was confirmed by FST analysis (Table 3). A very low FST value of 0.04876 was obtained, which suggests that the SVR sequences were derived from the same population. There were no fixed differences between the two species, and there were 99 shared mutations.
DISCUSSION
C. coli and C. jejuni are closely related bacterial species that cause a large number of clinical cases of gastroenteritis worldwide. It is therefore important to understand their molecular epidemiology and evolution. Central to this goal is the availability of a reliable approach to isolate typing that provides data for all strains which can be compared among laboratories and over time. An MLST scheme was developed for C. coli by extending an existing scheme for C. jejuni (3) and was validated with 68 C. coli isolates from different sources, locations, and years and of different serotypes (Table 2). MLST of all isolates tested confirmed the conservation of the primer binding sites. In addition, SVR sequences from the flaA genes were obtained. A high level of resolution was achieved, indicating the suitability of the approach for investigation of the molecular epidemiology of C. coli.
Although a diverse range of C. coli isolates was examined, the species showed less diversity than C. jejuni at each of the MLST loci. This agrees with findings obtained by amplified fragment length polymorphism analysis (AFLP), in which C. coli strains from poultry were less variable than C. jejuni strains (6). However, the identification of a single divergent C. coli isolate (ST-868) suggests that greater diversity that has not been sampled to date may exist. Both species contained similar levels of sequence diversity within the 321 bp of the flaA SVRs.
The results of biochemical tests used to distinguish C. coli and C. jejuni can be ambiguous. Hybridization, multilocus enzyme electrophoresis, AFLP, and fluorescent AFLP studies have confirmed that they are separate species with 22 to 49% homology (1, 6, 14, 15). As expected, the nucleotide sequences of the MLST loci segregated according to microbiological species (Fig. 1A), confirmed by 300 fixed nucleotide differences and a high FST value of 0.93170. The two species were closely related, sharing approximately 86.5% identity at the nucleotide sequence level, and the level of identity rose to approximately 95.0% at the amino acid sequence level.
In contrast, no evidence of segregation by species was detected within the C. jejuni or C. coli SVR sequences (Fig. 3). No fixed nucleotide differences were detected in this locus (Table 3), with SVR allele 16 found in both species. The low FST value of 0.04876 also implies that these sequences represent a single population. These observations agree with those of a previous study (2) in which flaA typing, conducted by enzyme digestion of a PCR product, could not distinguish C. coli and C. jejuni. Both C. coli and C. jejuni are naturally competent to take up DNA. Both intragenomic recombination and intergenomic recombination have been demonstrated within the flagellin locus of C. jejuni (13). Thus, frequent interspecies recombination appears to explain the common gene pool for flaA shared by these species. These data indicate the unsuitability of the flaA SVR (when used alone) as a marker for the molecular epidemiology of C. coli and C. jejuni. However, the diversity of this locus can allow closely related strains with the same MLST ST to be distinguished.
Unambiguous evidence of interspecies recombination within the housekeeping genes was confined to one C. jejuni isolate which contained a C. coli sequence in one of seven loci (ST-61) (Fig. 2). This genotype has now been described in many isolates by multiple laboratories (see the database at http://pubmlst.org/campylobacter/). C. coli ST-868 may have a history of recombination with C. jejuni, since 19 of 441 polymorphic sites within this genotype were characteristic of this species (Fig. 2B) and the STRUCTURE program assigns the C. jejuni ancestry to five short runs of its DNA. These sequences could have entered the C. coli population by recombination of larger gene fragments (similar to that observed in ST-61), followed by extensive recombination within other C. coli strains, which could have resulted in the observation of only short fragments in the extant population.
However, there is an alternative phylogenetic explanation for why ST-868 shares nucleotides with C. jejuni, which is that the sequence may represent the ancestral state, with the mutation observed in the remaining C. coli isolates occurring subsequent to the divergence of ST-868. This explanation predicts that the shared nucleotides will be distributed at random among the polymorphisms that distinguish C. coli and C. jejuni. Recombination, on the other hand, would lead to a nonrandom distribution, with adjacent polymorphic nucleotides giving the same ancestral signal.
The distribution of shared nucleotides showed some evidence of being nonrandom. On average, 0.8 runs [(19/411) × (19/441) × 443] of two adjacent such polymorphisms would be expected. Five adjacent pairs of polymorphisms (the single run of three counts as two pairs), the occurrence of which has a probability of 0.0004, were observed. However, the evidence is weakened by the fact that two of the pairs of changes in tkt cause amino acid changes. These nucleotide changes may have occurred in quick succession due to natural selection, which provides an alternative explanation to recombination for their clustering on the chromosome. Thus, while the import of nucleotides from the housekeeping genes of C. jejuni to C. coli seems likely, it is not proven by the present data; and the contributions of recombination and mutation to the divergence of ST-868 from the other C. coli strains remain unknown.
