Abstract
Purpose
The purpose of this study is to determine the profile of extracellular microRNAs (exmiRNAs) in follicular fluid (FF) and explore their association with fertilization potential and embryo quality.
Methods
We collected FF from single follicles containing mature oocytes from 40 women undergoing IVF and we screened for the expression of 754 exmiRNAs in FF using the TaqMan OpenArray® qPCR platform. To determine the association of exmiRNAs and IVF outcomes, we compared their expression levels in FF samples that differ by fertilization status (normally, abnormally, and failed to fertilize) and embryo quality (top vs. non-top).
Results
We detected 207 exmiRNAs, of which miR-30d-5p, miR-320b, miR-10b-3p, miR-1291, and miR-720 were most prevalent. We identified four exmiRNAs with significant fold change (FC) when FF that contained normally fertilized was compared to failed to fertilize oocytes [miR-202-5p (FC = 1.82, p = 0.01), miR-206 (FC = 2.09, p = 0.04), miR-16-1-3p (FC = 1.88, p = 0.05), and miR-1244 (FC = 2.72, p = 0.05)]. We also found four exmiRNAs to be significantly differentially expressed in FF that yielded top quality versus non-top quality embryos [(miR-766-3p (FC = 1.95, p = 0.01), miR-663b (FC = 0.18, p = 0.02), miR-132-3p (FC = 2.45, p = 0.05), and miR-16-5p (FC = 3.80, p = 0.05)]. In-silico analysis revealed that several of these exmiRNAs are involved in pathways implicated in reproductive system diseases, organismal abnormalities, and organ development.
Conclusions
Our findings suggest that exmiRNAs in the follicular fluid can lead to downstream events that will affect fertilization and day 3 embryo morphology. We encourage further observational and experimental studies to confirm our findings and to determine the role of exmiRNAs in human reproduction.
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-017-0876-8) contains supplementary material, which is available to authorized users.
Keywords: Follicular fluid, MicroRNAs, Extracellular vesicles, Exosomes, Fertilization, Reproduction
Background
Bidirectional communication between the oocyte and granulosa compartment within the ovarian follicle is critical for normal follicular differentiation, oocyte competence, fertilization, and embryonic development [1–3]. Prevailing evidence indicates that interactions between the oocyte and granulosa cells in the follicular microenvironment involve a network of gap junctions and autocrine, paracrine, and endocrine signaling factors [1, 2, 4–6].
Extracellular RNAs (exRNAs) have recently been identified as a new mechanism of intercellular communication, as well as source of biomarkers. In biofluids, exRNAs can be found encapsulated in extracellular vesicles (EVs), in protein-bound complexes, and bound to Argonaute 2 protein [7]. Recent evidence shows that exRNAs encapsulated in EVs can be transferred between cells [8–10]. EVs are nanosized (<1000 nm) double-lipid membrane vesicles that are released by various cell types into human biofluids, including granulosa cells, under both physiological and pathological conditions [9]. Emerging evidence shows that intercellular communication mediated by EVs may support oocyte maturation, sperm penetration into the oocyte, and embryo implantation, although in many cases, the functional roles of EVs in the reproductive tract remain largely unexplored [11].
exRNAs contain considerable amounts of non-coding RNAs, of which microRNAs (miRNAs) are the most abundant [7]. MicroRNAs can regulate gene expression in both normal and disease states by direct targeting of the messenger RNA and inhibition of its translation to a protein, and/or by interfering with the epigenome [12–17]. More specifically, extracellular miRNAs (exmiRNAs) in follicular fluid (FF) are predicted to act on targets that regulate the WNT, epidermal growth factor receptor (ErbB), mitogen-activated protein kinase (MAPK), and transforming growth factor beta (TGFβ) signaling pathways, all of which contribute to follicular development, meiotic resumption, and ovulation [18–22]. Follicular fluid, a product of blood exudate and secretions of granulosa cells, is a unique source of biochemical factors and secreted molecules. Recent studies showed that EVs are present in the FF, in granulosa and cumulus cells, and postulate that EVs can serve as a vehicle for exchanging proteins, lipids, and miRNAs between the oocyte and the granulosa cells around it [18, 23]. However, whether specific exmiRNAs that circulate in follicular fluid encrypt important information about fertilization and embryo quality is currently unknown.
In this study, we characterized the profile of exmiRNAs in FF from which mature oocytes were obtained, and in an exploratory analysis, we examined their association with fertilization potential and embryo quality in patients undergoing IVF.
Methods
Participants
Women 19 to 37 years old undergoing IVF/ICSI in a tertiary university-affiliated hospital from May 2013 through July 2014 with ≤6 previous attempts were eligible for recruitment. Diagnoses for inclusion were male factor, mechanical, or unexplained infertility, as well as preimplantation genetic diagnosis for a known genetic mutation. We excluded women with risk factors or diseases that could influence oocyte quality, such as women with body mass index ≥35 kg/m2, a diagnosis of polycystic ovarian syndrome or endometriosis, a diminished ovarian reserve, a known chromosomal translocation, recurrent abortions, or those carrying Fragile X. To avoid potential confounding bias by different stimulation regimens, we only included women using only one controlled ovarian stimulation protocol: gonadotropin releasing hormone antagonist and Ovidrelle (Merck Serono) for final oocyte maturation. This study was approved by our institution in accordance with the Declaration of Helsinki. All participants provided written informed consent.
IVF protocols and sample collection
Otherwise discarded, FF from a single follicle >18 mm was collected and centrifuged at 1500×g for 15 min to separate the serum fraction debris; 500 μL aliquots of the supernatant were stored immediately at −80 °C. Each oocyte, its corresponding embryo, and FF sample were separately tracked in the clinical IVF laboratory.
