Abstract
The Mycobacterium tuberculosis Beijing family isolates may cause more than a quarter of all tuberculosis cases worldwide, are emerging in some areas, and are often associated with drug resistance. Early recognition of transmission of this genotype is therefore important. To evaluate the usefulness of variable-number tandem-repeat (VNTR) typing to discriminate and recognize strains of the Beijing family, M. tuberculosis isolates from Hong Kong were subjected to VNTR analysis, spoligotyping, and IS6110 restriction fragment length polymorphism (RFLP) typing. The allelic diversity of the 14 VNTR loci included in the analysis varied from 0 to 0.618 among Beijing strains. The discriminatory power of VNTR analysis was slightly lower than that of IS6110 RFLP. Our analysis shows that VNTR typing, which has many practical advantages over RFLP typing, can be used for epidemiological studies of Beijing strains. However, VNTR-defined clusters should be subtyped with IS6110 RFLP for maximal resolution.
Tuberculosis (TB) remains a major public health threat. Globally, the number of TB cases is rising, and it is estimated that about 12 million new TB cases and 4 million deaths will be recorded annually by the year 2010. Many outbreaks of multidrug-resistant (MDR) TB, with poor response to treatment and high disease and death rates, have been reported in, for instance, former republics of the USSR and several provinces of China. In the United States, large outbreaks of MDR TB were caused by strains of the W family, which form a minor subgroup of the Mycobacterium tuberculosis Beijing family (2-4). Recent reviews of the literature published on this family of strains (3, 10) suggest that the M. tuberculosis Beijing family may account for more than a quarter of all TB cases worldwide.
Members of the M. tuberculosis Beijing family constitute a genetically homogeneous group of bacteria, presumably due to recent clonal expansion favored by selective advantages (19, 22, 27). In China, more than 85% of the M. tuberculosis isolates belong to this genotype family, and this family is also highly prevalent in other Asian countries (39). In many countries, this genotype is found to be associated with multidrug resistance (2-4, 10). In 1995, it was hypothesized that the Mycobacterium bovis BCG-induced immunological defense may not protect against infection by strains of the Beijing family (39). Recent studies have consistently indicated a lower efficacy of BCG vaccination against infection of Beijing family strains with the BALB/c mouse model (19). Furthermore, it was recently reported that Beijing strains carry mutations in putative mutator genes, and this may explain, in part, an increased rate of adaptation of these bacteria when exposed to antituberculosis drugs or the hostile intracellular environment (22), although no higher mutation rate was found in the presence of rifampin in Beijing strains in vitro.
Members of the clonal Beijing family can be recognized easily by a number of molecular techniques. They are characterized by highly similar multibanded IS6110 restriction fragment length polymorphism (RFLP) patterns and spoligo patterns showing hybridization to spacers 35 to 43 (39). These strains have at least two characteristic insertions of IS6110, one in the dnaA-dnaN region and one in the NTF region, enabling recognition with region A RFLP or a multiplex PCR (3, 39). A molecular definition of the Beijing lineage has recently been established (14).
The molecular markers currently used to study the epidemiology of tuberculosis have greatly enhanced our understanding of the epidemiology of tuberculosis, but they are impractical and, in some theoretical terms, not ideal. The most standardized typing method, IS6110 RFLP typing, shows the highest level of discrimination. However, Beijing strains often carry 20 to 25 copies of IS6110 and exhibit highly similar RFLP patterns. The computer-assisted analysis of these patterns is therefore cumbersome. If two such multibanded patterns differ in only one or a few bands, it is difficult to determine whether these patterns reflect direct transmission of a given strain that underwent a small variation or whether this matching is the result of accidental disclosure of two closely related strains. Moreover, RFLP typing remains a time-consuming and technically demanding technique which requires large quantities of DNA.
Typing based on variable numbers of tandem repeats (VNTRs) is a promising PCR-based molecular typing method for M. tuberculosis isolates (8, 20, 28, 31). With this multilocus method, the isolates can be typed for practical epidemiological purposes, and simultaneously, the evolutionary relationships between strains can be defined to identify the genotype family to which the strain belongs (31). The discriminatory power of VNTR typing with mycobacterial interspersed repetitive units (MIRUs) (32, 33) is close to that of IS6110 RFLP (20, 31), and the method is highly reproducible (15, 31) and has been adapted to achieve a high throughput (31). Furthermore, the results can be expressed in a numerical code, which considerably facilitates the exchange of results between laboratories (8, 20, 31).
Several studies have investigated the usefulness of different VNTR loci for typing of M. tuberculosis (8, 17, 31, 33) or M. bovis (23, 26). These studies demonstrated the usefulness of VNTR typing for subtyping IS6110 RFLP-defined clusters (16, 20) and for epidemiological typing (1, 6, 9, 18). Furthermore, VNTR loci proved to be very useful for studying the population structure of M. tuberculosis (29, 34). However, there has not been an extensive comparison of the usefulness of VNTR loci for typing of Beijing family strains. In the reports focusing on Beijing and W family strains, the authors used only the five exact tandem repeats (ETRs) A to E (2, 4, 7, 21).
We performed a literature search for VNTR loci that showed polymorphism among M. tuberculosis Beijing family strains and tested these variable loci in addition to the ETR loci A to E. We used 14 VNTR loci to type 69 M. tuberculosis isolates from Hong Kong, where 70% of the isolates belong to the Beijing family (5). The polymorphism obtained by VNTR typing was compared with that obtained by IS6110 RFLP and spoligotyping.