Two previous studies by fluorescent AFLP have indicated that different C. coli strains are associated with particular animal hosts or environmental sources (15, 17). In the present study, the relationship between C. coli genotype and isolation source was examined by using a radial neighbor-joining tree (Fig. 1C), but no clustering of the STs by source was apparent. However, chickens and pigs located on the same farm were colonized with different STs (Table 2); this may indicate a host preference by certain C. coli genotypes, and analysis of further isolates by MLST may clarify this issue.
C. coli MLST and flaA SVR sequencing provide sufficient resolution to be useful in future studies for the investigation of isolates from cases of human disease and the potential sources of human infection. As all MLST data are directly comparable, this approach could allow accurate assessments of the contributions that different infection sources make to the burden of human disease to be made. The inability to distinguish the C. coli and C. jejuni species by use of the flaA SVR sequence calls into question the use of the flaA locus alone in any method aimed at studying the epidemiology of the diseases caused by these organisms. Further extension of this MLST scheme to additional Campylobacter species will aid in providing an understanding of their evolutionary relationships.
Acknowledgments
We thank David Wareing, previously based at Preston Public Health Laboratory Service, •••, United Kingdom, and now located at Dynal Ltd., •••, United Kingdom, for supplying DNA from the C. coli Penner serotype reference isolates. The Campylobacter fetus strain included in the analysis whose results are shown in Fig. 1A was kindly provided by Jaap Wagenaar, Institute for Animal Science and Health (ID-Lelystad), Lelystad, The Netherlands.
This work was funded by the United Kingdom Department for Environment, Food and Rural Affairs (contract number OZ0604). M.C.J.M. is a Wellcome Trust Senior Research Fellow in Basic Biological Sciences.
REFERENCES
- 1.Aeschbacher, M., and J. Piffaretti. 1989. Population genetics of human and animal enteric Campylobacter strains. Infect. Immun. 57:1432-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Boer, P., B. Duim, A. Rigter, J. van der Plas, W. F. Jacobs-Reitsma, and J. A. Wagenaar. 2000. Computer-assisted analysis and epidemiological value of genotyping methods for Campylobacter jejuni and Campylobacter coli. J. Clin. Microbiol. 38:1940-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dingle, K. E., F. M. Colles, D. R. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. Willems, R. Urwin, and M. C. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dingle, K. E., N. Van Den Braak, F. M. Colles, L. J. Price, D. L. Woodward, F. G. Rodgers, H. P. Endtz, A. Van Belkum, and M. C. Maiden. 2001. Sequence typing confirms that Campylobacter jejuni strains associated with Guillain-Barré and Miller-Fisher syndromes are of diverse genetic lineage, serotype, and flagella type. J. Clin. Microbiol. 39:3346-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dingle, K. E., F. M. Colles, R. Ure, J. A. Wagenaar, B. Duim, F. J. Bolton, A. J. Fox, D. R. Wareing, and M. C. Maiden. 2002. Molecular characterization of Campylobacter jejuni clones: a basis for epidemiologic investigation. Emerg. Infect. Dis. 8:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duim, B., T. M. Wassenaar, A. Rigter, and J. A. Wagenaar. 1999. High-resolution genotyping of Campylobacter strains isolated from poultry and humans with amplified fragment length polymorphism fingerprinting. Appl. Environ. Microbiol. 65:2369-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duim, B., P. C. Godschalk, N. van den Braak, K. E. Dingle, J. R. Dijkstra, E. Leyde, J. van der Plas, F. M. Colles, H. P. Endtz, J. A. Wagenaar, M. C. Maiden, and A. van Belkum. 2003. Molecular evidence for dissemination of unique Campylobacter jejuni clones in Curacao, Netherlands Antilles. J. Clin. Microbiol. 41:5593-5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Embley, T. M. 1991. The linear PCR reaction: a simple and robust method for sequencing amplified rRNA genes. Lett. Appl. Microbiol. 13:171-174. [DOI] [PubMed] [Google Scholar]
- 9.Falush, D., M. Stephens, and J. K. Pritchard. 2003. Inference of population structure from multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman, C. J., J. Neiman, H. C. Wegener, and R. V. Tauxe. 2000. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations, p. 121-138. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter. ASM Press, Washington, D.C.