Oocytes were stripped and assessed for maturity 1–2 h after retrieval. Since follicles of different maturation status might contain different miRNA profiles [24], only FF from single follicles that contained mature oocytes (i.e., those exhibiting one polar body) were analyzed. Oocytes and embryos were maintained at 37 °C in a humidified atmosphere of 5% CO2 [25]. The fertilization check was performed 16–18 h after ICSI, and zygotes with two pronuclei were cultured individually in 25 μL drops of Sydney IVF cleavage medium K-SICM-20 (Cook, Ireland) until embryo transfer. Embryo morphology was assessed on day 3 using standard criteria based on the number of blastomeres, symmetry, and the extent of fragmentation [26].
RNA extraction from FF
Follicular fluid aliquots (500 μL) were thawed on ice, centrifuged for 15 min at 3000×g at 4 °C, and subsequently filtered using a 0.8 μm membrane unit (Millipore Corp., Bedford, MA, USA) to remove any remaining cellular debris and large aggregates. We extracted total RNA from FF using the exoRNeasy Serum/Plasma Maxi kit (Qiagen, Valencia, CA, USA) [27]. FF samples were processed following the manufacturer’s instructions, except that we modified the recommended XBP buffer/sample volume ratio from 1:1 to 2:1 to optimize use with the FF. We evaluated the concentration, quality, and size distribution of the total RNA extracted from FF using the RNA 6000 Nano Kit on Agilent’s 2100 Bioanalyzer instrument (Agilent Technologies, Foster City, CA) (Figure S1; Supplemental Material).
Expression analysis of exmiRNAs in FF
We screened for the levels of 754 miRNAs using the TaqMan OpenArray® technology on the QuantStudio™ 12K Flex Real-Time PCR System (Life Technologies, Carlsbad, CA). A volume of 6 μL for each sample was prepared, and all samples were reverse-transcribed and pre-amplified (16 cycles) using the Megaplex™ Reverse Transcription Primers, Human Pool A v2.1, Human Pool B v3.0 and Megaplex™ PreAmp Primers, Human Pool A v2.1, and Human Pool B v3.0. Quantitative PCR (qPCR) was performed on the QuantStudio™ 12K Flex Real-Time PCR System. Expression levels were calculated in relative cycle threshold values (Crt), which estimates the amplification cycle at which the fluorescence levels for each of the analyzed miRNAs exceeded the background fluorescence threshold [27].
Outcome variable assessment
We defined normally fertilized oocytes as those exhibiting two pro-nuclei and two polar bodies at the fertilization check, failed to fertilize oocytes as those not exhibiting any pronuclei, and abnormally fertilized oocytes as those presenting one or three pronuclei. Embryo morphology was assessed on day 3 using the standard criteria of the number of blastomeres and extent of fragmentation and blastomere asymmetry [28, 29]. Top quality embryos on day 3 were designated as embryos with 7–8 cells, ≤10% fragmentation, and symmetric blastomeres. Those were the groups we used for our exmiRNAs relative expression comparisons.
Data analysis
We used the Thermo Fisher Cloud Relative Quantification software to extract the miRNA qPCR data. Due to the lack of standard endogenous controls for extracellular RNAs, we decided to use the Delta Crt values (ΔCrt) and global mean method to normalize our data (ΔCrt_miRNAi = (Crt_miRNAi − Crt_miRNAi_global_mean)), as suggested by Mestdagh et al. [30]. We then calculated relative expression levels between the groups by using the fold change method and the 2-ΔΔCt formula [31]. For this analysis, we included only exmiRNAs that exerted a Crt value <35, and an amplification score ≥1.24 and quantification cycle (Cq) confidence score ≥0.8, as recommended by the manufacturer for reliable data. Data that did not meet these criteria were considered as missing values and therefore not included in the analysis. One sample was excluded from the analysis because the sperm was abnormal and all the oocytes retrieved failed to fertilize. Standard descriptive statistics were used to explore the characteristics of the study participants and determine any differences between the compared groups. For all exmiRNAs relative expression group comparisons, the Student’s t test for statistical significance was performed. All statistical analyses were performed in SAS 9.4 (SAS Institute Inc., Cary, NC, USA). Lastly, we used TargetScan Release 7.0 [32] and Ingenuity Pathway Analysis (Ingenuity Systems®, Redwood City, CA, USA) software to identify messenger RNA (mRNA) putative targets of the differentially expressed exmiRNAs (p < 0.05) found in FF, and to in silico explore relevant downstream biological pathways.
Results
Participants
We collected a single FF sample from 46 women, of whom 40 contained a mature oocyte. These women were 30.7 ± 3.7 (mean ± standard deviation [SD]) years old, had body mass index of 22.7 ± 3.5 kg/m2 (range 16.8–31.8), had undergone through 1.8 ± 1.3 (range 1–6 cycles) of IVF prior to enrollment, and had 9.1 ± 3.8 (range 3–21) oocytes retrieved during IVF process. No differences were observed for any of these characteristics between the groups we used for our relative expression comparisons (all p values >0.05).
Profile of exmiRNAs in FF
We screened for 754 exmiRNAs and detected 207 in one or more of 40 samples analyzed. The five most prevalently detected exmiRNAs were miR-30d-5p (40/40 samples), miR-320b (39/40 samples), miR-10b-3p (36/40 samples), miR-1291 (35/40 samples), and miR-720 (34/40 samples). Other prevalently detected exmiRNAs were miR-572 (33/40), miR-7-1-3p (33/40), miR-942-5p (32/40), miR-1226-5p (31/40), miR-1255b-5p (31/40), miR-136-3p (31/40), miR-380-5p (31/40), miR-126-5p (30/40), miR-30a-5p (30/40), and miR-663b (30/40) (Table 1). The full list of detected exmiRNAs in all analyzed samples and by different groups, together with expression levels, is provided in Table S1 (Supplemental Material). In silico analysis of the predicted targets of the five most prevalent exmiRNAs in FF containing mature oocytes—miR-30d-5p, miR-320b, miR10b-3p, miR-1291, and miR-720—revealed their association with biological pathways such as ubiquitin, the tumor necrosis factor (TNF) receptor superfamily, TGFβ, and MAPK pathways; biological processes such as meiosis and embryonic ectoderm development; and biological factors such as cytochrome P450, IZUMO family member 4, zona pellucida binding protein 2, and StAR-related lipid transfer proteins. Notably, ubiquitin and MAPK pathways are associated with oocyte maturation, spindle assembly, and polar body emission [33, 34].
