Abstract
Purpose
The purpose of this study is to explore which part of the trophectoderm best represents the inner cell mass after aCGH analysis.
Methods
Fifty-one preimplantation genetic diagnosis/preimplantation genetic screening of abnormal blastocysts diagnosed by array comparative genomic hybridization were included in this study. Blastocysts were thawed, incubated for 3 to 4 h, and then biopsied. Four regions were biopsied per blastocyst, including the inner cell mass (ICM), trophectoderm (TE) cells opposite the ICM, TE cells at the upper right of the ICM, and TE cells at the lower right of the ICM. The biopsied pieces were processed through multiple annealing and looping-based amplification cycle sequenced for 24-chromosome aneuploidy screening. The aneuploidy results were compared among the ICM and the different regional trophectoderm cells from the same blastocyst.
Results
Fifty of 51 (98.04%) ICM samples were concordant with at least one of the TE biopsies derived from the same embryos. There were 43 blastocysts in which ICM and the other three TE pieces were consistent. Discordance among the four pieces occurred in eight blastocysts. Only one blastocyst was discordant between the ICM and the other three TE pieces, while seven blastocysts were discordant between one of TE and the other three biopsied pieces. There was no special region that the mosaic TE was located.
Conclusions
Our findings indicate that TE aneuploidy is an excellent predictor of ICM aneuploidy. The blastocyst mosaic cells are inclined to be located in TE. Moreover, the mosaic TE was not limited to the special region.
Keywords: Blastocyst, 24-chromosome aneuploidy screening, Multiple annealing and looping-based amplification cycle sequencing, Mosaic, Preimplantation genetic screening
With the robust growth of genetic test platforms, preimplantation genetic screening (PGS) has developed rapidly [1–3]. About 10 years ago, many in vitro fertilization (IVF) laboratories performed PGS by fluorescent in situ hybridization (FISH) to select euploid embryos [4]. However, FISH is limited because only a few chromosomes can be detected simultaneously in a single biopsied cell. FISH is also less reliable, and some studies show PGS via FISH analysis failed to increase pregnancy or live birth rates [5]. Currently, PGS by the FISH technique is not recommended at cleavage stages. Wilton reported the first successful clinical application of comparative genomic hybridization (CGH)-PGS in 2001 [6], which detects all chromosomes in one biopsied blastomere. Subsequently, 24-chromosome aneuploidy screening has almost replaced FISH. Several years later, the extended technologies, namely array CGH [7, 8] and single nucleotide polymorphism array [9, 10], are widely applied to PGS cycles. Next-generation sequencing (NGS) is another new technique that has been introduced into PGS cycles. Treff and his colleagues [3] evaluated semiconductor-based NGS for genetic analysis of human embryos. Huang [11, 12] validated multiple annealing and looping-based amplification cycle (MALBAC) sequencing for 24-chromosome aneuploidy screening of cleavage-stage embryos and blastocysts.
In addition to the genetic test platforms, there is another key step in PGS: the biopsy. The biopsy procedure should acquire enough cells to gain accurate genetic information, while resulting in the least harm to embryos (oocytes). At present, there are three stages of the biopsy procedure: polar body biopsy from the oocyte, blastomere biopsy of cleavage-stage embryos, and trophectoderm (TE) cell biopsy of blastocysts. The polar body can only predict the genetic information of maternal genome. The aneuploidy rate of oocytes is very high, ranging from 22 to 72% [13]. The blastomere can provide the genetic information for both maternal and paternal genomes, but the mosaic rate of cleavage-stage embryos is between 30 and 85% [11, 14–16]. With the development of embryos into blastocysts, the mosaic rate decreases. Currently, an increasing number of researchers prefer the blastocyst stage as the optimal time to perform biopsies for PGS [17].
It is very difficult to decide if a mosaic aneuploid embryo should be transferred or not. Some scholars reported healthy live births after the transfer of mosaic aneuploid blastocysts [18]. It is unclear whether the genetic information of TE cells reflect that of the inner cell mass (ICM). Furthermore, it is unclear which part of the TE best reflects the ICM. To clarify these aspects, we investigated blastocyst mosaics with an ICM and different regional TE cells.
Materials and methods
This study was approved by the Institutional Review Board of Peking University Third Hospital, China. Written informed consent was obtained from each couple.
A total of 51 abnormal preimplantation genetic diagnosis (PGD)/PGS blastocysts were donated by 23 couples. The ages of the female patients ranged from 24 to 44. The indications of PGD/PGS were carriers of balanced translocations and recurrent miscarriage. The genetic test technique of these PGD/PGS cycles was array CGH.