MATERIALS AND METHODS
Mycobacterial strains
Sixty-nine M. tuberculosis isolates were randomly selected from M. tuberculosis strains isolated in Hong Kong in 2001. Patient information was insufficient to validate epidemiological linkage between patients. DNA was prepared from colonies by either boiling for 10 min at 100°C with subsequent immediate ice-bath cooling for 5 min or DNA isolation according to the method of van Soolingen et al. (38).
Molecular typing methods.
IS6110 RFLP typing and spoligotyping were performed at the National Institute of Public Health and the Environment, Bilthoven, The Netherlands, according to standardized protocols (13, 36, 38). In addition to spoligotyping with a commercial membrane containing the standard set of 43 spacer oligonucleotides (Isogen Bioscience BV, Maarssen, The Netherlands) (13), spoligotyping was also performed on a homemade membrane containing oligonucleotides of five novel spacers (spacers 56, 57, 66, 67, and 68). These spacers have previously shown a dichotomy among M. tuberculosis Beijing family strains (35).
VNTR typing was performed by using AmpliTaq Gold polymerase (Applied Biosystems, Foster City, Calif.) at the Public Health Laboratory Center, Department of Health, Hong Kong according to the method of Frothingham and Meeker-O'Connell (8), except for loci QUB26 and QUB11a, which were amplified from a few samples at the National Institute of Public Health and the Environment by using the Hotstart Taq DNA polymerase kit (QIAGEN, Hilden, Germany) as described by Supply et al. (31). The interpretation of the PCR products of these PCRs was too difficult due to the high number of repeats present at these loci (14 and 10, respectively) and the consequent additional (incomplete) PCR bands obtained by using the protocol of Frothingham and Meeker-O'Connell (8). VNTR typing exploits the variability in the numbers of tandem repeats at particular loci in the M. tuberculosis genome. These VNTR loci are amplified by PCR with primers directed to the regions flanking the repeats. Because the sizes of the repeat units are known, the number of repeats can be determined from the size of the PCR product after gel electrophoresis. The loci that were used in this study included the ETR loci A to E (8), loci containing MIRUs (31), and loci characterized at the Queens University of Belfast (QUB) (23, 26). ETR-D and ETR-E are identical to MIRU VNTR loci 4 and 31, respectively (8, 32, 33). The literature on VNTR analysis (1, 2, 4, 6, 7, 11, 15, 16, 21, 23, 24, 26, 29, 31) and unpublished data on QUB analysis of 90 M. tuberculosis complex isolates from the study by Kremer et al. (15) were searched for VNTR loci that showed polymorphism among Beijing family strains, and these loci were included in the study. The VNTR loci and the primers used to amplify them are listed in Table 1. PCR products were analyzed on a 2 to 3% NuSieve agarose gel (Applied Biosystems). VNTR allele-naming tables used in this study were the same as in the original studies characterizing the respective VNTR sets (8, 26, 33). Differences between 77- and 53-bp repeats in ETR-D/MIRU VNTR locus 4 (32) were not considered here, and 53-bp repeats were therefore included in the VNTR count, as in reference 8.
TABLE 1.
Primer sequence and repeat unit size in M. tuberculosis H37Rv of the VNTR loci used in this study
| VNTR locus | VNTR locus alias | VNTR locus convention (28) | PCR primer sequence (5′-3′) | Repeat unit size (bp) × no. in H37Rva |
|---|---|---|---|---|
| ETR-A | VNTR 2165 | CGA AGC CTG GGG TGC CCG CGA TTT | (75 × 3) + 23 | |
| AAA TCG GTC CCA TCA CCT TCT TAT | ||||
| ETR-B | VNTR 2461 | GCG AAC ACC AGG ACA GCA TCA TG | (57 × 3) + 8 | |
| GGC ATG CCG GTG ATC GAG TGG | ||||
| ETR-C | VNTR 0577 | GTG AGT CGC TGC AGA ACC TGC AG | (58 × 4) − 21 | |
| GGC GTC TTG ACC TCC ACG AGT G | ||||
| ETR-D | MIRU4 | VNTR 0580 | CAG GTC ACA ACG AGA GGA AGA GC | (77 × 3) + 7 |
| GCG GAT CGG CCA GCG ACT CCT C | ||||
| ETR-Eb | MIRU31 | VNTR 3192 | CTT CGG CGT CGA AGA GAG CCT C | (53 × 3) − 4 |
| CGG AAC GCT GGT CAC CAC CTA AG | ||||
| MIRU10 | VNTR 0960 | GTT CTT GAC CAA CTG CAG TCG TCC | 53 × 3 | |
| GCC ACC TTG GTG ATC AGC TAC CT | ||||
| MIRU16 | VNTR 1644 | TCG GTG ATC GGG TCC AGT CCA AGT A | 53 × 2 | |
| CCC GTC GTG CAG CCC TGG TAC | ||||
| MIRU20c | VNTR 2059 | TCG GAG AGA TGC CCT TCG AGT TAG | 77 × 2 | |
| GGA GAC CGC GAC CAG GTA CTT GTA | ||||
| MIRU26 | VNTR 2996 | TAG GTC TAC CGT CGA AAT CTG TGA C | 51 × 3 | |
| CAT AGG CGA CCA GGC GAA TAG | ||||
| MIRU31b | ETR-E | VNTR 3192 | ACT GAT TGG CTT CAT ACG GCT TTA | 53 × 3 |
| GTG CCG ACG TGG TCT TGA T | ||||
| MIRU39 | VNTR 4348 | CGC ATC GAC AAA CTG GAG CCA AAC | 53 × 2 | |
| CGG AAA CGT CTA CGC CCC ACA CAT | ||||
| MIRU40 | VNTR 0802 | GGG TTG CTG GAT GAC AAC GTG T | 54 × 1 | |
| GGG TGA TCT CGG CGA AAT CAG ATA | ||||
| QUB11a | VNTR 2163a | TTC AGG GGG GAT CCG GGA | (69 × 2) + 8 | |
| CCC ATC CCG CTT AGC ACA TTC GTA | ||||
| QUB11b | VNTR 2163b | CGT AAG GGG GAT GCG GGA AAT AGG | (69 × 5) + 9 | |
| CGA AGT GAA TGG TGG CAT | ||||
| QUB26 | VNTR 4052 | GGC CAG GTC CTT CCC GAT | (111 × 5) + 24 | |
| AAC GCT CAG CTG TCG GAT | ||||
| QUB1895 | VNTR 1895 | GGT GCA CGG CCT CGG CTC C | (57 × 4) + 11 | |
| AAG CCC CGC CGC CAA TCA A | ||||
| QUB3232c | VNTR 3232 | CAG ACC CGG CGT CAT CAA C | (56 × 3) + 50 | |
| CCA AGG GCG GCA TTG TGT T | ||||
| QUB3336c | VNTR 3336 | ATC CCC GCG GTA CCC ATC | 59 × 5 | |
| GCC AGC GGT GTC GAC TAT CC |
Some loci have several complete repeats and one partial repeat. For example, the ETR-A locus contains three complete 75-bp repeats followed by an additional 23 bp of repetitive sequence.