- 11.Gillespie, I. A., S. J. O'Brien, J. A. Frost, G. K. Adak, P. Horby, A. V. Swan, M. J. Painter, K. R. Neal, and Campylobacter Sentinel Surveillance Scheme Collaborators. 2002. A case-case comparison of Campylobacter coli and Campylobacter jejuni infection: a tool for generating hypotheses. Emerg. Infect. Dis. 8:937-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanninen, M. L., M. Hakkinen, and H. Rautelin. 1999. Stability of related human and chicken Campylobacter jejuni genotypes after passage through chick intestine studied by pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 65:2272-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrington, C. S., F. M. Thomson-Carter, and P. E. Carter. 1997. Evidence for recombination in the flagellin locus of Campylobacter jejuni: implications for the flagellin gene typing scheme. J. Clin. Microbiol. 35:2386-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harvey, S. M., and J. R. Greenwood. 1983. Relationships among catalase-positive campylobacters determined by deoxyribonucleic acid-deoxyribonucleic acid hybridization. Int. J. Syst. Bacteriol. 33:275-284. [Google Scholar]
- 15.Hopkins, K. L., M. Desai, J. A. Frost, J. Stanley, and J. M. Logan. 2004. Fluorescent amplified fragment length polymorphism genotyping of Campylobacter jejuni and Campylobacter coli strains and its relationship with host specificity, serotyping and phage typing. J. Clin. Microbiol. 42:229-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 17.Leatherbarrow, A. J., C. A. Hart, R. Kemp, N. J. Williams, A. Ridley, M. Sharma, P. J. Diggle, E. J. Wright, J. Sutherst, and N. P. French. 2004. Genotypic and antibiotic susceptibility characteristics of a Campylobacter coli population isolated from dairy farmland in the United Kingdom. Appl. Environ. Microbiol. 70:822-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manning, G., C. G. Dowson, M. C. Bagnall, I. H. Ahmed, M. West, and D. G. Newell. 2003. Multilocus sequence typing for comparison of veterinary and human isolates of Campylobacter jejuni. Appl. Environ. Microbiol. 69:6370-6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mead, P. S., L. Slutsker, V. Deitz, F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meinersmann, R. J., L. O. Helsel, P. I. Fields, and K. L. Heitt. 1997. Discrimination of Campylobacter jejuni isolates by fla gene sequencing. J. Clin. Microbiol. 35:2810-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Penner, J. L., J. N. Hennessy, and R. V. Congi. 1983. Serotyping of Campylobacter jejuni and Campylobacter coli on the basis of thermostable antigens. Eur. J. Clin. Microbiol. 2:378-383. [DOI] [PubMed] [Google Scholar]
- 22.Rozas, J., J. C. Sánchez-DelBarrio, X. Messeguer, and R. Rozas. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496-2497. [DOI] [PubMed] [Google Scholar]
- 23.Sails, A. D., B. Swaminathan, and P. I. Fields. 2003. Utility of multilocus sequence typing as an epidemiological tool for investigation of outbreaks of gastroenteritis caused by Campylobacter jejuni. J. Clin. Microbiol. 41:4733-4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steinbrueckner, B., F. Ruberg, and M. Kist. 2001. Bacterial genetic fingerprint: a reliable factor in the study of the epidemiology of human Campylobacter enteritis? J. Clin. Microbiol. 39:4155-4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suerbaum, S., M. Lohrengel, A. Sonnevend, F. Ruberg, and M. Kist. 2001. Allelic diversity and recombination in Campylobacter jejuni. J. Bacteriol. 183:2553-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tam, C. C., S. J. O'Brien, G. K. Adak, S. M. Meakins, and J. A. Frost. 2003. Campylobacter coli—an important foodbourne pathogen. J. Infect. 47:28-32. [DOI] [PubMed] [Google Scholar]
- 27.Wang, G., C. G. Clark, T. M. Taylor, C. Pucknell, C. Barton, L. Price, D. L. Woodward, and F. Rodgers. 2002. Colony multiplex PCR assay for the identification and differentiation of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis, and C. fetus subsp. fetus. J. Clin. Microbiol. 40:4744-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wareing, D. R., R. Ure, F. M. Colles, F. J. Bolton, A. J. Fox, M. C. Maiden, and K. E. Dingle. 2003. Reference isolates for the clonal complexes of Campylobacter jejuni. Lett. Appl. Microbiol. 36:106-110. [DOI] [PubMed] [Google Scholar]
- 29.Wassenaar, T. M., B. Geilhausen, and D. G. Newell. 1998. Evidence of genomic instability in Campylobacter jejuni isolated from poultry. Appl. Environ. Microbiol. 64:1816-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]