In all of these PGD/PGS cycles, fertilization was performed by intracytoplasmic sperm injection (ICSI) on the day of oocyte retrieval. Zona pellucida drilling was conducted on day 3 of ICSI, followed by blastocyst culture. All of the cycles were subjected to trophectoderm-cell-biopsy by laser. The biopsied blastocysts were vitrificated individually. The morphological criterion for a blastocyst biopsy was a score above 5 BC according to Gardner’s criterion [19]. Array CGH was performed on 24 sure-plus chips (Illumina). The Sure Plex DNA amplification system was used for whole genome amplification (WGA). Samples and control DNA were labeled with Cy3 and Cy5 fluorophores and then hybridized overnight. After laser scanning, Blue Fuse software was used to analyze microarray data concerning chromatin loss/gain across all 24 chromosomes.
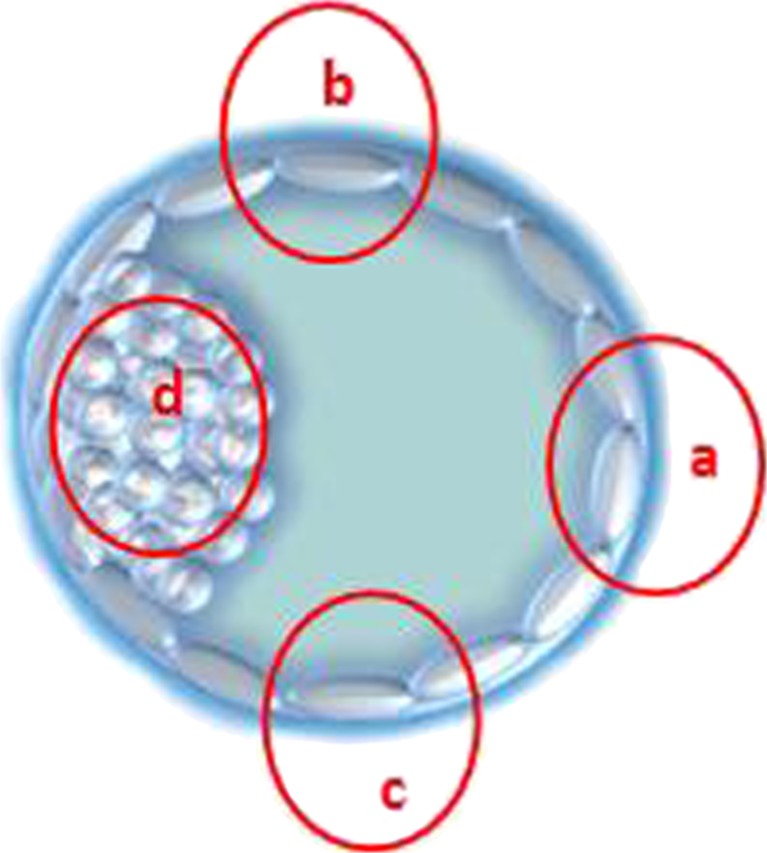
The 51 frozen blastocysts were thawed and incubated for 3 to 4 h. When the thawed blastocysts had expanded, the biopsy was performed. First, the ICM was held at the 9 o’clock position. Then, three regions of TE cells were biopsied, including the TE cells opposite the ICM (at 3 o’clock, region “a”), TE cells at the upper right area of the ICM (at 12 o’clock, region “b”), and TE cells at the lower right area of the ICM (at 6 o’clock, region “c”). Last, ICM cells were biopsied (region “d”) (Fig. 1). TE cells from region “b” or “c” are closer to the ICM, while TE cells from region “d” are farther to the ICM.
Fig. 1.
Different biopsied regions of the blastocyst. a TE cells opposite to the ICM; b TE cells right upper of the ICM; c right down of the ICM; d ICM
The biopsied cells of all four regions were whole genomic amplified by MALBAC. The amplification was initiated by a pool of random primers, each of which had a common 27 nucleotide sequence and eight variable nucleotides. MALBAC can generate the micrograms of DNA required for NGS. Using an Illumina HiSeq 2500 platform, the amplified genome of each biopsied cell was sequenced at an approximate 0.04× genome depth. Therefore, we sequenced a total of approximately 40 million bases, obtaining an average genome coverage of 3% for each single cell [11, 12].
We compared the MALBAC sequencing results of the biopsied cells with the array CGH results of the same blastocyst. Furthermore, we compared the MALBAC sequencing results of the ICM cells and the three regions of TE cells from the same blastocyst.