Table 1.
Levels of extracellular miRNAs detected in ≥50% of the analyzed follicular fluid samples from mature oocytes
| miRNA | Mean Crta | SDb | N c |
|---|---|---|---|
| hsa-miR-30d-5p | 20.71 | 3.09 | 40 |
| hsa-miR-320b | 23.87 | 1.36 | 39 |
| hsa-miR-10b-3p | 25.36 | 2.91 | 36 |
| hsa-miR-1291 | 21.61 | 1.71 | 35 |
| hsa-miR-720 | 18.85 | 0.84 | 34 |
| hsa-miR-572 | 23.25 | 1.31 | 33 |
| hsa-miR-7-1-3p | 22.69 | 1.35 | 33 |
| hsa-miR-942-5p | 23.37 | 1.87 | 32 |
| hsa-miR-1226-5p | 21.68 | 1.53 | 31 |
| hsa-miR-1255b-5p | 24.29 | 2.57 | 31 |
| hsa-miR-136-3p | 26.8 | 2.01 | 31 |
| hsa-miR-380-5p | 26.78 | 1.68 | 31 |
| hsa-miR-126-5p | 24.11 | 2.42 | 30 |
| hsa-miR-30a-5p | 17.43 | 2.2 | 30 |
| hsa-miR-663b | 17.42 | 3.03 | 30 |
| hsa-miR-31-3p | 24.77 | 1.42 | 29 |
| hsa-miR-629-3p | 21.99 | 1.97 | 29 |
| hsa-miR-1300 | 26.26 | 1.6 | 28 |
| hsa-miR-424-3p | 22.09 | 1.35 | 28 |
| hsa-miR-590-3p | 27.47 | 1.97 | 28 |
| hsa-miR-202-5p | 26.56 | 1.4 | 27 |
| hsa-miR-206 | 27.13 | 1.74 | 27 |
| hsa-miR-601 | 26.49 | 1.2 | 27 |
| hsa-miR-1290 | 25.85 | 1.76 | 26 |
| hsa-miR-505-5p | 28.1 | 1.52 | 26 |
| hsa-miR-622 | 26.44 | 2.1 | 26 |
| hsa-miR-1303 | 23.71 | 1.33 | 25 |
| hsa-miR-144-5p | 28.17 | 1.77 | 25 |
| hsa-miR-340-3p | 28.1 | 1.14 | 25 |
| hsa-miR-93-3p | 28.42 | 1.63 | 25 |
| hsa-miR-99a-3p | 26.64 | 1.62 | 25 |
| hsa-miR-409-3p | 21.09 | 1.46 | 24 |
| hsa-miR-27a-5p | 27.23 | 1.68 | 23 |
| hsa-miR-661 | 17.75 | 1.88 | 23 |
| hsa-miR-1244 | 29.13 | 1.04 | 22 |
| hsa-miR-20a-3p | 28.21 | 1.41 | 22 |
| hsa-miR-34a-3p | 24.91 | 1.46 | 22 |
| hsa-miR-645 | 27.9 | 1.47 | 22 |
| hsa-miR-1274A | 20.49 | 1.48 | 21 |
| hsa-miR-151-3p | 24.5 | 1.79 | 21 |
| hsa-miR-223-5p | 27.26 | 1.28 | 21 |
| hsa-miR-22-5p | 26.9 | 2.08 | 21 |
| hsa-miR-26a-1-3p | 31.48 | 1.54 | 21 |
| hsa-miR-497-5p | 28.72 | 1.63 | 21 |
| hsa-miR-1274B | 15.09 | 1.34 | 20 |
| hsa-miR-378a-3p | 26.63 | 0.95 | 20 |
aValues are expressed in relative cycle threshold (Crt) values
bStandard deviation
cNumber of samples (out of a total of 40 analyzed) in which each miRNA was detected
Exploratory analysis of exmiRNAs in FF and oocyte fertilization potential
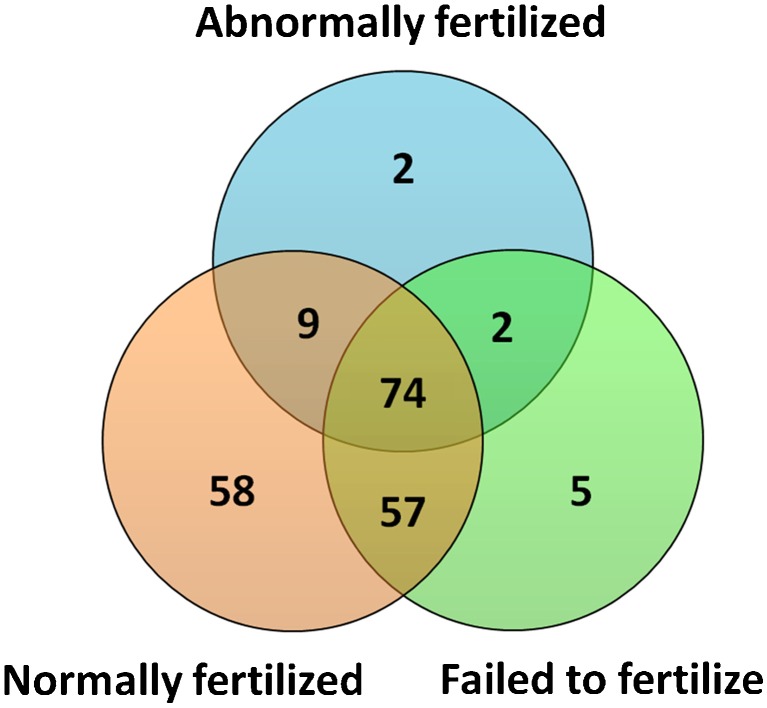