Note that ETR-E and MIRU31 designate the same locus.
The amplification of MIRU20, QUB3232, and QUB3336 was not successful by using the protocol of Frothingham and Meeker-O'Connell (8).
Patterns were considered clustered if they were 100% identical.
Computer-assisted and statistical analysis.
The BioNumerics software (version 3.5, Applied Maths, Sint-Martens-Latum, Belgium) was used to analyze molecular typing results. IS6110 RFLP and spoligo patterns were analyzed as fingerprint types, and VNTR types were analyzed as a character type. Similarities between VNTR types were calculated by using the categorical multistate coefficient, and the dendrogram was constructed according to the unpaired group method using arithmetic averages.
The level of discrimination of each typing method was calculated by using the Hunter-Gaston discriminatory index (HGI) (12), by using the following equation:
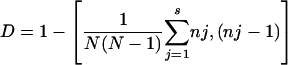 |
where D is the numerical index of discrimination, N is the total number of strains in the typing scheme, s is the total number of different strain types, and nj is the number of strains belonging to the jth type.
The allelic diversity (h), indicating the probability of observing a given allele at a particular locus in a population, was determined according to the method of Selander et al. (25).
RESULTS
Spoligotyping.
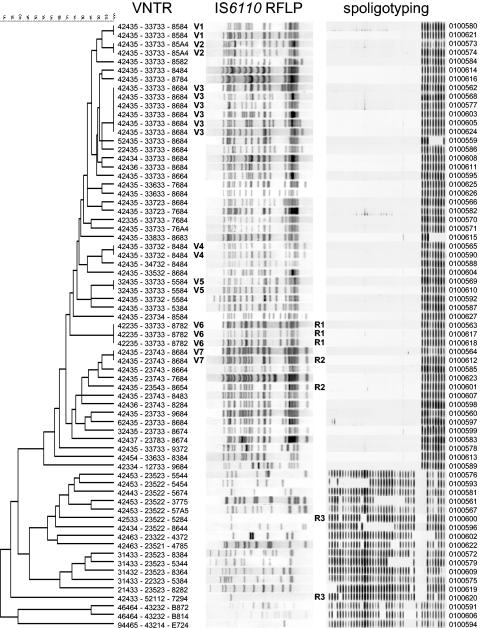
Fifty-one (74%) isolates showing a Beijing-characteristic spoligotype were identified from a collection of 69 strains recovered from Hong Kong (14, 39); 49 isolates showed the nine-spacer spoligotype and two strains showed hybridization to three and four spacers, respectively (Fig. 1). This percentage of tuberculosis cases attributable to the Beijing lineage is close to that observed by Chan et al. (5) in another sample of M. tuberculosis isolates collected in the same country in 1998 and 1999.
FIG. 1.
DNA fingerprint results for 69 M. tuberculosis isolates from Hong Kong ordered by similarity on the basis of VNTR typing results, as determined by the unpaired group method using arithmetic averages algorithm (dendrogram with similarity indicated on top, utmost left). The VNTR profiles consist of ETRs A to E, followed by MIRUs 10, 16, 26, 39, and 40, and by QUBs 11a, 11b, 26, and 1895. ETR-E and ETR-D are identical to MIRU31 and MIRU4, respectively. The VNTR profile consists of digits (when nine or less repeats were detected at a particular locus) and letters; the letter A represents 10 repeats, the letter B represents 11 repeats, and so on. Refer to Materials and Methods for allele conventions for ETR-D/MIRU4. The IS6110 RFLP patterns are shown from high to low molecular weight (left to right), and the spoligo patterns are shown from spacer 1 to 43 (left to right). The isolate numbers are depicted to the right of the DNA fingerprint patterns. Isolates clustered by VNTR are marked with a capital letter V (clusters V1 to V7). Isolates clustered by RFLP are marked with a capital letter R (clusters R1 to R3).