Results
A total of 204 MALBAC sequencing results were obtained from 51 blastocysts. There were 50 blastocysts whose MALBAC sequencing results were concordant with those of the CGH diagnosis.
Fifty of 51 (98.04%) ICM samples were concordant with at least one of the TE biopsies derived from the same embryos. There were 43 blastocysts (84.31%, 43/51) in which the ICM and the other three TE pieces were consistent. That is, the MALBAC sequencing and array CGH (aCGH) results were consistent among these 43 blastocysts. Table 1 shows the details of these 43 blastocysts.
Table 1.
The blastocysts which ICM and the other three TE pieces were consistent
| Embryo no. | PGD/PGS indication | Biopsy region | MALBAC sequencing results (research results) | aCGH results (PGD/PGS results) |
|---|---|---|---|---|
| 1 | 46,XY,t(10,17) | a, b, c, d | +15,XY | +15,XY |
| 2 | 46,XY,t(3,12) | a, b, c, d | −22,XY | −22,XY |
| 3 | 45,XY,rob(15,21) | a, b, c, d | +8q,+17,XX | +8q,+17,XX |
| 5 | 46,XY,t(7,22) | a, b, c, d | −7p,XY | −7p,XY |
| 7 | 45,XY,rob(13,14) | a, b, c, d | −13XX | −13XX |
| 8 | 45,XY,rob(13,14) | a, b, c, d | +13,+14,XX | +13,+14,XX |
| 9 | 46,XX,t(1,20) | a, b, c, d | −1p, XX | −1p,XX |
| 12 | PGS recurrent miscarriage | a, b, c, d | −19,−21,−22,XY | −19,−21,−22,XY |
| 15 | 46,XY,t(11,13) | a, b, c, d | −11q,+13q,XY | −11q,+13q,XY |
| 16 | 46,XY,t(11,13) | a, b, c, d | +13,XY | +13,XY |
| 17 | 45,XY,rob(13,22) | a, b, c, d | −22,XY | −22,XY |
| 18 | 46,XX,t(2,10) | a, b, c, d | −22,XX | −22,XX |
| 19 | 46,XX,t(2,10) | a, b, c, d | −10q,+15,+16,+21,XX | −10q,+15,+16,+21,XX |
| 20 | 46,XX,t(2,10) | a, b, c, d | +2q,−10q,XY | +2q,−10q,XY |
| 21 | 46,XX,t(3,16) | a, b, c, d | +3q,−16,XX | +3q,−16,XX |
| 22 | 46,XX,t(3,16) | a, b, c, d | +1,XX | +1,XX |
| 24 | 46,XY,t(3,5) | a, b, c, d | −3q,+5q,XY | −3q,+5q,XY |
| 25 | 46,XY,t(3,5) | a, b, c, d | +5,XY | +5,XY |
| 26 | 46,XY,t(3,5) | a, b, c, d | −3q,+5q,XY | −3q,+5q,XY |
| 27 | 46,XX,t(5,21) | a, b, c, d | XO | XO |
| 29 | 46,XX,t(6,12) | a, b, c, d | −16,−22,XY | −16,−22,XY |
