Abstract
Bartonella quintana is a worldwide fastidious bacterium of the Alphaproteobacteria responsible for bacillary angiomatosis, trench fever, chronic lymphadenopathy, and culture-negative endocarditis. The recent genome sequencing of a B. quintana isolate allowed us to propose a genome-wide sequence-based typing method. To ensure sequence discrimination based on highly polymorphic areas, we amplified and sequenced 34 spacers in a large collection of B. quintana isolates. Six of these exhibited polymorphisms and allowed the characterization of 4 genotypes. However, the strain variants suggested by the noncoding sequences did not correlate with the results of pulsed-field gel electrophoresis (PFGE), which suggested a higher degree of variability. Modification of the PFGE profile of one isolate after nine subcultures confirmed that rearrangement frequencies are high in this species, making PFGE unreliable for epidemiological purposes. The low extent of sequence heterogeneity in the species suggests a recent emergence of this bacterium as a human pathogen. Direct typing of natural samples allowed the identification of a fifth genotype in the DNA extracted from a human body louse collected in Burundi. We have named the typing technique herein described multispacer typing.
Bartonella quintana is a fastidious gram-negative rod that infects humans and belongs to the alpha subgroup of the Proteobacteria (20, 29, 39). Recent reports suggest that humans are the natural reservoir of B. quintana (13) and that the human body louse is the vector (14, 36). Trench fever, occurring in allied and German troops during World War I, was the first disease recognized to be caused by B. quintana (24). It was described as a relapsing fever or quintan fever characterized by attacks of fever associated with headaches, skin pain, and dizziness, recurring every 4 to 6 days (24). B. quintana is also known to be responsible for endocarditis (9, 15, 34, 42, 43) and bacillary angiomatosis, which occur in both human immunodeficiency virus-infected and immunocompetent patients (23, 29, 36, 37).
More recently, chronic asymptomatic bacteremias and relapsing febrile illness have been reported in homeless populations (5, 13, 32, 44, 45). Up to 14% of bacteremic subjects have been retrieved among homeless people tested in the university hospital in Marseilles, France (5), suggesting an epidemic in this population. It is not known whether the epidemic is due to a single, a few, or many different strains. Some of the patients have persistent bacteremias (duration of up to 78 weeks) (13). The mechanisms for persistence of the infection are currently unknown, but since some patients have a concomitantly high level of antibodies and bacteremia, reinfection by other strains may occur. It was previously demonstrated that specific antibodies did not protect from reinfection by another serotype or genotype in Bartonella henselae-infected cats (47). The occurrences of specific pathovars have not been investigated.
The 1.6-Mb genome of B. quintana was recently sequenced (2) and found to be a derivate of the larger 1.9-Mb genome of B. henselae, with the main difference among the two species residing in the absence of genomic islands in the trench fever agent. The availability of complete genome sequence information for B. quintana now allows the rational design of typing methods. To date, only pulsed-field gel electrophoresis (PFGE) has been used for B. quintana typing on a limited number of strains (40). This method was applied to seven different isolates of B. quintana, and a comparison of the genomic fingerprints showed polymorphisms in DNA restriction patterns, with a specific profile for each isolate (40). Surprisingly, the locations of restriction sites of the cutting endonucleases used for PFGE did not match perfectly with the sizes of fragments obtained with PFGE (unpublished data). This emphasizes the need for more reproducible and convenient typing methods.
Multilocus sequence typing is a new typing method based on the comparison of nucleotide sequences of 450- to 500-bp internal fragments of a number (usually seven) of housekeeping genes (12). For each gene, the different sequences obtained are assigned to alleles; alleles at the seven loci provide an allelic profile, allowing nonambiguous determination of sequence type (12). Multilocus sequence typing was first developed for Neisseria meningitidis (12, 27) and Streptococcus pneumoniae (11, 12). It has by now been described for several human pathogens, including B. henselae (19), enabling the identification of seven sequences types among a total of 37 human and feline isolates.
The aim of this study was to develop a simple and reproducible typing method for B. quintana based on highly polymorphic sequence regions. Since spacers (intergenic and pseudogene sequences, also called junk DNA) are more variable than gene sequences, we have here examined a molecular typing method based on the sequences of such noncoding zones rather than of housekeeping genes. This approach was highly successful and we have named the technique MST, which stands for multispacer typing (10).
MATERIALS AND METHODS
B. quintana isolates and growth conditions.
The 71 isolates analyzed in the present study are listed in Table 1. PFGE analysis of the Toulouse, Fuller, Grenoble, Oklahoma, SH-perm, Marseille, and Paris strains have already been described elsewhere (40). Methods used for primary isolation of B. quintana from clinical samples have been described elsewhere (26). Subcultures were performed on blood agar (BioMerieux, Marcy l'Etoile, France) at 37°C in 5% CO2 (Genbag CO2 system; BioMerieux). Four agar plates were obtained for each isolate. Bacteria were harvested and suspended in 500 μl of sterile water after 3 to 5 days of culture.
TABLE 1.