To determine the association between FF exmiRNAs and fertilization, we compared FF samples from oocytes that were normally fertilized (i.e., 2PN) (N = 30) to those that failed to fertilize (i.e., no PN) (N = 5) and to those that were abnormally fertilized (n = 4). Of the 207 exmiRNAs detected, 74 were commonly detected in all groups, 58 were detected only in FF that contained normally fertilized oocytes, 5 were detected only in FF that contained failed to fertilize oocytes, and 2 were detected only in FF that contained abnormally fertilized oocytes (Fig. 1, Tables S2 and S3; Supplemental Material). When we compared the expression levels of FF exmiRNAs in normally fertilized (n = 30) versus failed to fertilize (n = 5) oocytes, we found significantly higher expression levels of miR-202-5p (FC = 1.82, p = 0.01), miR-206 (FC = 2.09, p = 0.04), miR-16-1-3p (FC = 1.88, p = 0.05), and miR-1244 (FC = 2.72, p = 0.05) (Table 2, Table S4; Supplemental Material). In addition, when we compared the expression levels of FF exmiRNAs in normally fertilized (n = 30) versus abnormally (n = 4) fertilized oocytes, we found lower levels of miR-454-5p (FC = 0.25, p < 0.0001), miR-425-3p (FC = 0.32, p = 0.001), miR-16-5p (FC = 0.38, p = 0.003), and miR-222-3p (FC = 0.34, p = 0.04) (Table 3, Table S5; Supplemental Material).
Fig. 1.
Extracellular miRNAs detected in follicular fluid that contained normally fertilized, abnormally fertilized, and failed to fertilize oocytes. The Venn diagram shows overlapping and non-overlapping extracellular miRNAs between the groups
Table 2.
Fold change expression of extracellular miRNAs in follicular fluid containing normally fertilized versus failed to fertilized oocytes
| miRNAa | Fold change | Normally fertilized (n = 30) | Failed to fertilize (n = 5) | P value |
|---|---|---|---|---|
| hsa-miR-202-5p | 1.82 | 22 | 4 | 0.01 |
| hsa-miR-206 | 2.09 | 21 | 3 | 0.04 |
| hsa-miR-16-1-3p | 1.88 | 13 | 4 | 0.05 |
| hsa-miR-1244 | 2.72 | 18 | 2 | 0.05 |
| hsa-miR-340-3p | 1.91 | 18 | 4 | 0.07 |
| hsa-miR-30a-3p | 1.77 | 14 | 3 | 0.08 |
| hsa-miR-614 | 2.50 | 2 | 2 | 0.08 |
| hsa-miR-99a-3p | 2.12 | 18 | 5 | 0.10 |
| hsa-miR-1291 | 1.84 | 27 | 5 | 0.12 |
| hsa-miR-141-5p | 2.93 | 3 | 2 | 0.13 |
| hsa-miR-601 | 1.50 | 21 | 4 | 0.13 |
| hsa-miR-26a-1-3p | 0.48 | 15 | 3 | 0.15 |
| hsa-miR-592 | 1.71 | 5 | 3 | 0.17 |
| hsa-miR-29c-5p | 0.65 | 14 | 2 | 0.21 |
| hsa-miR-1179 | 2.31 | 4 | 3 | 0.27 |
| hsa-miR-10b-3p | 0.53 | 26 | 5 | 0.28 |
| hsa-miR-151a-5p | 2.06 | 11 | 4 | 0.30 |
| hsa-miR-16-5p | 0.68 | 13 | 2 | 0.33 |
| hsa-miR-20a-3p | 1.52 | 15 | 5 | 0.34 |
| hsa-miR-497-5p | 1.73 | 16 | 4 | 0.34 |
aWe only show the top-20 miRNAs by p value. The full list of miRNAs is shown in Table S3 (Supplemental Material)
Table 3.