| 30 | 46,XX,t(6,12) | a, b, c, d | −6p,+12p,XY | −6p,+12p,XY |
| 31 | 47,XYY | a, b, c, d | −22,XY | −22,XY |
| 32 | 47,XYY | a, b, c, d | −16,XY | −16,XY |
| 33 | 47,XYY | a, b, c, d | +5,XY | +5,XY |
| 34 | 46,XY,t(4,16) | a, b, c, d | +4q,−16q,XX | +4q,−16q,XX |
| 35 | 46,XY,t(4,16) | a, b, c, d | +4q,−16q,XY | +4q,−16q,XY |
| 36 | 46,XY,t(4,16) | a, b, c, d | −4q,+16q,XX | −4q,+16q,XX |
| 37 | 46,XY,t(4,16) | a, b, c, d | +4q,−16q,XX | +4q,−16q,XX |
| 38 | 46,XY,t(4,16) | a, b, c, d | −4q,+16q,XY | −4q,+16q,XY |
| 39 | PGS recurrent miscarriage | a, b, c, d | +17,XY | +17,XY |
| 40 | PGS recurrent miscarriage | a, b, c, d | −18,XX | −18,XX |
| 41 | 46,XY,t(4,16) | a, b, c, d | −6q,+9q,XY | −6q,+9q,XY |
| 42 | 46,XY,t(5,15) | a, b, c, d | +5q,−15q,XY | +5q,−15q,XY |
| 43 | 46,XY,t(5,15) | a, b, c, d | +5q,XX | +5q,XX |
| 44 | 45,XY,rob(13,14) | a, b, c, d | −14,XY | −14,XY |
| 45 | 45,XY,rob(13,14) | a, b, c, d | −19,XY | −19,XY |
| 46 | 45,XY,rob(13,14) | a, b, c, d | +13,+14,XX | +13,+14,XX |
| 47 | 45,XY,rob(13,14) | a, b, c, d | +13,XY | +13,XY |
| 48 | 46,XY,t(8,19) | a, b, c, d | −8p,XX | −8p,XX |
| 49 | 46,XY,t(5,15) | a, b, c, d | −5q,XY | −5q,XY |
| 50 | 46,XY,t(5,15) | a, b, c, d | +5q,XX | +5q,XX |
| 51 | 46,XY,t(5,15) | a, b, c, d | +5q,XX | +5q,XX |
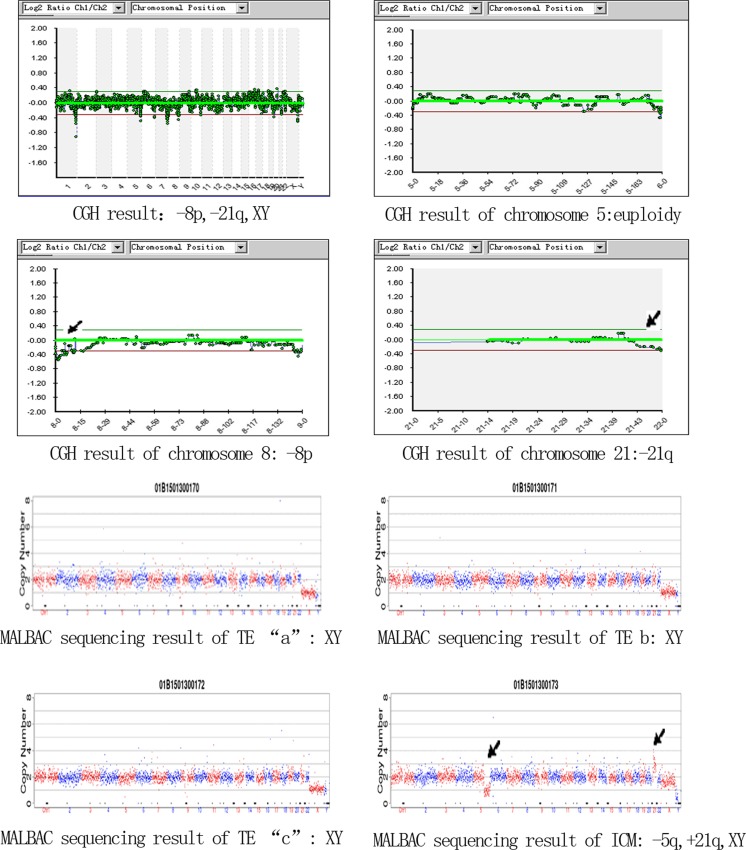
There were eight blastocysts with discordance among the four pieces (15.69%, 8/51). Only one blastocyst was discordant between the ICM and the other three TE pieces, while seven blastocysts were discordant between one of the TE regions and the other three biopsied pieces. Table 2 shows the details of these eight blastocysts. Blastocyst No. 28 was a typical mosaic embryo in which aCGH result indicated “−8p,−21q,XY,” while the MALBAC sequencing results showed that TE regions “a,” “b,” and “c” were “XY,” but the ICM was “−5q,+21q,XY” (Fig. 2). Blastocyst No. 6, 11, and 23 had multiple abnormalities in one of TE biopsy by MALBAC sequencing, which means we detected not only the same aberrations among the other TE cells and ICM but also some other aberrations.
Table 2.