List of B. quintana isolates tested in this work and repartitioned according to genotype for spacers 471 and 894
| Isolate | Yr | Sample source | Disease | Origin | Sequence type for spacer:
|
|
|---|---|---|---|---|---|---|
| 471 | 894 | |||||
| 1084 SDF 2000 | 2000 | Blood culture | Bacteremiaa | Marseilles, France | NDb | 2 |
| 22 | 2001 | Blood culture | Bacteremia | Marseilles, France | ND | 2 |
| 41 647 | 2000 | Blood culture | Bacteremia | Marseilles, France | 2 | 2 |
| 44 235 | 2000 | Blood culture | Bacteremia | Marseilles, France | 2 | 2 |
| 57 | 2000 | Louse | Marseilles, France | ND | 2 | |
| Fuller | 1948 | Blood culture | Trench fever | Yugoslavia | 3 | 3 |
| Grenoble | 1994 | Strain | Bacillary angiomatosis | Grenoble, France | 2 | 2 |
| Marseille | 1993 | Blood culture | Lymphadenopathy | Marseilles, France | 2 | 2 |
| Oklahoma | 1992 | Blood culture | Bacillary angiomatosis | Oklahoma | 2 | 2 |
| Paris | 1993 | Blood culture | Endocarditis | Paris, France | ND | 2 |
| SH-perm | Louse | Russia | 2 | 2 | ||
| Toulouse | 1992 | Strain | Bacillary angiomatosis | Toulouse, France | 3 | 3 |
| UR.BQ.M.NHP.140 | 2001 | Louse | Marseilles, France | ND | 2 | |
| UR.BQ.M.NHP.145A | 2001 | Louse | Marseilles, France | ND | 1 | |
| UR.BQ.M.TD.148 | 2001 | Blood culture | Bacteremia | Marseilles, France | ND | 1 |
| UR.BQ.M.TF.141 | 2001 | Blood culture | Bacteremia | Marseilles, France | 1 | 1 |
| UR.BQ.M.TF.141C | 2001 | Blood culture | Bacteremia | Marseilles, France | 1 | 1 |
| UR.BQ.M.64 | 1999 | Lymph node | Lymphadenopathy | Marseilles, France | ND | 2 |
| UR.BQ.M.AS.13 | 1997 | Blood culture | Bacteremia | Marseilles, France | 2 | 2 |
| UR.BQ.M.AS.15 | 1997 | Blood culture | Bacteremia | Marseilles, France | ND | 2 |
| UR.BQ.M.IE.48 | 1999 | Blood culture | Endocarditis | Marseilles, France | ND | 2 |
| UR.BQ.M.L.Y.1.5 | 1994 | Bone marrow | Lymphadenopathy | Marseilles, France | ND | 2 |
| UR.BQ.M.NHP.108 | 2000 | Louse | Marseilles, France | ND | 2 | |
| UR.BQ.M.NHP.147C | 2001 | Louse | Marseilles, France | ND | 2 | |
| UR.BQ.M.NHP.151E | 2001 | Louse | Marseilles, France | ND | 1 | |
| UR.BQ.M.NHP.158B | 2001 | Louse | Marseilles, France | ND | 2 | |
| UR.BQ.M.NHP.91 | 2000 | Louse | Marseilles, France | 1 | 1 | |
| UR.BQ.M.TF.101 | 2000 | Blood culture | Bacteremia | Marseilles, France | 2 | 2 |
| UR.BQ.M.TF.105 | 2000 | Blood culture | Bacteremia | Marseilles, France | 1 | 1 |
| UR.BQ.M.TF.122 | 2000 | Blood culture | Bacteremia | Marseilles, France | ND | 2 |
| UR.BQ.M.TF.131 | 2000 | Blood culture | Bacteremia | Marseilles, France | ND | 2 |
| UR.BQ.M.TF.138 | 2001 | Blood culture | Bacteremia | Marseilles, France | ND | 1 |
| UR.BQ.M.TF.139 | 2001 | Blood culture | Bacteremia | Marseilles, France | ND | 2 |
| UR.BQ.M.TF.141C | 2001 | Blood culture | Bacteremia | Marseilles, France | 1 | 1 |
| UR.BQ.M.TF.142 | 2001 | Blood culture | Bacteremia | Marseilles, France | ND | 2 |
| UR.BQ.M.TF.142B | 2001 | Blood culture | Bacteremia | Marseilles, France | ND | 2 |
| UR.BQ.M.TF.146 | 2001 | Blood culture | Bacteremia | Marseilles, France | ND | ND |
| UR.BQ.M.TF.146B | 2001 | Blood culture | Bacteremia | Marseilles, France | ND | 2 |
| UR.BQ.M.TF.146D | 2001 | Blood culture | Bacteremia | Marseilles, France | ND | 2 |
| UR.BQ.M.TF.146E | 2001 | Blood culture | Bacteremia | Marseilles, France | 2 | 2 |
| UR.BQ.M.TF.148B | 2001 | Blood culture | Bacteremia | Marseilles, France | 1 | 1 |
| UR.BQ.M.TF.149 | 2001 | Blood culture | Bacteremia | Marseilles, France | 1 | 1 |
| UR.BQ.M.TF.150 | 2001 | Blood culture | Bacteremia | Marseilles, France | ND | 2 |
| UR.BQ.M.TF.152 | 2001 | Blood culture | Bacteremia | Marseilles, France | ND | 1 |
| UR.BQ.M.TF.153 | 2001 | Blood culture | Bacteremia | Marseilles, France | ND | 2 |
| UR.BQ.M.TF.16 | 1997 | Blood culture | Bacteremia | Marseilles, France | 2 | 2 |
| UR.BQ.M.TF.20 | 1997 | Blood culture | Bacteremia | Marseilles, France | ND | 2 |
| UR.BQ.M.TF.201B | 2002 | Blood culture | Bacteremia | Marseilles, France | ND | ND |
| UR.BQ.M.TF.202A | 2002 | Blood culture | Bacteremia | Marseilles, France | ND | ND |
| UR.BQ.M.TF.202F | 2002 | Blood culture | Bacteremia | Marseilles, France | ND | ND |
| UR.BQ.M.TF.21 | 1997 | Blood culture | Bacteremia | Marseilles, France | ND | 2 |
| UR.BQ.M.TF.22 | 1998 | Blood culture | Bacteremia | Marseilles, France | ND | 1 |
| UR.BQ.M.TF.24 | 1998 | Blood culture | Bacteremia | Marseilles, France | ND | 2 |
| UR.BQ.M.TF.36 | 1998 | Blood culture | Bacteremia | Marseilles, France | ND | 2 |
| UR.BQ.M.TF.38 | 1999 | Blood culture | Bacteremia | Marseilles, France | ND | 1 |
| UR.BQ.M.TF.39 | 1999 | Blood culture | Bacteremia | Marseilles, France | ND | 2 |
| UR.BQ.M.TF.41 | 1998 | Blood culture | Bacteremia | Marseilles, France | ND | 2 |
| UR.BQ.M.TF.42 | 1998 | Blood culture | Bacteremia | Marseilles, France | ND | 2 |
| UR.BQ.M.TF.43 | 1998 | Blood culture | Bacteremia | Marseilles, France | ND | 2 |
| UR.BQ.M.TF.44 | 1998 | Blood culture | Bacteremia | Marseilles, France | ND | 2 |
| UR.BQ.M.TF.47 | 1999 | Blood culture | Bacteremia | Marseilles, France | ND | 2 |
| UR.BQ.M.TF.60 | 1999 | Blood culture | Bacteremia | Marseilles, France | ND | 2 |
| UR.BQ.M.TF.61 | 1999 | Blood culture | Bacteremia | Marseilles, France | ND | 2 |
| UR.BQ.M.TF.65 | 1999 | Blood culture | Bacteremia | Marseilles, France | ND | 2 |
| UR.BQ.M.TF.66 | 1999 | Blood culture | Bacteremia | Marseilles, France | ND | 2 |
| UR.BQ.M.TF.85 | 2000 | Blood culture | Bacteremia | Marseilles, France | ND | 2 |
| UR.BQ.M.TF.88 | 2000 | Blood culture | Bacteremia | Marseilles, France | 2 | 2 |
| UR.BQ.P.BA.A.7 | 1995 | Strain | Bacillary angiomatosis | Paris, France | 2 | 2 |
| UR.BQ.P.IE.10 | 1996 | Blood culture | Endocarditis | Paris, France | ND | 2 |
| UR.BQ.P.IE.62 | 1999 | Cardiac valve | Endocarditis | Paris, France | ND | 2 |
| UR.BQ.P.TF.162 | 2001 | Blood culture | Bacteremia | Paris, France | ND | 2 |
All cases of bacteremia were found in homeless people.