Fold change expression of extracellular miRNAs in follicular fluid containing normally versus abnormally fertilized oocytes
| miRNAa | Fold change | Normally fertilized (n = 30) | Abnormally fertilized (n = 4) | P value |
|---|---|---|---|---|
| hsa-miR-454-5p | 0.25 | 13 | 2 | <0.0001 |
| hsa-miR-425-3p | 0.32 | 10 | 2 | 0.001 |
| hsa-miR-16-5p | 0.38 | 13 | 2 | 0.003 |
| hsa-miR-222-3p | 0.34 | 4 | 2 | 0.04 |
| hsa-miR-520f-3p | 9.18 | 9 | 3 | 0.09 |
| hsa-miR-30d-5p | 0.52 | 30 | 4 | 0.12 |
| hsa-miR-19b-3p | 0.36 | 8 | 2 | 0.12 |
| hsa-miR-223-5p | 0.47 | 16 | 3 | 0.14 |
| hsa-miR-10b-3p | 15.84 | 26 | 4 | 0.17 |
| hsa-miR-1255b-5p | 0.36 | 22 | 4 | 0.18 |
| hsa-miR-1226-5p | 0.65 | 22 | 4 | 0.18 |
| hsa-miR-663b | 0.16 | 22 | 3 | 0.21 |
| hsa-miR-380-5p | 0.52 | 21 | 4 | 0.24 |
| hsa-miR-720 | 0.64 | 25 | 3 | 0.32 |
| hsa-miR-151a-5p | 1.93 | 11 | 3 | 0.47 |
| hsa-miR-206 | 0.28 | 21 | 2 | 0.50 |
| hsa-miR-26a-1-3p | 0.36 | 15 | 2 | 0.54 |
| hsa-miR-572 | 0.53 | 25 | 2 | 0.59 |
| hsa-miR-7-1-3p | 1.19 | 24 | 4 | 0.62 |
| hsa-miR-649 | 0.42 | 11 | 2 | 0.62 |
aWe only show the top-20 miRNAs by p value. The full list of miRNAs is shown in Table S4 (Supplemental Material)
Exploratory analysis of exmiRNAs in FF and day 3 embryo quality
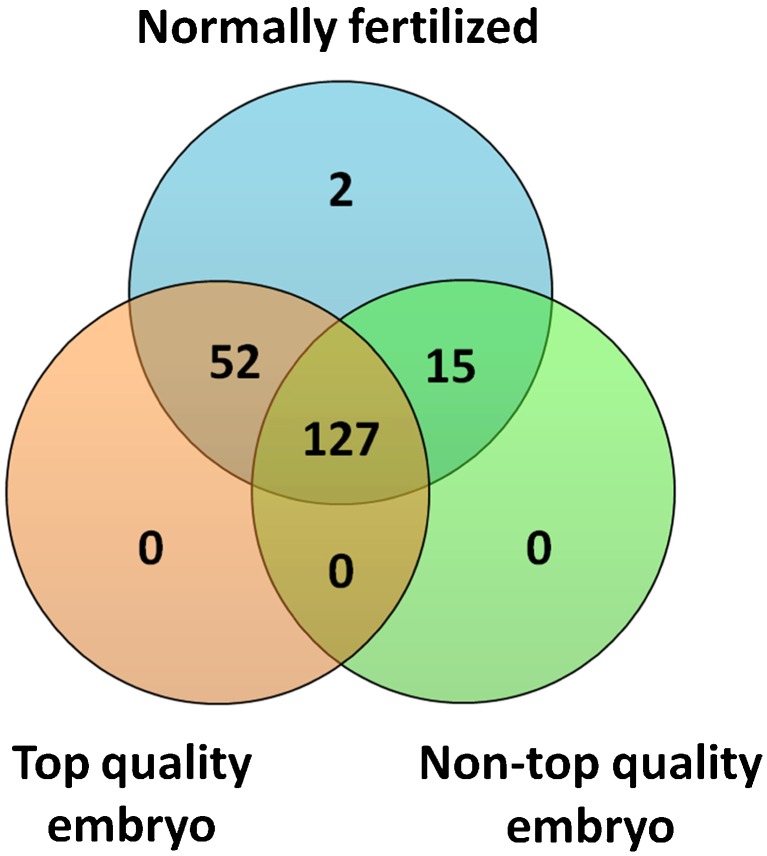
We compared the expression levels of exmiRNAs in FF that contained normally fertilized oocytes and that developed into top quality embryos on day 3 (N = 19) relative to FF that contained oocytes that ultimately developed into embryos of impaired quality (N = 10). One case was excluded due to embryo transfer on day 2. Out of 194 exmiRNAs detected in these samples, 127 were commonly found in both groups, 52 exmiRNAs were only found in FF samples that yielded top quality embryos, and 15 miRNAs in FF that yielded non-top quality embryos (Fig. 2, Table S6; Supplemental Material). Interestingly, FF samples that yielded top quality embryos shared a larger number of exmiRNAs (n = 52) with FF samples that contained normally fertilized oocytes, as compared to FF samples that yielded non-top quality embryos (n = 15) (Fig. 2). When we compared the exmiRNAs between the two groups, we found that miR-766-3p (FC = 1.95, p = 0.01), miR-663b (FC = 0.18, p = 0.02), miR-132-3p (FC = 2.45, p = 0.05), and miR-16-5p (FC = 3.80, p = 0.05) had significantly different expression levels (Table 4, Table S7; Supplemental Material).
Fig. 2.
Extracellular miRNAs detected in follicular fluid that contained normally fertilized oocytes, which yielded top quality or non-top quality embryos. The Venn diagram shows overlapping and non-overlapping extracellular miRNAs between the groups
Table 4.
Fold change expression of extracellular miRNAs in follicular fluid containing normally fertilized oocytes that yielded top quality versus non-top quality embryos
| miRNAa | Fold change | Top quality (n = 19) | Non-top quality (n = 10) | P value |
|---|---|---|---|---|
| hsa-miR-766-3p | 1.95 | 14 | 6 | 0.01 |
| hsa-miR-663b | 0.18 | 11 | 2 | 0.02 |
| hsa-miR-132-3p | 2.45 | 4 | 2 | 0.05 |
| hsa-miR-16-5p | 3.80 | 8 | 4 | 0.05 |
| hsa-miR-7-2-3p | 3.81 | 2 | 2 | 0.06 |
| hsa-miR-99b-3p | 1.83 | 8 | 4 | 0.09 |
| hsa-miR-144-5p | 3.22 | 13 | 9 | 0.10 |
| hsa-miR-7-1-3p | 2.52 | 11 | 4 | 0.11 |
| hsa-miR-320b | 2.05 | 12 | 3 | 0.12 |
| hsa-miR-661 | 0.38 | 14 | 9 | 0.14 |
| hsa-miR-590-3p | 0.51 | 17 | 10 | 0.16 |
| hsa-miR-24-3p | 0.17 | 10 | 5 | 0.16 |
| hsa-miR-572 | 0.73 | 3 | 2 | 0.17 |
| hsa-miR-454-5p | 1.96 | 7 | 5 | 0.17 |
| hsa-miR-1274A | 0.66 | 14 | 5 | 0.21 |
| hsa-miR-20a-3p | 1.91 | 8 | 3 | 0.21 |
| hsa-miR-34b-3p | 0.41 | 11 | 2 | 0.24 |
| hsa-miR-223-5p | 1.55 | 11 | 4 | 0.24 |
| hsa-miR-193b-5p | 7.35 | 15 | 8 | 0.25 |
| hsa-miR-26a-1-3p | 0.55 | 10 | 5 | 0.25 |
aWe only show the top-20 miRNAs by p value. The full list of miRNAs is shown in Table S6 (Supplemental Material)
In silico pathway analysis of differentially expressed exmiRNAs in FF
To explore the biological relevance of exmiRNAs that were differentially expressed in relation to fertilization status or embryo quality (Tables 2, 3, and 4), we included all 11 exmiRNAs that had a p value <0.05—miR-1244, miR-132-3p, miR-16-1-3p, miR-16-5p, miR-202-5p, miR-206, miR-222-3p, miR-30a-3p, miR-425-3p, miR-454-5p, miR-663b, miR-7-2-3p, and miR-766-3p—for additional in silico analysis using Ingenuity Pathway Analysis (IPA). We found that several of these exmiRNAs were implicated in relevant diseases and cellular functions to reproduction and embryo development (Table S8; Supplemental Material). Common targets were proteins involved in cell cycle regulation and members of the MAPK, TGFβ, and WNT signaling pathways that contribute to follicular development, meiotic resumption, and ovulation [18–22].