It has previously been shown that novel spacers disclosed by van Embden et al. (37) could be used to improve the discrimination of strains of the Beijing lineage (35). The use of spacers 56, 57, 66, 67, and 68 in spoligotyping increased the number of spoligotype patterns among the Beijing lineage strains in this study from three to five spoligotypes. The HGI for the 51 Beijing strains thereby increased from 0.078 to 0.253 (Table 2).
TABLE 2.
Comparison of the discriminatory power of various genetic markers to type M. tuberculosis strains from Hong Kong
| Strains typed (n) | Typing method | Total no. of type patterns | No. of unique types (%) | No. of clusters | No. of clustered isolates (%) | Maximum no. of isolates in a cluster | Average cluster size | HGI | Ha |
|---|---|---|---|---|---|---|---|---|---|
| All strains (69)c | Spoligo (43 spacers) | 19 | 16 (23) | 3 | 53 (77) | 49 | 17.7 | 0.498 | NAd |
| Spoligo (5 spacers) | 5 | 0 (0) | 5 | 69 (100) | 49 | 13.8 | 0.467 | NA | |
| Spoligo (48 spacers) | 21 | 17 (25) | 4 | 52 (75) | 44 | 13.0 | 0.593 | NA | |
| ETR VNTR (5 loci) | 22 | 13 (19) | 9 | 56 (81) | 35 | 6.2 | 0.738 | 0.285 | |
| MIRU VNTR (5 loci) | 21 | 12 (17) | 9 | 57 (83) | 27 | 6.3 | 0.829 | 0.350 | |
| MIRU VNTR (7 loci) | 30 | 21 (30) | 9 | 48 (70) | 25 | 5.3 | 0.861 | 0.411 | |
| QUB VNTR (4 loci) | 36 | 24 (35) | 12 | 45 (65) | 16 | 3.8 | 0.937 | 0.532 | |
| VNTR of all locib | 57 | 50 (72) | 7 | 19 (28) | 6 | 2.7 | 0.990 | 0.377 | |
| IS6110 RFLP | 65 | 62 (90) | 3 | 7 (10) | 3 | 2.3 | 0.998 | NA | |
| Beijing family strains (51) | Spoligo (43 spacers) | 3 | 2 (4) | 1 | 49 (96) | 49 | 49 | 0.078 | NA |
| Spoligo (5 spacers) | 3 | 0 (0) | 3 | 51 (100) | 45 | 17 | 0.218 | NA | |
| Spoligo (48 spacers) | 5 | 3 (6) | 2 | 48 (94) | 44 | 24 | 0.253 | NA | |
| ETR VNTR (5 loci) | 12 | 8 (16) | 4 | 43 (84) | 35 | 10.8 | 0.528 | 0.117 | |
| MIRU VNTR (5 loci) | 13 | 7 (14) | 6 | 44 (86) | 27 | 7.3 | 0.705 | 0.192 | |
| MIRU VNTR (7 loci) | 21 | 14 (27) | 7 | 37 (73) | 13 | 5.3 | 0.905 | 0.196 | |
| QUB VNTR (4 loci) | 20 | 10 (20) | 10 | 41 (80) | 16 | 4.1 | 0.886 | 0.383 | |
| VNTR of all locib | 39 | 32 (63) | 7 | 19 (37) | 6 | 2.7 | 0.982 | 0.218 | |
| IS6110 RFLP | 48 | 46 (90) | 2 | 5 (10) | 3 | 2.5 | 0.997 | NA |
The mean allelic diversity (H) is the average of the allelic diversity values of the according loci.
Combined result of ETR, MIRU (5 non-overlapping loci) and QUB.
Fifty-one strains belonged to the Beijing family, and 18 strains were non-Beijing family strains.
NA, not applicable.
VNTR typing.
A search of the literature identified 17 VNTR loci (Table 1) that had shown polymorphism among Beijing family strains. One locus, alternatively designated ETR-E and MIRU31, was amplified by using two different primer sets described in the original studies (8, 31) to investigate the reproducibility of the method in our laboratory. With these different primer sets, the results for this locus were 100% reproducible, confirming previous studies on the intra- and interlaboratory reproducibility of VNTR typing with ETRs (15) and MIRUs (6, 24, 31). The results for MIRU31 are therefore identical to those of ETR-E. Similarly, ETR-D and MIRU4 designate the same locus, and this locus was solely amplified by using the primers described by Frothingham and Meeker-O'Connell (8). Three loci (MIRU20, QUB3232, and QUB3336) were discarded because we were unable to amplify them by using the protocol of Frothingham and Meeker-O'Connell (8). The final VNTR patterns and a dendrogram indicating the mutual similarities between the VNTR patterns are shown in Fig. 1.
In total, VNTR typing differentiated 57 genotypes among the 69 isolates (HGI, 0.990) (Table 2). The 51 Beijing family isolates were divided into 39 VNTR types (Table 2). Among the Beijing family strains, VNTR typing identified seven clusters with isolates that invariably had the nine-spacer spoligotype (Fig. 1, clusters V1 to V7). Most of the Beijing strains could be recognized by a VNTR signature (Fig. 1) even though we used VNTR loci that had already shown variability among Beijing strains. The most common VNTR patterns among Beijing strains were 42435 for the ETR profile (n = 35, 68.6%), 33733 for the MIRU profile (n = 27, 52.9%), and 8684 for the QUB profile (n = 16, 31.4%). The largest VNTR cluster (V3), consisting of six isolates, also represented this combined VNTR type 42435-33733-8684. The predominant ETR, MIRU, and QUB profiles among Beijing strains were not observed among any of the non-Beijing strains. The alleles at MIRU26 (seven repeats) and ETR-E/MIRU31 (five repeats) were the most specific for Beijing in this collection, as these were not observed (MIRU26) or observed only once (ETR-E/MIRU31) among non-Beijing strains.