ND, not done.
Amplification, sequencing, and PFGE conditions.
A total of 1,427 open reading frames were annotated in the complete genome of B. quintana (Toulouse strain) (2). To propose a typing method based on the sequencing of a fragment amplified from a single PCR run, we retained for analyses only noncoding sequences of 500 to 1,500 bp. We analyzed 178 such sequences, of which 34 were selected for amplification and sequencing with primers (Table 2) designed by using the primer design tool (CyberGene AB).
TABLE 2.
List of primers used for amplification and sequencing of spacers used in this study
| Spacer | Size (bp)a | Forward primer | Reverse primer |
|---|---|---|---|
| 81 | 639 | TCTTGAGTGGTGGAAGCAA | TGACCACAATAATCTAATTC |
| 85 | 693 | AATCTTCTGGCTTTTAATGAAATG | ATTCAATGACATATCAAAATGATC |
| 91 | 702 | GCAATTATTAAGGCCACAGT | ACCGCAATGAAAGCTCCATA |
| 112 | 793 | TGTTGCTCCTGCTGCCAAAA | GCTCAATGTGTTCTTTATCA |
| 128 | 1,077 | GCTGTAATTGGTGAAGTTCT | TCTTCGCAGGACTATAAATA |
| 129 | 1,136 | AGTGATAGGCAGCAATCTTG | TCTGATATTCATGCTTATGATACG |
| 136 | 654 | ATTGTAAGCGGAGTTCTCAAAATC | TTACATTGGTGGTCAGCATCTTTC |
| 144 | 1,158 | CAAGTGGCTAAGATTATGTT | TGACTTACAGATTCTATGCC |
| 170 | 1,375 | CAAAATTGGCGAGCTTCAC | ATAACCTGGCGTTTGCTCAT |
| 178 | 908 | AAGCCTTTTGCGATTCTACCC | TCAGCCCAATCAATGCGAAG |
| 192 | 726 | TTCTCGTTATCCTCTCGTATG | CCTGCAATGCTGAATTTTGG |
| 219 | 702 | GTTTTTAGTACGCCTCCCAC | CTCTATCCAAAGCACAAACT |
| 253 | 716 | TTTATTCCCGGTAACATGCC | AGAAAGCGGCGCATATCGTT |
| 303 | 746 | AACAGCATCAACGAACAAGC | GGAGTTGTCTAAAGGAAGGA |
| 322 | 1,158 | AGATACTCCTTTCGTGCTGC | TTTTACGTGAGGCGGGAAAG |
| 336 | 740 | GCTAAGAAAGAAAGCGAAGC | ATGCTAACCCACTAAAACGG |
| 339 | 1,501 | CCGCACTGAGAATTTTAGAG | GGCGAAAGGGCATCATAATC |
| 352 | 1,733 | CGAAACATCTATCGCCAAAC | GAGAAAAAGCACTGGCTATT |
| 355 | 1,194 | TATATTCAGCGCGGTAATCG | CTTAGGAAGAGTGTAGATGA |
| 385 | 999 | AACACATCGCAATGAATCCG | GCAACATCTTCAGCAAGACG |
| 405 | 676 | TTTCTATCTGATAATGACTCAAACC | ATTCCCGCTACGAGAAAGAAAAC |
| 437 | 848 | GATAAAGGAAGCATTCATCG | ATCGTCATCAAGGCAAAAAC |
| 438 | 837 | AGTAAAGCTACTCTAAAGCC | CCTTCCCATTGTTTAATCAG |
| 470 | 1,468 | TAACATCTCTTGCGTTAAGC | TGAGGTCGATATTTACGAGC |
| 471 | 869 | AGCCTTACAGGCAAGAACAAATC | ATGATAGTAAAAGCAATGATAAACC |
| 472 | 826 | AGGATAGTGCATTATTTTAGC | TTTATGACGAATGAGCGCAA |
| 507 | 1,419 | TGATCCAGCAAGTGCTTCAG | GTTCGTCTACGAGCGTTGTT |
| 531 | 948 | GATATTATGAAAAACCATCAG | AAGATAAATGGTGCGAGCAC |
| 581 | 740 | ACCAGAAGCAAGTGACGTTC | GATATGAGTGCTTCTGTTGT |
| 593 | 690 | ATTTTTCTTTTGAACAAATGC | GCGGCATCATAGTCATTACC |
| 597 | 827 | GCCGCTTATTGTTGTCGATC | CGGCGTCTCAGTATAGAATT |
| 598 | 821 | CCATAAGCAGCAAGTGCGAT | AGCAATGATGGCACTCTTGC |
| 894 | 683 | CGGTTTGTAACGCTCTCAATGGA | TCAACGTAATCGTTCTCTGTGTC |
| 895 | 809 | ATATGCAAAAAGGCAAATGACCTG | TTAGGAATATCAACCAAAACTGATC |
Sizes refer to estimated sizes of PCR products according to the genome sequence of the B. quintana Toulouse strain (2).
DNA was extracted by using the Chelex procedure (7). PCR conditions for all amplification reactions were as follows: initial denaturation at 94°C for 3 min, followed by 44 cycles (94°C for 30 s, 50 to 61°C [according to the melting temperatures of the primers] for 30 s, and 68°C for 90 s), and final extension at 68°C for 7 min. Reactions were performed in 25-μl volumes with buffers and Elongase from Invitrogen Life Technologies (Cergy Pontoise, France). PCR products were visualized under UV illumination after electrophoresis migration on a 1% agarose gel stained with ethidium bromide. PCR products were purified by using the MultiScreen PCR filter plate (Millipore, Saint-Quentin en Yvelines, France) as recommended by the manufacturer.