The blastocysts which discordance among the four pieces
| Embryo no. | PGD/PGS indication | Biopsy region | MALBAC sequencing results | aCGH results |
|---|---|---|---|---|
| 4 | 46,XY,t(7,22) | a | 45,XO,+7q | XO |
| b, c, d | 45,XO | |||
| 6 | 46,XY,t(15,19) | a, c, d | 46,XY,+19q | +19q,XY |
| b | Multi-chromosomes abnormal | |||
| 10 | PGS recurrent miscarriage | a, b, d | 47,XX,+22 | +22,XX |
| c | 47,XX,+9q,+22 | |||
| 11 | PGS recurrent miscarriage | a | Multi-chromosomes abnormal | −21,XY |
| b, c, d | 45,XY,−21 | |||
| 13 | 46,XX,t(7,13) | a, c, d | 46,XX,+1,−22 | +1,−22,XX |
| b | 45,XX,−22 | |||
| 14 | 46,XY,t(11,13) | a, b, d | −11q,+13q,XY | −11q,+13q,XY |
| c | −11q,+13q,+16p,XY | |||
| 23 | 45,XX,rob(14,21) | a, b, d | +21,XY | +21,XY |
| c | Multi-chromosomes abnormal | |||
| 28 | 46,XX,t(5,21) | a, b, c | XY | −8p,−21q,XY |
| d | −5q,+21q,XY |
Fig. 2.
The aCGH result and the four biopsied cells’ MALBAC sequencing results of No. 28 blastocyst (the aCGH results show that this blastocyst is “−8p,−21q,XY”; the MALBAC sequencing results show that TE regions a, b , and c are “XY,” but ICM is “−5q,+21q,XY”)
Further analysis of the other seven mosaic blastocysts showed no special region of mosaic cells. The mosaic cells were located among the TE cells in these seven blastocysts, including mosaic cells in region “a” of two blastocysts, region “b” of two blastocysts, and region “c” of three blastocysts. The mosaic cells were multi-chromosome abnormal in three blastocysts and had either duplication or deletion of some segments in some chromosomes of the other four blastocysts.
Discussion
Chromosome imbalance is one of the major factors affecting the success of human IVF. Thus, PGS has become an embryo selection method in clinics. The blastocyst stage may be the optimal time to perform biopsies for PGS. Several TE cells can be obtained from a blastocyst biopsy, which improves the stability of WGA and generates more reliable genetic results [2, 7]. Another important advantage of blastocyst biopsy is that the mosaic rate of blastocysts is lower than that of cleavage-stage embryos, which objectively reflects the genetic information of the embryo.
The mosaic rate of cleavage-stage embryos is relatively high. Our previous study showed that the mosaic rate of D3 embryos is more than 50% [11], which is similar to the results in other studies [20, 21]. Some studies have focused on blastocyst mosaicism. Bradley examined discarded embryos with poor morphology. Thirteen karyotypically abnormal blastocysts were uniform (nonmosaic) [22]. Another study revealed that high proportions of aneuploid blastocysts (69.2%) were mosaic, including an aneuploid TE and euploid ICM, inconsistent anomalies between the ICM and TE or euploid TE cells, and an aneuploid ICM in the same blastocyst [23]. A study by Johnson indicated that approximately 80% of blastocysts are euploid [24].
In our study, we analyzed donated PGD/PGS abnormal blastocysts. ICM cells and three selected regions of TE cells were biopsied per blastocyst. To eliminate the effects of different technologies on the results, only MALBAC sequencing was performed in this study. Our results showed that 50 blastocysts had MALBAC sequencing results that were concordant with the CGH diagnosis, and eight (15.69%) blastocysts were mosaic.
Mosaicism is a major limitation for PGS. There are several reasons that the mosaic proportion of blastocysts is much lower than that of cleavage-stage embryos. First, not all cleavage-stage embryos can develop to the blastocyst stage, especially chromosomally abnormal embryos [25]. Second, mosaic cells in cleavage-stage embryos may develop into TE cells and not the ICM. In prenatal diagnosis, it has been found that some fetuses have normal chromosomes, while the placenta has both normal and abnormal chromosomes [26–28]. There may be a self-correction mechanism during embryonic development. Some abnormal mosaic blastomeres fail to incorporate into the blastocyst. We have previously reported an interesting translocation-PGD case [29]. An embryo was diagnosed as abnormal at the cleavage stage but was normal at the blastocyst stage according to TE cell diagnosis. This embryo was transferred with the couple’s consent, and the patient delivered a healthy baby carrying a chromosomally balanced translocation (tested by amniocentesis).
Studies on mosaics between the TE and ICM are very limited. In 1983, Kalousek and Dill showed chromosomal mosaicism in natural conceptions, and the existence of chromosomal mosaics was strictly confined to tissues of extraembryonic origin [26]. It has been estimated that approximately 2% of viable pregnancies have this type of mosaicism [30]. In a blastocyst biopsy, the TE cells that will develop into the placental tissue are biopsied. Whether the genetic information of the TE can reflect that of the ICM is unknown. A study by Fragouli indicated that all TE and ICM cells were consistent in ten blastocysts according to CGH aneuploidy screening [25]. Liu obtained different results [23]. In 13 blastocysts, four blastocysts had an abnormal TE but a normal ICM. Johnson showed that 96.1% of ICM samples were concordant with TE biopsies derived from the same embryos [24]. Our data showed that 50 of 51 (98.04%) ICM samples were concordant with the other TE biopsies. Therefore, it can be concluded that the TE karyotype is an excellent predictor of the ICM karyotype.