The allelic diversity differed significantly among VNTR loci. The highest allelic diversity (h) among all strains was observed for QUB11b (0.735), and the lowest allelic diversity was observed for MIRU16 (0.097) (Table 3). Comparing the allelic diversity among Beijing and non-Beijing strains showed that, for 12 of the 14 VNTR loci, the allelic diversity among Beijing strains was lower, consistent with the clonal relationship between these strains (39). The two loci that showed more diversity among the Beijing strains were ETR-C and MIRU10. Locus ETR-B was conserved among all Beijing isolates (h = 0.000), and the next most conserved loci were ETR-D/MIRU4 (h = 0.019) and MIRU16 (h = 0.058) (Table 3). The most diverse loci among Beijing strains were MIRU10 (h = 0.377), QUB11a (h = 0.384), and QUB11b (h = 0.618). These findings suggest that, in both Beijing and non-Beijing strains, some ETR and MIRU loci evolve at a slower rate compared to some QUB loci.
TABLE 3.
Allelic numbers and diversity of each VNTR locus in 69 M. tuberculosis strains from Hong Kong
| VNTR locus | VNTR locus alias | VNTR convention | No. of alleles | Allelic diversity (h) of (n):
|
||
|---|---|---|---|---|---|---|
| Beijing strains (51) | Non- Beijing strains (18) | All strains (69) | ||||
| ETR-A | VNTR 2165 | 6 | 0.201 | 0.471 | 0.296 | |
| ETR-B | VNTR 2461 | 6 | 0.000 | 0.575 | 0.201 | |
| ETR-C | VNTR 0577 | 5 | 0.165 | 0.052 | 0.151 | |
| ETR-Da | MIRU4 | VNTR 0580 | 5 | 0.019 | 0.654 | 0.272 |
| ETR-E | MIRU31 | VNTR 3192 | 6 | 0.200 | 0.412 | 0.504 |
| MIRU10 | VNTR 0960 | 5 | 0.377 | 0.327 | 0.546 | |
| MIRU16 | VNTR 1644 | 3 | 0.058 | 0.150 | 0.097 | |
| MIRU26 | VNTR 2996 | 7 | 0.200 | 0.484 | 0.522 | |
| MIRU39 | VNTR 4348 | 5 | 0.320 | 0.333 | 0.540 | |
| MIRU40 | VNTR 0802 | 4 | 0.196 | 0.549 | 0.397 | |
| QUB11a | VNTR 2163a | 10 | 0.384 | 0.755 | 0.568 | |
| QUB11b | VNTR 2163b | 7 | 0.618 | 0.804 | 0.735 | |
| QUB26 | VNTR 4052 | 9 | 0.299 | 0.817 | 0.510 | |
| QUB1895 | VNTR 1895 | 4 | 0.229 | 0.471 | 0.315 | |
The difference between short and long alleles is not considered.
IS6110 RFLP typing.
To further evaluate the usefulness of VNTR typing, all isolates were also subjected to the internationally standardized IS6110 RFLP typing method (36). IS6110 RFLP typing of the 69 M. tuberculosis isolates resulted in 65 RFLP types (HGI, 0.998), 48 types of which were observed among the Beijing family strains (Table 2). Two RFLP clusters were observed among Beijing strains (Fig. 1, clusters R1 and R2). The three Beijing isolates of RFLP cluster R1 also had identical spoligo and VNTR patterns (VNTR cluster V6). The two Beijing family strains of RFLP cluster R2 exhibited identical spoligo patterns, but they had VNTR patterns that differed by two repeats at MIRU26 and three repeats at QUB26.
Of the seven clusters identified among Beijing family strains by VNTR, six were subdivided by IS6110 RFLP. Three VNTR clusters consisted of two isolates with highly similar IS6110 RFLP patterns, differing in one, two, and three IS6110 bands, respectively (clusters V2, V4, and V7). Two clusters consisted of two isolates that showed more diversity in their RFLP patterns, differing by five (V5) or seven (V1) bands. One cluster (V3) consisted of six isolates with IS6110 RFLP patterns that shared a lower degree of similarity (78%).
DISCUSSION
Application of DNA fingerprinting has disclosed that the current worldwide tuberculosis epidemic is for a significant part determined by the worldwide spread of the Beijing family of M. tuberculosis (3, 10). Therefore, knowledge on the applicability of the recently described and increasingly used VNTR typing method to characterize and differentiate this group of strains is of the utmost importance. So far, limited information was available about the utility of VNTR typing to differentiate Beijing family isolates.
In this study, 69 M. tuberculosis isolates from Hong Kong, 74% of which were members of the Beijing family, were analyzed by VNTR typing by using 14 loci (including ETRs, MIRUs, and QUBs), IS6110 RFLP typing, and spoligotyping. Overall, the correlation between the three sets of genetic markers was high. All strains that were defined as belonging to the Beijing genotype on the basis of spoligotyping (14) showed the Beijing-characteristic multicopy IS6110 RFLP patterns (>60.2% similarity) described by van Soolingen et al. (39) and related VNTR patterns (>55.5% similarity). Therefore, the three methods scored equally in recognizing Beijing strains. However, spoligotyping yielded the most unambiguous identification of Beijing genotypes in the fastest and easiest way. As expected, the ability to differentiate between the 51 Beijing isolates varied between the three typing techniques. IS6110 RFLP (48 types; HGI, 0.997) was slightly superior to VNTR (39 types; HGI, 0.982), and both were highly superior to spoligotyping (5 types; HGI, 0.253).