The PCR products were sequenced in both directions by using the d-rhodamine terminator cycle sequencing ready reaction kit (PerkinElmer, Coignières, France) according to the manufacturer's recommendations. Sequencing products were resolved in an Applied Biosystems automatic sequencer model 3100 (PerkinElmer). As a negative control of each amplification reaction, we used sterile water processed as described above.
PFGE after BstZI restriction endonuclease digestion of B. quintana strains was performed as previously described (40).
Sequence analysis and data deposition.
The nucleotide sequences were edited with the Autoassembler package (PerkinElmer). Multiple alignment of sequences was carried out by using the CLUSTALW webware (http://pbil.ibcp.fr) (46). The sequences of the different genotypes for the six discriminating spacers have been deposited in the EMBL/GenBank databases and given accession numbers, as shown in Table 3.
TABLE 3.
GenBank accession numbers of sequences of different genotypes for the six discriminating spacers
RESULTS
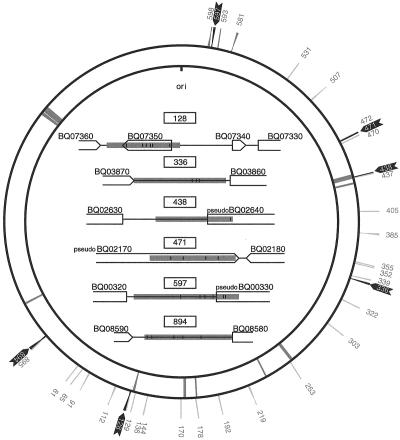
We have here examined the variability of 71 B. quintana isolates (Table 1) based on noncoding sequences as inferred from the complete genome sequence of B. quintana (2). Since the aim was to develop a typing method based on single PCR runs, the first selection criteria were that the sizes of the noncoding sequences should range from 500 to 1,500 bp. Of 178 sequences examined, primer pairs were designed for 34 noncoding segments. These were subjected to several rounds of screenings and selections with the aim of identifying the most useful set of sequences for genotyping purposes (Table 2). Both gene and pseudogene sequences were used as anchors for primer design. Three of the selected sequences contain internal tRNA genes (128, 129, and 339) and another two pseudogenes (144 and 178) in between the flanking gene sequences. Finally, two regions contain internal short open reading frames (128 and 352) putatively coding for proteins of unknown function. The position of the 34 PCR-amplified segments in the genome of the Toulouse strain of B. quintana are shown in Fig. 1.
FIG. 1.
Position and organization of noncoding sequence segments with polymorphisms in B. quintana strains. The outer circle shows the positions of the noncoding sequences analyzed by PCR. Polymorphic sequences are highlighted with black arrows, and sequences found only in one variant are shown in grey. The inner circle indicates the location of remnants of the prophage and genomic islands present in the B. henselae genome (2). The organization of the junk sequences with polymorphisms is displayed inside the two circles. Grey areas show the locations of the sequenced regions. The positions of the polymorphic sites, including both nucleotide substitutions (bars) and deletions (triangles), are shown within the sequenced regions.
Screening for spacers displaying sequence variants.
We hypothesized that the most polymorphic sequences may be those under no or weak selective constraints, i.e., sequences with the lowest similarity to their homologous sequences in B. henselae. For 10 segments, less than 10% of the sequence showed ≥80% similarity with its counterpart in B. henselae. These were selected for a first round of PCR amplifications in the B. quintana isolates Toulouse, Fuller, and Oklahoma, i.e., strains that are well discriminated by PFGE (40). Segment 471, which is flanked on one side by the pseudogene BQ02170 and on the other by the gene BQ02180, and segment 894, which consists of a spacer flanked by the genes BQ08590 and BQ08580, allowed the differentiation of the Toulouse and Fuller strains from the Oklahoma strain (Fig. 1). In total, 4 and 192 nucleotide differences were revealed for segments 471 and 894, respectively. To examine whether additional sequence variants exist, we sequenced region 471 in 18 strains and region 894 in 64 B. quintana isolates, and this allowed the determination of a third genotype. Those three different variants were observed for each of the two segments and were always associated. Following these results, we amplified and sequenced all of the remaining segments for the B. quintana strains UR.BQ.M.TF.141, Oklahoma, and Fuller. Among these, we identified four segments (128, 336, 438, and 597) that displayed sequence variability among the three isolates (Fig. 1). With a total of 17, 6, 10, and 9 nucleotide differences, respectively, these were retained as four additional, putative targets for MST.
Variability in noncoding sequences of selected isolates.
To examine the extent of variability in a broader set of strains, we sequenced spacers 128, 336, 438, 597, and 894 in 15 isolates (Table 4). These strains were selected on the basis of differences in PFGE profiles (40), clinical manifestations (homeless people with trench fever, endocarditis, and bacillary angiomatosis), and geographic origins. In this broader analysis, spacer 128 revealed three sequence types, consisting of single-nucleotide polymorphisms (SNPs) at five positions and one deletion of 12 bp. Spacer 336 distinguished the Fuller strain from all other strains based on SNPs at two positions and a deletion of 4 bp. Two sequence types were associated with spacer 438; these differed by a 10-bp deletion in the pseudogene sequence of BQ02640. Spacer 597 classified the isolates into three types on the basis of SNPs at 4 positions, one deletion of 1 bp, and a deletion of 5 bp. Finally, the sequence variants of spacer 894 included two SNPs, one single-nucleotide deletion, and a large deletion of 189 bp. Spacers 128, 597, and 894 were redundant in the sense that they yielded identical classification results. Taken together, the data suggest the presence of four genotypic variants among the 15 isolates (Table 4).
TABLE 4.