Discussion
In this study, we screened for 754 exmiRNAs in FF that contained mature oocytes from 40 women who have undergone IVF treatment, and evaluated each follicle separately. We detected an overall of 207 exmiRNAs and identified several that were prevalently present in most of the analyzed samples. In an exploratory analysis, we identified four exmiRNAs (miR-202-5p, miR-206, miR-16-1-3p, and miR-1244) that were expressed in higher levels in FF that contained normally fertilized oocytes versus those that failed to fertilize. We also detected four exmiRNAs (miR-766-3p, miR-663b, miR-132-3p, and miR-16-5p) that were differentially expressed in FF that contained oocytes that yielded top quality embryos versus those that did not yield top quality embryos.
Our findings that miR-320b, miR-720, miR-10b-3p, miR-126-5p, miR-30a-5p, miR-202-5p, and miR-1274A are among the most prevalent and highly expressed in follicular fluid are in line with findings reported by Sang et al. [35]. In addition, Moreno et al. recently compared the expression levels of miRNAs present in FF from women who underwent IVF treatment, and found several that were differentially expressed according to oocyte maturation status, including miR-720 which was found to be up-regulated in FF containing mature oocytes compared to FF containing immature oocytes [36]. In another study, Santonocito et al. compared the miRNA profile of exosomes in FF with the profile of free-circulating miRNAs in FF and found that miR10b-3p was up-regulated in the exosome fraction, but not in the free-circulating miRNA fraction in FF [18]. In agreement with these findings by Santonocito et al., we also detected miR-10b-3p in FF samples in 37 out of 40 analyzed samples. However, we did not find any significant differential expression in the levels of miR-10b-3p inFF associated with normally fertilized versus failed to fertilize oocytes. These findings suggest that miR-10b-3p is one of the most commonly detected miRNAs in FF, but its role and biological relevance to oocyte fertilization during IVF remains to be elucidated by further studies. Moreover, the most prevalently detected exmiRNAs in our study (i.e., miR-30d-5p, miR-320b, miR10b-3p, miR-1291, and miR-720) were previously found in mural granulosa and/or cumulus cells [23, 33, 34, 37]. This provides possible evidence that exmiRNAs in FF originate from granulosa and cumulus cells, which come in contact with the FF, and therefore they can contribute to the intercellular communication in the ovarian follicle.
Fertilization is a process that involves the degradation of maternal RNAs and zygotic genome activation [38–40]. In humans, the molecular mechanisms for this process remain largely unknown, but studies in zebrafish have identified miRNAs as a potential link between the decay of maternal mRNAs and the activation of the zygotic genome [41]. Interestingly, we found that miR-202-5p, miR-206, miR-16-1-3p, and miR-1244 were significantly up-regulated in follicles whose oocytes were successfully fertilized. Previous studies have shown that miR-202-5p not only is highly expressed in the cumulus cells surrounding mature oocytes and up-regulated in cumulus granulosa cells compared with mural granulosa cells but it is also detected in EVs isolated from FF [35, 37]. In addition, miR-202-5p regulates genes of the phosphoinositide signaling cascade, which plays a role in the generation of Ca2+ increase and oocyte activation during fertilization [42]. The exact mechanism that mediates oocyte activation at this stage remains largely unexplored, and it is possible that exmiRNAs may be implicated in this process. Furthermore, miR-202-5p targets genes that participate in the MAPK pathway, which is crucial for the resumption of the meiosis process, as well as following formation of the male and female pronuclei [43]. Lastly, miR-202-5p is known for its regulation of centrosomal proteins, which are critical for successful fertilization [44]. Although miR-202-5p was studied in several studies in relation to reproduction, to the best of our knowledge, this is the first study to show associations between miR-206, miR-16-1-3p, and miR-1244 with fertilization potential. In silico IPA analysis revealed that these miRNAs target genes that participate in the AMP-activated protein kinase (AMPK), which is a pathway that plays a crucial role in the regulation of cell growth and metabolism [45].