This is a study to investigate the relationship between mosaic cells and the regions of a blastocyst. In our study, seven blastocysts were discordant between one of the TE cell regions and the other three biopsied pieces. There was no special region with mosaic cells. Mosaic cells were randomly located among the TE cells. This study may serve as a guide for blastocyst biopsy. Any region of TE cells can be biopsied, as long as damage to the ICM is avoided.
A rapidly developing technique, NGS, has been applied to PGD/PGS in recent years. Treff and his colleagues [3] investigated the applicability of NGS for PGD and PGS. Their study evaluated semiconductor-based NGS for genetic analysis of human embryos. In our study, we used MALBAC sequencing, which is a combination of WGA and NGS. MALBAC is a newly developed amplification method. Hou [31], Lu [32], and Zong [33] introduced MALBAC for genomic analysis of single human oocytes and single human sperm cells. We used MALBAC to amplify the DNA of the biopsied cells. Then, the amplified genome of each sample was sequenced at an approximate 0.04× genome depth using the Illumina HiSeq 2500 platform. Our previous study validated MALBAC sequencing as a satisfactory method for 24-chromosome aneuploidy screening of cleavage-stage embryos and blastocysts [11, 12]. In this study, MALBAC sequencing showed that 98.04% of blastocysts (50 of 51) had MALBAC sequencing results that were concordant with the aCGH diagnosis. The only discordant blastocyst was a mosaic embryo (no. 28 blastocyst).
Of course, the present study had limitations. The embryos in this study were aCGH diagnosed as imbalanced blastocysts that were donated by clinical PGD/PGS couples. The mosaic proportion of these abnormal blastocysts cannot completely represent that of all blastocysts. However, euploidy blastocysts are the available embryos in clinical IVF, and it is very difficult to use these embryos for research. Second, the blastocysts in this study were chromosomally abnormal embryos, but they had high-quality morphology (the morphological criterion for a blastocyst biopsy was a score above 5 BC according to Gardner’s criterion in this study, as mentioned before). Therefore, the results of this study cannot represent all blastocysts. Moreover, when an expanding blastocyst is biopsied, the blastocyst will immediately collapse, which causes TE cells to cluster tightly. We only biopsied the ICM and three selected TE regions in each blastocyst.
In conclusion, we performed 24-chromosome screening of the ICM and various regions of TE cells in discarded imbalanced blastocysts by MALBAC sequencing. Our findings indicate that the TE karyotype is an excellent predictor of the ICM karyotype. Mosaic cells in blastocysts are inclined to be located in TE, and the mosaic cells were not limited to a specific region. These results not only reveal the genetic information of blastocysts but also provide beneficial information for PGD/PGS genetic counseling and embryo biopsy.
Compliance with ethical standards
This study was approved by the Institutional Review Board of Peking University Third Hospital, China. Written informed consent was obtained from each couple.
Funding
This study was supported by the grants from National High Technology Research and Development Program (2015AA020407), Beijing Municipal Science and Technology Commission (Z131100005213006, CBXM2015-036), the National Natural Science of China (31522034), research fund of National Health and Family Planning Commission of China (201402004) and special funds of Guangxi-distinguished experts, China.
Footnotes
Capsule Re-analysis of the aneuploidy blastocysts reveals that TE is an excellent predictor of ICM. And the mosaic TE was not limited to the special region.