Repartitioning of 15 selected B. quintana isolates according to results of PFGE and MST
| Isolate | Yr | Sample source | Pathology | Origin | PFGE type | Sequence type for spacer:
|
Geno- type | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 128 | 336 | 438 | 597 | 894 | |||||||
| UR.BQ.M.TF.141 | 2001 | Blood culture | Bacteremiaa | Marseilles, France | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| UR.BQ.M.TD.148 | 2001 | Blood culture | Bacteremia | Marseilles, France | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| UR.BQ.M.NHP.145A | 2001 | Louse | Marseilles, France | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Marseille | 1993 | Blood culture | Lymphadenopathy | Marseilles, France | 5 | 2 | 1 | 2 | 2 | 2 | 2 |
| UR.BQ.M.LY.I.5 | 1994 | Bone marrow | Lymphadenopathy | Marseilles, France | 2 | 2 | 1 | 2 | 2 | 2 | 2 |
| UR.BQ.P.BA.A.7 | 1995 | Blood culture | Bacillary angiomatosis | Paris, France | 4 | 2 | 1 | 2 | 2 | 2 | 2 |
| Paris | 1993 | Blood culture | Endocarditis | Paris, France | 5 | 2 | 1 | 2 | 2 | 2 | 2 |
| UR.BQ.P.IE.62 | 1999 | Cardiac valve | Endocarditis | Paris, France | 4 | 2 | 1 | 2 | 2 | 2 | 2 |
| UR.BQ.M.TF.146B | 2001 | Blood culture | Bacteremia | Marseilles, France | 1 | 2 | 1 | 2 | 2 | 2 | 2 |
| UR.BQ.M.NHP.140 | 2001 | Louse | Marseilles, France | 4 | 2 | 1 | 2 | 2 | 2 | 2 | |
| Oklahoma | 1992 | Blood culture | Bacillary angiomatosis | Oklahoma | 2 | 2 | 1 | 2 | 2 | 2 | 2 |
| SH-perm | Unknown | Unknown | Trench fever | Russia | 3 | 2 | 1 | 2 | 2 | 2 | 2 |
| Grenoble | 1994 | Blood culture | Bacillary angiomatosis | Grenoble, France | 2 | 2 | 1 | 2 | 2 | 2 | 2 |
| Toulouse | 1992 | Blood culture | Bacillary angiomatosis | Toulouse, France | 4 | 3 | 1 | 2 | 3 | 3 | 3 |
| Fuller | 1948 | Blood culture | Trench fever | Yugoslavia | 3 | 3 | 2 | 2 | 3 | 3 | 4 |
All cases of bacteremia were found in homeless people.
Direct MST of B. quintana from louse and human samples.
The use of MST was tested on recently isolated samples that previously tested positive for B. quintana by PCR methods described elsewhere (41). These included five lice collected from homeless people in Marseilles (France), four lice collected in Burundi, and one positive cardiac valve sent to our laboratory after cardiac valve surgery in a man from London (England) presenting with endocarditis (Table 5). With the aid of segments 336 and 438, the 10 samples were classified into two types for each segment. Interestingly, four different sequence variants were identified for segment 894, including the three previously observed plus a new sequence variant present in one of the four lice isolated in Burundi. Thus, four different genotypes were identified among the nine louse samples. These encompass all of the previously observed genotypes, with the exception of the sequence variant in the bacillary angiomatosis patient from Toulouse plus a novel, fifth genotype (Table 5).
TABLE 5.
Results of MST performed on samples
| Isolate | Yr | Sample source | Origin | Sequence type for spacer:
|
Geno- type | ||
|---|---|---|---|---|---|---|---|
| 336 | 438 | 894 | |||||
| 15469 | 2002 | Louse | Marseilles, France | 1 | 1 | 1 | 1 |
| 16763 | 2002 | Louse | Marseilles, France | 1 | 2 | 2 | 2 |
| 21007 | 2003 | Louse | Marseilles, France | 1 | 2 | 2 | 2 |
| 21034 | 2003 | Louse | Marseilles, France | 1 | 2 | 2 | 2 |
| 16311 | 2002 | Louse | Marseilles, France | 1 | 2 | 2 | 2 |
| 12733 | 2001 | Louse | Burundi | 2 | 2 | 3 | 4 |
| 12735 | 2001 | Louse | Burundi | 2 | 2 | 3 | 4 |
| 12737 | 2001 | Louse | Burundi | 2 | 2 | 3 | 4 |
| 12739 | 2001 | Louse | Burundi | 1 | 1 | 4 | 5 |
| 10529 | 2000 | Cardiac valve | London, England | 1 | 2 | 2 | 2 |
Patterns of nucleotide changes and insertion/deletion mutations.
Among the noncoding sequences examined here, we identified 12 SNPs and seven insertions/deletions affecting a total of 222 nucleotides. All but one of the single-base substitutions represents GC-AT changes, and only one is an A-T mutation. The insertion/deletion mutations range from single-base deletions to a large deletion of 189 nucleotides. The two single-base deletions are in homopolymeric tracts of 4 to 6 A's or T's in a row. Three indels of sizes 4 to 10 bp are direct repeats of 4 to 10 nucleotides, and the large deletion of 189 nucleotides is flanked by a repeated sequence of 12 bp, in accordance with previous studies which have suggested a role for repeated sequences in the generation of deletion mutations (15).
Lack of correlation between sequence and structure.
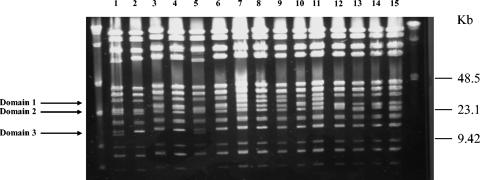
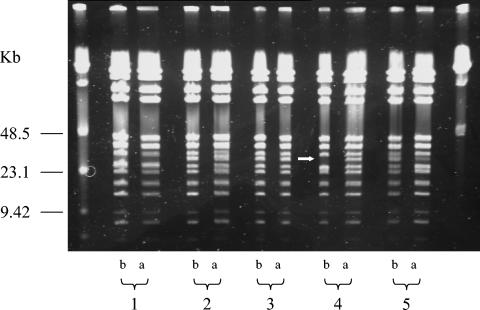
After digestion with BstZI restriction endonuclease, we found five different patterns among the 15 selected B. quintana isolates (Fig. 2; Table 3). The isolates UR.BQ.M.TF.141, UR.BQ.M.TD.148, UR.BQ.M.TF.146B, and UR.BQ.M.NHP.145A were similar to each other and were classified into PFGE type 1 (Fig. 2, domain 1). The UR.BQ.M.LY.I.5 isolate and the Oklahoma and Grenoble strains demonstrated similar patterns (PFGE type 2) (Fig. 2, domain 2), as did the SH-perm and Fuller isolates (PFGE type 3) (Fig. 1, domain 2). Likewise, strains UR.BQ.P.BA.A.7, Toulouse, UR.BQ.P.IE.62, and UR.BQ.M.NHP.140 were similar to each other and categorized into PFGE type 4 (Fig. 2, domain 2). Finally, the Marseille and Paris isolates exhibited identical profiles, referred to here as PFGE type 5 (Fig. 2, domain 3). Curiously, no correlation was retrieved between noncoding sequence types and PFGE types (Table 3). For example, for the BQ.M.TF.146D isolate, the PFGE pattern demonstrated differences before and after subcultures, whereas the MST profile remained the same (Fig. 3).