Our exploratory analysis also showed that several exmiRNAs (miR-663b, miR-766-3p, miR-132-3p, and miR-16-5p) were up-regulated in FF that yielded top quality embryos on day 3. According to the in silico IPA analysis, all these miRNAs target genes that participate in the MAPK pathway, which also plays a central role in the control of cell proliferation and is essential for the successful progression of mitosis in sea urchin embryos [46, 47]. In a similar study, Feng et al. compared exmiRNAs from FF that yielded top quality and non-top quality embryos [48]. Similar to our findings, they found that miR-132-3p was up-regulated in FF containing oocytes that yielded top quality embryos; however, this finding was not statistically significant. Several of the top differentially expressed exmiRNAs in our study (e.g., miR-16-5p, miR-24-3p, and miR-320b) were also identified by Feng et al. in relation to embryo quality, despite the fact that statistical significance was not reached. Interestingly, in both studies, these miRNAs were always found to be down-regulated in the non-top quality versus the top quality embryos. In addition, Kropp and Khatib found that increased levels of miR-24 in bovine IVF culture media of the embryo were associated with a decline in embryo development. In our study, miR-24 was also detected in higher levels in FF from samples that yielded non-top versus top quality embryos [49].
Our study has a few limitations. The small and unbalanced sample size in the failed to fertilize or abnormally fertilized vs. normally fertilized groups may have limited our ability to detect stronger associations. We attempted to overcome some of the confounding in the design by including women of similar characteristics between the groups (e.g., age, BMI), and by including only FF samples that were associated with mature oocytes. Our findings must be interpreted with caution as confounding by other variables we could not adjust for in our study such as different incubators, time of zygote, and embryo morphology assessment outside the incubators, and handling by different embryologists may be present. Last, although we used a method that extracts total RNA enriched in EVs, we cannot exclude the possibility that protein complex-associated (e.g., Argonaute 2), or lipid-bound (e.g., HDL) miRNAs might be present. However, the fact that several of our most prevalently detected miRNAs (e.g., miR-30d-5p, miR-10b-3p, and miR-720) were also found by others to be present in EVs provides some evidence that these miRNAs are enriched in EVs [50–54]. Despite the limitations, our study is one of the largest to date that examined the association between human FF exmiRNAs and fertilization potential and embryo quality.
Conclusions
We found that extracellular miR-202-5p, miR-206, miR-16-1-3p, and miR-1244 in human FF are associated with fertilization potential in IVF and that miR-663b, miR-766-3p, miR-132-3p, and miR-16-5p are associated with embryo quality. We encourage further research to validate our findings and to determine any biological effects of FF exmiRNAs on oocytes, cumulus, and granulosa cells. These effects can modify oocyte quality to affect these downstream events as fertilization and observational quality assessment of the embryo.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(DOCX 146 kb)
(XLSX 49 kb)
(XLSX 13 kb)
(XLSX 13 kb)
(XLSX 20 kb)
(XLSX 18 kb)
(XLSX 14 kb)
(XLSX 20 kb)
(XLSX 11 kb)
Acknowledgements
We thank the patients that donated the follicular fluid and the IVF team contributing to this study.
Compliance with ethical standards
Funding
This study was funded by Grant Award no. RPGA1301 from the Environmental and Health Fund (EHF), Israel and by grants P30ES00002 and R21ES024236 from the National Institute of Environmental Health Sciences, USA.
Footnotes
Ronit Machtinger and Rodosthenis S. Rodosthenous contributed equally to this work.
References
- 1.Eppig JJ. Oocyte control of ovarian follicular development and function in mammals. Reproduction. 2001;122(6):829–38. doi: 10.1530/rep.0.1220829. [DOI] [PubMed] [Google Scholar]
- 2.Eppig JJ, Chesnel F, Hirao Y, O’Brien MJ, Pendola FL, Watanabe S, et al. Oocyte control of granulosa cell development: how and why. Hum Reprod. 1997;12(11 Suppl):127–32. [PubMed] [Google Scholar]
- 3.Buccione R, Schroeder AC, Eppig JJ. Interactions between somatic cells and germ cells throughout mammalian oogenesis. Biol Reprod. 1990;43(4):543–7. doi: 10.1095/biolreprod43.4.543. [DOI] [PubMed] [Google Scholar]
- 4.Kidder GM, Vanderhyden BC. Bidirectional communication between oocytes and follicle cells: ensuring oocyte developmental competence. Can J Physiol Pharmacol. 2010;88(4):399–413. doi: 10.1139/Y10-009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winterhager E, Kidder GM. Gap junction connexins in female reproductive organs: implications for women’s reproductive health. Hum Reprod Update. 2015 doi: 10.1093/humupd/dmv007. [DOI] [PubMed] [Google Scholar]
- 6.Matzuk MM, Burns KH, Viveiros MM, Eppig JJ. Intercellular communication in the mammalian ovary: oocytes carry the conversation. Science. 2002;296(5576):2178–80. doi: 10.1126/science.1071965. [DOI] [PubMed] [Google Scholar]
- 7.Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borras FE, Buzas EI, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracellular Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Da Silveira JC, Sessions DR, Veeramachaneni DNR, Winger QA, Carnevale EM, Bouma GJ. MiRNAs within the ovarian follicle: Identification of cell-secreted vesicles as miRNA carriers. Biol Reprod. 2011;85(1).