It should be noted that the relative discriminatory powers of VNTR and RFLP typing may vary, depending on the strain collection. In this study, six of the seven VNTR-defined clusters were subdivided by RFLP, and two (including one cluster of isolates containing one copy of IS6110) of the three RFLP-defined clusters were split up by VNTR typing. The epidemiological significance of these differences is unknown because data on epidemiological linkage between patients was not available. As previously observed, at least some of the patterns differing by one to three IS6110 bands, but clustered by VNTR, may nevertheless correspond to epidemiologically related patients (11, 20, 24). However, in this study, three VNTR clusters showed M. tuberculosis Beijing isolates with IS6110 RFLP patterns that appeared too divergent to correspond to recent epidemiological links. This suggests that VNTR alone may overestimate recent transmission of M. tuberculosis Beijing strains. Further studies with detailed patient information are needed to investigate the relation between clustering of VNTR patterns and recent transmission of tuberculosis in more detail. Nevertheless, because of the advantages of VNTR typing over RFLP, VNTR typing can be used as a first screening method followed by additional typing in case Beijing strains with identical VNTR patterns are found.
The allelic diversity of the VNTR loci differed significantly per locus. Among the loci investigated in this study, MIRU10, QUB11a, and QUB11b were the most discriminative among Beijing strains and ETR-B, ETR-D, and MIRU16 were the least discriminative for this group of strains. The most common VNTR patterns for the Beijing strains in this study were 8684 for the QUB loci; 33733 for MIRU loci 10, 16, 26, 39, and 40; and 42435 for ETR loci A to E. This ETR profile is also the most common among members of the W family, which accounted for large outbreaks of (MDR) tuberculosis in the United States (2) and among strains in Russia (7, 21) and in various other countries (15). For all but two of the VNTR loci, the allelic diversity per locus was lower among the Beijing strains, consistent with their close genetic relationship.
Recently, Spurgiesz et al. (30) have subjected 34 M. tuberculosis Beijing family strains representing 14 different IS6110 RFLP types to VNTR typing with nine novel loci with very short repeats, and they were able to discriminate 7 VNTR types among these. It would be interesting to investigate whether some of these novel loci would increase the combined discriminatory power of the VNTR set used in this study or those used in previous studies. Le Fleche et al. (17) evaluated eight other loci and tested these on 90 M. tuberculosis complex strains, including 5 Beijing isolates. However, these loci are not likely to contribute significantly to the discriminatory power of the combined VNTR loci, as these investigators found only little polymorphism in both Beijing and non-Beijing strains. Because different investigators have used different VNTR loci, the interlaboratory comparison of VNTR results is hampered. Ideally, VNTR typing should therefore be standardized internationally. In a standardized procedure, we envisage that a limited number of loci would be amplified as a first screening, followed by a secondary set of VNTR loci in case of clustered isolates or isolates belonging to certain genotypes.
Besides the utility of VNTR typing to discriminate Beijing strains, the utility of spoligotyping with five recently disclosed additional spacers in the direct repeat region (37) was also investigated. Among the 49 Beijing isolates that exhibited the nine-spacer spoligotype in traditional spoligotyping, three types were discriminated by using the novel spacers. Combining the data from this study and that of Van Der Zanden et al. (35), who also subjected Beijing strains to spoligotyping with novel spacers, at least seven spoligotypes can be found among Beijing lineage strains that show the nine-spacer spoligotype in traditional spoligotyping. Thus, these findings indicate that these new spacers will be useful in studying the phylogeny of Beijing strains with greater resolution (35).
The percentage of Beijing strains that showed a spoligotype pattern consisting of less than nine spacers in traditional spoligotyping was comparable to that of another study conducted in Hong Kong (5) (4 and 5%, respectively). In our study, the IS6110 RFLP patterns of the two strains containing fewer than nine spacers in their spoligo pattern contained a high number of bands and matched those of the Beijing family, the modern lineage of the Beijing phylogenetic tree (14), suggesting that the corresponding deletions among the last nine spacers in the DR region of these strains were relatively recent events.
Especially for Beijing family strains, quick and easy simultaneous genotype recognition and epidemiologic typing is of the utmost importance, as these strains are often epidemic and MDR (3, 10). Our results indicate that VNTR typing can be used for epidemiological studies of Beijing strains. However, IS6110 RFLP should be used to subtype VNTR-defined clusters.
Acknowledgments
This study was carried out within the framework of the concerted action project “Next generation genetic markers and techniques to study the epidemiology and control of tuberculosis,” supported by European Union grant QLK2-CT-2000-00630. P.S. is a Researcher of the Centre National de la Recherche Scientifique (CNRS).
Annemarie van den Brandt, Mirjam Dessens, Mimount Enaimi, Petra de Haas, and Tridia van der Laan are gratefully acknowledged for excellent technical assistance. We are grateful to Richard Frothingham (Veterans Affairs Medical Center and Duke University Medical Center, Durham, N.C.) for sharing his VNTR protocol with us.