FIG. 2.
PFGE of BstZI restriction fragments of B. quintana DNAs. Lane 1, Marseille strain; lane 2, UR.BQ.M.LY.I.5 strain; lane 3, UR.BQ.P.BA.A.7 strain; lane 4, Toulouse strain; lane 5, Paris strain; lane 6, UR.BQ.P.IE.62 strain; lane 7, UR.BQ.M.TF.141 strain; lane 8, UR.BQ.M.TD.148 strain; lane 9, UR.BQ.M.TF.146B strain; lane 10, UR BQ.M.NHP.140 strain; lane 11, UR.BQ.M NHP.145A strain; lane 12, Oklahoma strain; lane 13, SH-perm strain; lane 14, Grenoble strain; lane 15, Fuller strain.
FIG. 3.
PFGE of BstZI restriction fragments of five randomly selected B. quintana DNAs of strains after nine subcultures (a) compared with the pattern of the initial strain (b). Lane 1, UR.BQ.M.TF.202.A isolate; lane 2, UR.BQ.M.TF.202.F isolate; lane 3, UR.BQ.M.TF.146 isolate; lane 4, UR.BQ.M.TF.146D isolate; lane 5, UR.BQ.M.TF.201.B isolate. The white arrow shows a variation in the PFGE profile of the BQ.M.TF.146D isolate before and after subcultures.
DISCUSSION
This study has revealed a surprisingly low level of sequence polymorphisms in the noncoding DNA of the examined B. quintana strains. Initially, we speculated that intergenic sequences including pseudogenes not under selective pressure should be a likely source of sequence variability among strains of the same species. Indeed, one of the most widely used intergenic spacers, the 16S-23S rRNA spacer region, exhibits extensive variability in many bacterial species, including Streptococcus sp. (18), Tropheryma whipplei (28), and mycobacteria (30). However, the low frequency of polymorphisms in B. quintana is in agreement with a previous study in which 16S-23S rRNA spacer sequencing allowed the identification of a specific sequence for each of the tested B. henselae isolates, while the B. quintana isolates fell into only two different groups (40).
Moreover, recent data obtained from complete genome sequencing of the two species suggest that B. quintana is a subset of B. henselae, with an evolutionary scenario that involves losses and rearrangements in the B. quintana genome (2). Such losses of sequences, associated with rearrangements, have been described previously for parasitic bacteria (16). For example, the genome of Rickettsia prowazekii (3), another louse-borne pathogen, is a subset of the genome of a close relative, Rickettsia conorii (31). Because the emergence of the human body louse may be no older than 100,000 years (21), its strict coevolving pathogens, such as R. prowazekii and B. quintana, are likely to represent recently evolved species. A recent adaptation of B. quintana to its unique human host and louse vector would provide an explanation for the low level of sequence variability in the genomes of the natural isolates of this species.
Another curious finding was the more extensive variation of the PFGE profiles than of the MST results. Our hypothesis is that this variability is due to frequent genome rearrangements in B. quintana, a scenario that is supported by the modification of the PFGE profile in one strain after nine subcultures while its MST profile remained the same. Such rearrangements have already been demonstrated with Yersinia pestis, in which it appeared as if different colonies obtained from the same strain displayed different PFGE patterns (17). It has recently been demonstrated that long repeated sequences are located in the genomic islands of the B. henselae genome and that the constituting remnants of these islands are associated with rearrangements in B. quintana (2). Genome rearrangements triggered by repeats have already been described for other bacteria, including T. whipplei (4, 35), Y. pestis (8, 22, 33), Mycoplasma spp. (38), and Anaplasma (1, 6). Gene degradations, gene duplications, and genome rearrangements are the main forces that allow evolution and niche adaptation of intracellular pathogens, as acquisition of genes from their eukaryotic host appear insignificant (25, 31, 35). Taken together, the data suggest that PFGE is not a convenient tool for molecular typing of bacteria with extensive genome rearrangements, like that of B. quintana. Based upon our data, DNA rearrangement does not compromise the genotyping utility of MST, as MST profiles were not found to be modified after several subcultures.
Finally, using MST based on five noncoding segments allowed us to identify four genotypes of B. quintana. This study did not, however, allow the association between a given genotype and the disease associated with B. quintana infection. We found that genotype 1 is observed in bacteremic homeless people only (three strains), that, in France, genotypes 1 (three strains), 2 (eight strains), and 3 (one strain) may be observed, that genotype 2 was found for the Oklahoma isolate, the sole New World strain tested, and that the oldest isolate (Fuller) was of the original genotype, genotype 4. Unfortunately, a full analysis of the results is hampered by the overrepresentation of strains from France and homeless people. This is mostly due the fastidiousness of B. quintana, especially in patients that are not bacteremic homeless people (26). MST needs to be applied to large collections of B. quintana isolates, so we may know whether it will provide relevant clinical, microbiological, and epidemiological data.
Diagnosis of bacillary angiomatosis and endocarditis are mostly based on the results of serology and/or results of PCR amplification of tissue samples (26), encouraging us to directly test spacer typing on natural samples. Interestingly, four different genotypes were recovered from clinical samples (lice and cardiac valves) by using MST. This allowed the characterization of an additional genotype (genotype 5). Two of the genotypes (1 and 2) were observed in lice collected from homeless people in Marseilles (France), and two others (4 and 5) were observed in lice collected in Burundi. The five genotypes depend on the determination of the sequences of only two spacers (336 and 894). With the exception of the Fuller strain, which is an old isolate, it can be concluded that the B. quintana genotypes now circulating in Africa are different from those observed in Europe. MST likely represents a novel approach for typing of bacteria with low levels of polymorphism and circumvents problems associated with recurrent genome rearrangements. MST allowed the definition of five genotypes within B. quintana species and will now be applied to large collections of isolates and samples from different areas in the world to investigate the geographical distribution of genotypes.
Acknowledgments
The project was funded in part by the Programme Hospitalier de Recherche Clinique 2003.