References
- 1.Forman EJ, Hong KH, Ferry KM, Tao X, Taylor D, Levy B, et al. In vitro fertilization with single euploid blastocyst transfer: a randomized controlled trial. Fertil Steril. 2013;100:100–7. doi: 10.1016/j.fertnstert.2013.02.056. [DOI] [PubMed] [Google Scholar]
- 2.Yang Z, Liu J, Collins GS, Salem SA, Liu X, Lyle SS, et al. Selection of single blastocysts for fresh transfer via standard morphology assessment alone and with array CGH for good prognosis IVF patients: results from a randomized pilot study. Mol Cytogenet. 2012;5:24. doi: 10.1186/1755-8166-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Treff NR, Fedick A, Tao X, Devkota B, Taylor D, Scott RT., Jr Evaluation of targeted next-generation sequencing-based preimplantation genetic diagnosis of monogenic disease. Fertil Steril. 2013;99:1377–84. doi: 10.1016/j.fertnstert.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 4.Munné S, Sandalinas M, Escudero T, Velilla E, Walmsley R, Sadowy S, et al. Improved implantation after preimplantation genetic diagnosis of aneuploidy. Reprod Biomed Online. 2003;7:91–7. doi: 10.1016/S1472-6483(10)61735-X. [DOI] [PubMed] [Google Scholar]
- 5.Mastenbroek S, Twisk M, van Echten-Arends J, Sikkema-Raddatz B, Korevaar JC, Verhoeve HR, et al. In vitro fertilization with preimplantation genetic screening. N Engl J Med. 2007;357:9–17. doi: 10.1056/NEJMoa067744. [DOI] [PubMed] [Google Scholar]
- 6.Wilton L, Williamson R, McBain J, Edgar D, Voullaire L. Birth of a healthy infant after preimplantation confirmation of euploidy by comparative genomic hybridization. New Engl J Med. 2001;345:1537–41. doi: 10.1056/NEJMoa011052. [DOI] [PubMed] [Google Scholar]
- 7.Gutiérrez-Mateo C, Colls P, Sánchez-García J, Escudero T, Prates R, Ketterson K, et al. Validation of microarray comparative genomic hybridization for comprehensive chromosome analysis of embryos. Fertil Steril. 2011;95:953–8. doi: 10.1016/j.fertnstert.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 8.Scriven PN, Ogilvie CM, Khalaf Y. Embryo selection in IVF: is polar body array comparative genomic hybridization accurate enough? Hum Reprod. 2012;4:951–3. doi: 10.1093/humrep/des017. [DOI] [PubMed] [Google Scholar]
- 9.Scott RT, Jr, Ferry K, Su J, Tao X, Scott K, Treff NR. Comprehensive chromosome screening is highly predictive of the reproductive potential of human embryos: a prospective, blinded, nonselection study. Fertil Steril. 2012;9:870–5. doi: 10.1016/j.fertnstert.2012.01.104. [DOI] [PubMed] [Google Scholar]
- 10.Schoolcraft WB, Treff NR, Stevens JM, Ferry K, Katz-Jaffe M, Scott RT., Jr Live birth outcome with trophectoderm biopsy, blastocyst vitrification, and single-nucleotide polymorphism microarray–based comprehensive chromosome screening in infertile patients. Fertil Steril. 2011;96:638–40. doi: 10.1016/j.fertnstert.2011.06.049. [DOI] [PubMed] [Google Scholar]
- 11.Huang J, Yan L, Fan W, Zhao N, Zhang Y, Tang F, et al. Validation of multiple annealing and looping-based amplification cycle sequencing for 24-chromosome aneuploidy screening of cleavage-stage embryos. Fertil Steril. 2014;102:1685–91. doi: 10.1016/j.fertnstert.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 12.Huang J, Yan L, Lu S, Zhao N, Xie XS, Qiao J. Validation of a next-generation sequencing-based protocol for 24-chromosome aneuploidy screening of blastocysts. Fertil Steril. 2016;105:1532–6. doi: 10.1016/j.fertnstert.2016.01.040. [DOI] [PubMed] [Google Scholar]
- 13.Fragouli E, Wells D, Thornhill A, Serhal P, Faed MJ, Harper JC, et al. Comparative genomic hybridization analysis of human oocytes and polar bodies. Hum Reprod. 2006;21:2319–28. doi: 10.1093/humrep/del157. [DOI] [PubMed] [Google Scholar]
- 14.Chow JF, Yeung WS, Lau EY. Array comparative genomic hybridization analyses of all blastomeres of a cohort of embryos from young IVF patients revealed significant contribution of mitotic errors to embryo mosaicism at the cleavage stage. Reprod Biol Endocrinol. 2014;12:105. doi: 10.1186/1477-7827-12-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wells D. Embryo aneuploidy and the role of morphological and genetic screening. Reprod Biomed Online. 2010;21:274–7. doi: 10.1016/j.rbmo.2010.06.035. [DOI] [PubMed] [Google Scholar]