REFERENCES
- 1.Barlow, R. E., D. M. Gascoyne-Binzi, S. H. Gillespie, A. Dickens, S. Qamer, and P. M. Hawkey. 2001. Comparison of variable number tandem repeat and IS6110-restriction fragment length polymorphism analyses for discrimination of high- and low-copy-number IS6110 Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 39:2453-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bifani, P., B. Mathema, M. Campo, S. Moghazeh, B. Nivin, E. Shashkina, J. Driscoll, S. S. Munsiff, R. Frothingham, and B. N. Kreiswirth. 2001. Molecular identification of streptomycin monoresistant Mycobacterium tuberculosis related to multidrug-resistant W strain. Emerg. Infect. Dis. 7:842-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bifani, P. J., B. Mathema, N. E. Kurepina, and B. N. Kreiswirth. 2002. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 10:45-52. [DOI] [PubMed] [Google Scholar]
- 4.Bifani, P. J., B. Mathema, Z. Liu, S. L. Moghazeh, B. Shopsin, B. Tempalski, J. Driscol, R. Frothingham, J. M. Musser, P. Alcabes, and B. N. Kreiswirth. 1999. Identification of a W variant outbreak of Mycobacterium tuberculosis via population-based molecular epidemiology. JAMA 282:2321-2327. [DOI] [PubMed] [Google Scholar]
- 5.Chan, M. Y., M. W. Borgdorff, C. W. Yip, P. E. W. de Haas, W. S. Wong, K. M. Kam, and D. van Soolingen. 2001. Seventy percent of the Mycobacterium tuberculosis isolates in Hong Kong represent the Beijing genotype. Epidemiol. Infect. 127:169-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cowan, L. S., L. Mosher, L. Diem, J. P. Massey, and J. T. Crawford. 2002. Variable-number tandem repeat typing of Mycobacterium tuberculosis isolates with low copy numbers of IS6110 by using mycobacterial interspersed repetitive units. J. Clin. Microbiol. 40:1592-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drobniewski, F., Y. Balabanova, M. Ruddy, L. Weldon, K. Jeltkova, T. Brown, N. Malomanova, E. Elizarova, A. Melentyey, E. Mutovkin, S. Zhakharova, and I. Fedorin. 2002. Rifampin- and multidrug-resistant tuberculosis in Russian civilians and prison inmates: dominance of the Beijing strain family. Emerg. Infect. Dis. 8:1320-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frothingham, R. and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 144:1189-1196. [DOI] [PubMed] [Google Scholar]
- 9.Gascoyne-Binzi, D. M., R. E. Barlow, R. Frothingham, G. Robinson, T. A. Collyns, R. Gelletlie, and P. M. Hawkey. 2001. Rapid identification of laboratory contamination with Mycobacterium tuberculosis using variable number tandem repeat analysis. J. Clin. Microbiol. 39:69-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glynn, J. R., J. Whiteley, P. J. Bifani, K. Kremer, and D. van Soolingen. 2002. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg. Infect. Dis. 8:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hawkey, P. M., E. G. Smith, J. T. Evans, P. Monk, G. Bryan, H. H. Mohamed, M. Bardhan, and R. N. Pugh. 2003. Mycobacterial interspersed repetitive unit typing of Mycobacterium tuberculosis compared to IS6110-based restriction fragment length polymorphism analysis for investigation of apparently clustered cases of tuberculosis. J. Clin. Microbiol. 41:3514-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunter, P. R., and M. A. Gaston. 1998. Numerical index of the discriminatory ability of typing systems: an application of Simpsons's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. Van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kremer, K., J. R. Glynn, T. Lillebaek, S. Niemann, N. E. Kurepina, B. N. Kreiswirth, P. J. Bifani, and D. van Soolingen. 2004. Definition of the Beijing/W lineage of Mycobacterium tuberculosis on the basis of genetic markers. J. Clin. Microbiol. 42:4040-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kremer, K., D. van Soolingen, R. Frothingham, W. H. Haas, P. W. Hermans, C. Martin, P. Palittapongarnpim, B. B. Plikaytis, L. W. Riley, M. A. Yakrus, J. M. Musser, and J. D. van Embden. 1999. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J. Clin. Microbiol. 37:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwara, A., R. Schiro, L. S. Cowan, N. E. Hyslop, M. F. Wiser, S. Roahen-Harrison, P. Kissinger, L. Diem, and J. T. Crawford. 2003. Evaluation of the epidemiologic utility of secondary typing methods for differentiation of Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 41:2683-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Fleche, P., M. Fabre, F. Denoeud, J. L. Koeck, and G. Vergnaud. 2002. High resolution, on-line identification of strains from the Mycobacterium tuberculosis complex based on tandem repeat typing. BMC Microbiol. 2:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lok, K. H., W. H. Benjamin, M. E. Kimerling, V. Pruitt, D. Mulcahy, N. Robinson, N. B. Keenan, and N. E. Dunlap. 2002. Molecular typing of Mycobacterium tuberculosis strains with a common two-band IS6110 pattern. Emerg. Infect. Dis. 8:1303-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez, B., D. Aguilar, H. Orozco, M. Burger, C. Espitia, V. Ritacco, L. Barrera, K. Kremer, P. R. Hernandez, K. Huygen, and D. van Soolingen. 2003. A marked difference in pathogenesis and immune response induced by different Mycobacterium tuberculosis genotypes. Clin. Exp. Immunol. 133:30-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mazars, E., S. Lesjean, A. L. Banuls, M. Gilbert, V. Vincent, B. Gicquel, M. Tibayrenc, C. Locht, and P. Supply. 2001. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc. Natl. Acad. Sci. USA 98:1901-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mokrousov, I., I. Filliol, E. Legrand, C. Sola, T. Otten, E. Vyshnevskaya, E. Limeschenko, B. Vyshnevskiy, O. Narvskaya, and N. Rastogi. 2002. Molecular characterization of multiple-drug-resistant Mycobacterium tuberculosis isolates from northwestern Russia and analysis of rifampin resistance using RNA/RNA mismatch analysis as compared to the line probe assay and sequencing of the rpoB gene. Res. Microbiol. 153:213-219. [DOI] [PubMed] [Google Scholar]