REFERENCES
- 1.Allred, D. R., T. C. McGuire, G. H. Palmer, S. R. Leib, T. M. Harkins, T. F. McElwain, and A. F. Barbet. 1990. Molecular basis for surface antigen size polymorphisms and conservation of a neutralization-sensitive epitope in Anaplasma marginale. Proc. Natl. Acad. Sci. USA 87:3220-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alsmark, U. C., A. C. Frank, E. O. Karlberg, B. Legault, B. Canbäck, D. Ardell, A. S. Eriksson, A. K. Näslund, S. Handley, S. Huvet, B. La Scola, M. Holmberg, and S. G. Andersson. 2004. The louse-borne human pathogen Bartonella quintana is a genomic derivate of the zoonotic agent Bartonella henselae. Proc. Natl. Acad. Sci. USA 101:9716-9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson, S. G., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Ponten, U. C. Alsmark, R. M. Podowski, A. K. Naslund, A. S. Eriksson, H. H. Winkler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133-140. [DOI] [PubMed] [Google Scholar]
- 4.Bentley, S. D., M. Maiwald, L. D. Murphy, M. J. Pallen, C. A. Yeats, L. G. Dover, H. T. Norbertczak, G. S. Besra, M. A. Quail, D. E. Harris, A. von Herbay, A. Goble, S. Rutter, R. Squares, S. Squares, B. G. Barrell, J. Parkhill, and D. A. Relman. 2003. Sequencing and analysis of the genome of the Whipple's disease bacterium Tropheryma whipplei. Lancet 361:637-644. [DOI] [PubMed] [Google Scholar]
- 5.Brouqui, P., B. Lascola, V. Roux, and D. Raoult. 1999. Chronic Bartonella quintana bacteremia in homeless patients. N. Engl. J. Med. 340:184-189. [DOI] [PubMed] [Google Scholar]
- 6.De La Fuente, J., J. C. Garcia-Garcia, E. F. Blouin, S. D. Rodriguez, M. A. Garcia, and K. M. Kocan. 2001. Evolution and function of tandem repeats in the major surface protein 1a of the ehrlichial pathogen Anaplasma marginale. Anim. Health Res. Rev. 2:163-173. [PubMed] [Google Scholar]
- 7.De Lamballerie, X., C. Zandotti, C. Vignoli, C. Bollet, and P. de Micco. 1992. A one-step microbial DNA extraction method using “Chelex 100” suitable for gene amplification. Res. Microbiol. 143:785-790. [DOI] [PubMed] [Google Scholar]
- 8.Deng, W., V. Burland, G. Plunkett III, A. Boutin, G. F. Mayhew, P. Liss, N. T. Perna, D. J. Rose, B. Mau, S. Zhou, D. C. Schwartz, J. D. Fetherston, L. E. Lindler, R. R. Brubaker, G. V. Plano, S. C. Straley, K. A. McDonough, M. L. Nilles, J. S. Matson, F. R. Blattner, and R. D. Perry. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drancourt, M., J. L. Mainardi, P. Brouqui, F. Vandenesch, A. Carta, F. Lehnert, J. Etienne, F. Goldstein, J. Acar, and D. Raoult. 1995. Bartonella (Rochalimaea) quintana endocarditis in three homeless men. N. Engl. J. Med. 332:419-423. [DOI] [PubMed] [Google Scholar]
- 10.Drancourt, M., V. Roux, L. V. Dang, L. Tran-Hung, D. Castex, V. Chenal, H. Ogata, E. Crubezy, and D. Raoult. 2004.. Genotyping, Orientalis-like Yersinia pestis, and plague pandemics. Emerg. Infect. Dis. 10:1585-1592. [DOI] [PMC free article] [PubMed]
- 11.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144(Pt 11):3049-3060. [DOI] [PubMed] [Google Scholar]
- 12.Enright, M. C., and B. G. Spratt. 1999. Multilocus sequence typing. Trends Microbiol. 7:482-487. [DOI] [PubMed] [Google Scholar]
- 13.Foucault, C., K. Barrau, P. Brouqui, and D. Raoult. 2002. Bartonella quintana bacteremia among homeless people. Clin. Infect. Dis. 35:684-689. [DOI] [PubMed] [Google Scholar]
- 14.Foucault, C., and D. Raoult. 2000. Louse-associated bacterial infections. Infect. Dis. Clin. Pract. 9:281-291. [Google Scholar]
- 15.Fournier, P. E., H. Lelievre, S. J. Eykyn, J. L. Mainardi, T. J. Marrie, F. Bruneel, C. Roure, J. Nash, D. Clave, E. A. James, C. Benoit-Lemercier, L. Deforges, H. Tissot-Dupont, and D. Raoult. 2001. Epidemiological and clinical characteristics of Bartonella quintana and Bartonella henselae endocarditis: a study of 48 patients. Medicine (Baltimore) 80:245-251. [DOI] [PubMed] [Google Scholar]
- 16.Frank, A. C., H. Amiri, and S. G. Andersson. 2002. Genome deterioration: loss of repeated sequences and accumulation of junk DNA. Genetica 115:1-12. [DOI] [PubMed] [Google Scholar]
- 17.Guiyoule, A., F. Grimont, I. Iteman, P. A. Grimont, M. Lefevre, and E. Carniel. 1994. Plague pandemics investigated by ribotyping of Yersinia pestis strains. J. Clin. Microbiol. 32:634-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hassan, A. A., I. U. Khan, A. Abdulmawjood, and C. Lammler. 2003. Inter- and intraspecies variations of the 16S-23S rDNA intergenic spacer region of various streptococcal species. Syst. Appl. Microbiol. 26:97-103. [DOI] [PubMed] [Google Scholar]
- 19.Iredell, J., D. Blanckenberg, M. Arvand, S. Grauling, E. J. Feil, and R. J. Birtles. 2003. Characterization of the natural population of Bartonella henselae by multilocus sequence typing. J. Clin. Microbiol. 41:5071-5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacomo, V., P. J. Kelly, and D. Raoult. 2002. Natural history of Bartonella infections (an exception to Koch's postulate). Clin. Diagn. Lab. Immunol. 9:8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kittler, R., M. Kayser, and M. Stoneking. 2003. Molecular evolution of Pediculus humanus and the origin of clothing. Curr. Biol. 13:1414-1417. [DOI] [PubMed] [Google Scholar]