- 16.Munné S, Weier HU, Grifo J, Cohen J. Chromosome mosaicism in human embryos. Biol Reprod. 1994;51:373–9. doi: 10.1095/biolreprod51.3.373. [DOI] [PubMed] [Google Scholar]
- 17.Scott KL, Hong KH, Scott RT., Jr Selecting the optimal time to perform biopsy for preimplantation genetic testing. Fertil Steril. 2013;100:608–14. doi: 10.1016/j.fertnstert.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 18.Greco E, Minasi MG, Fiorentino F. Healthy babies after intrauterine transfer of mosaic aneuploid blastocysts. N Engl J Med. 2015;373:2089–90. doi: 10.1056/NEJMc1500421. [DOI] [PubMed] [Google Scholar]
- 19.Gardner DK, Schoolcraft WB. In vitro culture of human blastocysts. In: Jansen R, Mortimer D, editors. Towards reproductive certainty: infertility and genetics beyond. Carnforth, UK: Parthenon Press; 1999. pp. 377–88. [Google Scholar]
- 20.Baart EB, Martini E, van den Berg I, Macklon NS, Galjaard RJ, Fauser BC, et al. Preimplantation genetic screening reveals a high incidence of aneuploidy and mosaicism in embryos from young women undergoing IVF. Hum Reprod. 2006;21:223–33. doi: 10.1093/humrep/dei291. [DOI] [PubMed] [Google Scholar]
- 21.Rius M, Daina G, Obradors A, Ramos L, Velilla E, Fernandez S, et al. Comprehensive embryo analysis of advanced maternal age-related aneuploidies and mosaicism by short comparative genomic hybridization. Fertil Steril. 2011;95:413–6. doi: 10.1016/j.fertnstert.2010.07.1051. [DOI] [PubMed] [Google Scholar]
- 22.Bradley CK, Peura T, Dumevska B, Jovasevic A, Chami O, Schmidt U, et al. Cell lines from morphologically abnormal discarded IVF embryos are typically euploid and unaccompanied by intrachromosomal aberrations. Reprod Biomed Online. 2014;28:780–8. doi: 10.1016/j.rbmo.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 23.Liu J, Wang W, Sun X, Liu L, Jin H, Li M, et al. DNA microarray reveals that high proportions of human blastocysts from women of advanced maternal age are aneuploid and mosaic. Biol Reprod. 2012;87:148. doi: 10.1093/biolreprod/87.s1.148. [DOI] [PubMed] [Google Scholar]
- 24.Johnson DS, Cinnioglu C, Ross R, Filby A, Gemelos G, Hill M, et al. Comprehensive analysis of karyotypic mosaicism between trophectoderm and inner cell mass. Mol Hum Reprod. 2010;16:944–9. doi: 10.1093/molehr/gaq062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fragouli E, Lenzi M, Ross R, Katz-Jaffe M, Schoolcraft WB, Wells D. Comprehensive molecular cytogenetic analysis of the human blastocyst stage. Hum Reprod. 2008;23:2596–608. doi: 10.1093/humrep/den287. [DOI] [PubMed] [Google Scholar]
- 26.Kalousek DK, Dill FJ. Chromosomal mosaicism confined to the placenta in human conception. Science. 1983;221:665–7. doi: 10.1126/science.6867735. [DOI] [PubMed] [Google Scholar]
- 27.Goldberg JD, Wohlferd MM. Incidence and outcome of chromosomal mosaicism found at the time of chorionic villus sampling. Am J Obstet Gynecol. 1997;176:1349–53. doi: 10.1016/S0002-9378(97)70356-9. [DOI] [PubMed] [Google Scholar]
- 28.Kalousek DK, Vekemans M. Confined placental mosaicism. J Med Genet. 1996;33:529–33. doi: 10.1136/jmg.33.7.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang J, Zhao N, Wang X, Qiao J, Liu P. Chromosomal characteristics at cleavage and blastocyst stages from the same embryos. J Assist Reprod Genet. 2015;32:781–7. doi: 10.1007/s10815-015-0450-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ledbetter DH, Zachary JM, Simpson JL, Golbus MS, Pergament E, Jackson L, et al. Cytogenetic results from the US collaborative study on CVS. Prenat Diagn. 1992;12:317–45. doi: 10.1002/pd.1970120503. [DOI] [PubMed] [Google Scholar]
- 31.Hou Y, Fan W, Yan L, Li R, Lian Y, Huang J, et al. Genome analyses of single human oocytes. Cell. 2013;155:1492–506. doi: 10.1016/j.cell.2013.11.040. [DOI] [PubMed] [Google Scholar]
- 32.Lu S, Zong C, Fan W, Yang M, Li J, Chapman AR, et al. Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science. 2012;338:1627–30. doi: 10.1126/science.1229112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–6. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]