- 22.Rad, M. E., P. Bifani, C. Martin, K. Kremer, S. Samper, J. Rauzier, B. Kreiswirth, J. Blazquez, M. Jouan, D. van Soolingen, and B. Gicquel. 2003. Mutations in putative mutator genes of Mycobacterium tuberculosis strains of the W-Beijing family. Emerg. Infect. Dis. 9:838-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roring, S., A. Scott, D. Brittain, I. Walker, G. Hewinson, S. D. Neill, and R. A. Skuce. 2002. Development of variable-number tandem repeat typing of Mycobacterium bovis: comparison of results with those obtained by using existing exact tandem repeats and spoligotyping. J. Clin. Microbiol. 40:2126-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Savine, E., R. M. Warren, G. D. van der Spuy, N. Beyers, P. D. van Helden, C. Locht, and P. Supply. 2002. Stability of variable-number tandem repeats of mycobacterial interspersed repetitive units from 12 loci in serial isolates of Mycobacterium tuberculosis. J. Clin. Microbiol. 40:4561-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Selander, R. K., D. A. Caugant, H. Ochman, J. M. Musser, M. N. Gilmour, and T. S. Whittan. 1986. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl. Environ. Microbiol. 51:873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skuce, R. A., T. P. McCorry, J. F. McCarroll, S. M. M. Roring, A. N. Scott, D. Brittain, S. L. Hughes, R. G. Hewinson, and S. D. Neill. 2002. Discrimination of Mycobacterium tuberculosis complex bacteria using novel VNTR-PCR targets. Microbiology 148(Part 2):519-528. [DOI] [PubMed] [Google Scholar]
- 27.Smith, N. H., J. Dale, J. Inwald, S. Palmer, S. V. Gordon, R. G. Hewinson, and J. M. Smith. 2003. The population structure of Mycobacterium bovis in Great Britain: clonal expansion. Proc. Natl. Acad. Sci. USA 100:15271-15275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smittipat, N., and P. Palittapongarnpim. 2000. Identification of possible loci of variable number of tandem repeats in Mycobacterium tuberculosis. Tuber. Lung Dis. 80:69-74. [DOI] [PubMed] [Google Scholar]
- 29.Sola, C., I. Filliol, E. Legrand, S. Lesjean, C. Locht, P. Supply, and N. Rastogi. 2003. Genotyping of the Mycobacterium tuberculosis complex using MIRUs: association with VNTR and spoligotyping for molecular epidemiology and evolutionary genetics. Infect. Genet. Evol. 3:125-133. [DOI] [PubMed] [Google Scholar]
- 30.Spurgiesz, R. S., T. N. Quitugua, K. L. Smith, J. Schupp, E. G. Palmer, R. A. Cox, and P. Keim. 2003. Molecular typing of Mycobacterium tuberculosis by using nine novel variable-number tandem repeats across the Beijing family and low-copy-number IS6110 isolates. J. Clin. Microbiol. 41:4224-4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Supply, P., S. Lesjean, E. Savine, K. Kremer, D. van Soolingen, and C. Locht. 2001. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39:3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Supply, P., J. Magdalena, S. Himpens, and C. Locht. 1997. Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol. Microbiol. 26:991-1003. [DOI] [PubMed] [Google Scholar]
- 33.Supply, P., E. Mazars, S. Lesjean, V. Vincent, B. Gicquel, and C. Locht. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 36:762-771. [DOI] [PubMed] [Google Scholar]
- 34.Supply, P., R. M. Warren, A. L. Banuls, S. Lesjean, G. D. van der Spuy, L. A. Lewis, M. Tibayrenc, P. D. van Helden, and C. Locht. 2003. Linkage disequilibrium between minisatellite loci supports clonal evolution of Mycobacterium tuberculosis in a high tuberculosis incidence area. Mol. Microbiol. 47:529-538. [DOI] [PubMed] [Google Scholar]
- 35.Van Der Zanden, A. G. M., K. Kremer, L. A. Schouls, K. Caimi, A. Cataldi, A. Hulleman, N. J. D. Nagelkerke, and D. van Soolingen. 2002. Improvement of differentiation and interpretability of spoligotyping for Mycobacterium tuberculosis complex isolates by introduction of new spacer oligonucleotides. J. Clin. Microbiol. 40:4628-4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, and P. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Embden, J. D., T. van Gorkom, K. Kremer, R. Jansen, B. A. Der-Zeijst, and L. M. Schouls. 2000. Genetic variation and evolutionary origin of the direct repeat locus of Mycobacterium tuberculosis complex bacteria. J. Bacteriol. 182:2393-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Soolingen, D., P. E. W. de Haas, and K. Kremer. 2001. Restriction fragment length polymorphism typing of Mycobacteria, p. 165-203. In T. Parisch and N. G. Stoker (ed.), Mycobacterium tuberculosis protocols. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 39.van Soolingen, D., L. Qian, P. E. de Haas, J. T. Douglas, H. Traore, F. Portaels, H. Z. Qing, D. Enkhsaikan, P. Nymadawa, and J. D. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J. Clin. Microbiol. 33:3234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]