- 22.Klevytska, A. M., L. B. Price, J. M. Schupp, P. L. Worsham, J. Wong, and P. Keim. 2001. Identification and characterization of variable-number tandem repeats in the Yersinia pestis genome. J. Clin. Microbiol. 39:3179-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koehler, J. E., F. D. Quinn, T. G. Berger, P. E. LeBoit, and J. W. Tappero. 1992. Isolation of Rochalimaea species from cutaneous and osseous lesions of bacillary angiomatosis. N. Engl. J. Med. 327:1625-1631. [DOI] [PubMed] [Google Scholar]
- 24.Kostrzewski, J. 1949. The epidemiology of trench fever. Bull. Acad. Pol. Sci. Med. 7:233-263. [PubMed] [Google Scholar]
- 25.Kurland, C. G., B. Canback, and O. G. Berg. 2003. Horizontal gene transfer: a critical view. Proc. Natl. Acad. Sci. USA 100:9658-9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.La Scola, B., and D. Raoult. 1999. Culture of Bartonella quintana and Bartonella henselae from human samples: a 5-year experience (1993 to 1998). J. Clin. Microbiol. 37:1899-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maiwald, M., P. W. Lepp, and D. A. Relman. 2003. Analysis of conserved non-rRNA genes of Tropheryma whipplei. Syst. Appl. Microbiol. 26:3-12. [DOI] [PubMed] [Google Scholar]
- 29.Maurin, M., and D. Raoult. 1996. Bartonella (Rochalimaea) quintana infections. Clin. Microbiol. Rev. 9:273-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mijs, W., P. de Haas, R. Rossau, L. T. Van der, L. Rigouts, F. Portaels, and D. van Soolingen. 2002. Molecular evidence to support a proposal to reserve the designation Mycobacterium avium subsp. avium for bird-type isolates and ‘M. avium subsp. hominissuis’ for the human/porcine type of M. avium. Int. J. Syst. Evol. Microbiol. 52:1505-1518. [DOI] [PubMed] [Google Scholar]
- 31.Ogata, H., S. Audic, P. Renesto-Audiffren, P. E. Fournier, V. Barbe, D. Samson, V. Roux, P. Cossart, J. Weissenbach, J. M. Claverie, and D. Raoult. 2001. Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science 293:2093-2098. [DOI] [PubMed] [Google Scholar]
- 32.Ohl, M. E., and D. H. Spach. 2000. Bartonella quintana and urban trench fever. Clin. Infect. Dis. 31:131-135. [DOI] [PubMed] [Google Scholar]
- 33.Podladchikova, O. N., G. G. Dikhanov, A. V. Rakin, and J. Heesemann. 1994. Nucleotide sequence and structural organization of Yersinia pestis insertion sequence IS100. FEMS Microbiol. Lett. 121:269-274. [DOI] [PubMed] [Google Scholar]
- 34.Raoult, D., P. E. Fournier, M. Drancourt, T. J. Marrie, J. Etienne, J. Cosserat, P. Cacoub, Y. Poinsignon, P. Leclercq, and A. M. Sefton. 1996. Diagnosis of 22 new cases of Bartonella endocarditis. Ann. Intern. Med. 125:646-652. [DOI] [PubMed] [Google Scholar]
- 35.Raoult, D., H. Ogata, S. Audic, C. Robert, K. Suhre, M. Drancourt, and J. M. Claverie. 2003. Tropheryma whipplei Twist: a human pathogenic Actinobacteria with a reduced genome. Genome Res. 13:1800-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raoult, D., and V. Roux. 1999. The body louse as a vector of reemerging human diseases. Clin. Infect. Dis. 29:888-911. [DOI] [PubMed] [Google Scholar]
- 37.Relman, D. A., J. S. Loutit, T. M. Schmidt, S. Falkow, and L. S. Tompkins. 1990. The agent of bacillary angiomatosis. An approach to the identification of uncultured pathogens. N. Engl. J. Med. 323:1573-1580. [DOI] [PubMed] [Google Scholar]
- 38.Rocha, E. P., and A. Blanchard. 2002. Genomic repeats, genome plasticity and the dynamics of Mycoplasma evolution. Nucleic Acids Res. 30:2031-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rolain, J. M., M. Maurin, and D. Raoult. 2000. Bactericidal effect of antibiotics on Bartonella and Brucella spp.: clinical implications. J. Antimicrob. Chemother. 46:811-814. [DOI] [PubMed] [Google Scholar]
- 40.Roux, V., and D. Raoult. 1995. Inter- and intraspecies identification of Bartonella (Rochalimaea) species. J. Clin. Microbiol. 33:1573-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roux, V., and D. Raoult. 1999. Body lice as tools for diagnosis and surveillance of reemerging diseases. J. Clin. Microbiol. 37:596-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spach, D. H., K. P. Callis, D. S. Paauw, Y. B. Houze, F. D. Schoenknecht, D. F. Welch, H. Rosen, and D. J. Brenner. 1993. Endocarditis caused by Rochalimaea quintana in a patient infected with human immunodeficiency virus. J. Clin. Microbiol. 31:692-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spach, D. H., A. S. Kanter, N. A. Daniels, D. J. Nowowiejski, A. M. Larson, R. A. Schmidt, B. Swaminathan, and D. J. Brenner. 1995. Bartonella (Rochalimaea) species as a cause of apparent “culture-negative” endocarditis. Clin. Infect. Dis. 20:1044-1047. [DOI] [PubMed] [Google Scholar]
- 44.Spach, D. H., A. S. Kanter, M. J. Dougherty, A. M. Larson, M. B. Coyle, D. J. Brenner, B. Swaminathan, G. M. Matar, D. F. Welch, and R. K. Root. 1995. Bartonella (Rochalimaea) quintana bacteremia in inner-city patients with chronic alcoholism. N. Engl. J. Med. 332:424-428. [DOI] [PubMed] [Google Scholar]
- 45.Stein, A., and D. Raoult. 1995. Return of trench fever. Lancet 345:450-451. [DOI] [PubMed] [Google Scholar]
- 46.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamamoto, K., B. B. Chomel, R. W. Kasten, C. C. Chang, T. Tseggai, P. R. Decker, M. Mackowiak, K. A. Floyd-Hawkins, and N. C. Pedersen. 1998. Homologous protection but lack of heterologous-protection by various species and types of Bartonella in specific pathogen-free cats. Vet. Immunol. Immunopathol. 65:191-204. [DOI] [PubMed] [Google Scholar]