- 9.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–83. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang M, Ouyang H, Xia G. The signal pathway of gonadotrophins-induced mammalian oocyte meiotic resumption. Mol Hum Reprod. 2009;15(7):399–409. doi: 10.1093/molehr/gap031. [DOI] [PubMed] [Google Scholar]
- 11.Machtinger R, Laurent LC, Baccarelli AA. Extracellular vesicles: roles in gamete maturation, fertilization and embryo implantation. Hum Reprod Update. 2016;22(2):182–93. doi:10.1093/humupd/dmv055. [DOI] [PMC free article] [PubMed]
- 12.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 14.Li SC, Tang P, Lin WC. Intronic microRNA: discovery and biological implications. DNA Cell Biol. 2007;26(4):195–207. doi: 10.1089/dna.2006.0558. [DOI] [PubMed] [Google Scholar]
- 15.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 16.Nagaraja AK, Andreu-Vieyra C, Franco HL, Ma L, Chen R, Han DY, et al. Deletion of Dicer in somatic cells of the female reproductive tract causes sterility. Mol Endocrinol. 2008;22(10):2336–52. doi: 10.1210/me.2008-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saetrom P, Snove O, Jr, Rossi JJ. Epigenetics and microRNAs. Pediatr Res. 2007;61(5 Pt 2):17R–23R. doi: 10.1203/pdr.0b013e318045760e. [DOI] [PubMed] [Google Scholar]
- 18.Santonocito M, Vento M, Guglielmino MR, Battaglia R, Wahlgren J, Ragusa M, et al. Molecular characterization of exosomes and their microRNA cargo in human follicular fluid: bioinformatic analysis reveals that exosomal microRNAs control pathways involved in follicular maturation. Fertil Steril. 2014;102(6):1751–U590. doi: 10.1016/j.fertnstert.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 19.da Silveira JC, de Andrade GM, Nogueira MF, Meirelles FV, Perecin F. Involvement of miRNAs and cell-secreted vesicles in mammalian ovarian antral follicle development. Reprod Sci. 2015;22(12):1474–83. doi: 10.1177/1933719115574344. [DOI] [PubMed] [Google Scholar]
- 20.Navakanitworakul R, Hung WT, Gunewardena S, Davis JS, Chotigeat W, Christenson LK. Characterization and small RNA content of extracellular vesicles in follicular fluid of developing bovine antral follicles. Sci Rep. 2016;6:25486. doi: 10.1038/srep25486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.da Silveira JC, Winger QA, Bouma GJ, Carnevale EM. Effects of age on follicular fluid exosomal microRNAs and granulosa cell transforming growth factor-beta signalling during follicle development in the mare. Reprod Fertil Dev. 2015;27(6):897–905. doi: 10.1071/RD14452. [DOI] [PubMed] [Google Scholar]
- 22.Sohel Md. Mahmodul H, Hoelker M, Noferesti SS, Salilew-Wondim D, Tholen E, Looft C et al. Exosomal and non-exosomal transport of extra-cellular microRNAs in follicular fluid: Implications for bovine oocyte developmental competence. PloS one. 2013;8(11). [DOI] [PMC free article] [PubMed]
- 23.da Silveira JC, Veeramachaneni DNR, Winger QA, Carnevale EM, Bouma GJ. Cell-secreted vesicles in equine ovarian follicular fluid contain mirnas and proteins: A possible new form of cell communication within the ovarian follicle. Biology of Reproduction. 2012;86(3). [DOI] [PubMed]
- 24.Kim YJ, Ku SY, Kim YY, Liu HC, Chi SW, Kim SH, et al. MicroRNAs transfected into granulosa cells may regulate oocyte meiotic competence during in vitro maturation of mouse follicles. Hum Reprod. 2013;28(11):3050–61. doi: 10.1093/humrep/det338. [DOI] [PubMed] [Google Scholar]
- 25.Machtinger R, Bormann CL, Ginsburg ES, Racowsky C. Is the presence of a non-cleaved embryo on day 3 associated with poorer quality of the remaining embryos in the cohort? J Assist Reprod Genet. 2015 doi: 10.1007/s10815-015-0455-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alpha Scientists in Reproductive M, Embryology ESIGo The Istanbul consensus workshop on embryo assessment: proceedings of an expert meeting. Hum Reprod. 2011;26(6):1270–83. doi: 10.1093/humrep/der037. [DOI] [PubMed] [Google Scholar]
- 27.Enderle D, Spiel A, Coticchia CM, Berghoff E, Mueller R, Schlumpberger M, et al. Characterization of RNA from exosomes and other extracellular vesicles isolated by a novel spin column-based method. PLoS One. 2015;10(8):e0136133. doi: 10.1371/journal.pone.0136133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Racowsky C, Combelles CM, Nureddin A, Pan Y, Finn A, Miles L, et al. Day 3 and day 5 morphological predictors of embryo viability. Reprod Biomed Online. 2003;6(3):323–31. doi: 10.1016/S1472-6483(10)61852-4. [DOI] [PubMed] [Google Scholar]
- 29.Machtinger R, Racowsky C. Morphological systems of human embryo assessment and clinical evidence. Reprod Biomed Online. 2013;26(3):210–21. doi: 10.1016/j.rbmo.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 30.Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F, et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10(6):R64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4. doi:10.7554/eLife.05005. [DOI] [PMC free article] [PubMed]
- 33.Assou S, Al-edani T, Haouzi D, Philippe N, Lecellier CH, Piquemal D, et al. MicroRNAs: new candidates for the regulation of the human cumulus-oocyte complex. Hum Reprod. 2013;28(11):3038–49. doi: 10.1093/humrep/det321. [DOI] [PubMed] [Google Scholar]
- 34.Liu FJ, Shen XF. Comparative analysis of human reproductive proteomes identifies candidate proteins of sperm maturation. Mol Biol Rep. 2012;39(12):10257–63. doi: 10.1007/s11033-012-1902-7. [DOI] [PubMed] [Google Scholar]
- 35.Sang Q, Yao Z, Wang H, Feng R, Wang H, Zhao X, et al. Identification of MicroRNAs in human follicular fluid: characterization of MicroRNAs that govern steroidogenesis in vitro and are associated with polycystic ovary syndrome in vivo. J Clin Endocrinol Metab. 2013;98(7):3068–79. doi: 10.1210/jc.2013-1715. [DOI] [PubMed] [Google Scholar]
- 36.Moreno JM, Nunez MJ, Quinonero A, Martinez S, de la Orden M, Simon C, et al. Follicular fluid and mural granulosa cells microRNA profiles vary in in vitro fertilization patients depending on their age and oocyte maturation stage. Fertil Steril. 2015;104(4):1037–46. doi: 10.1016/j.fertnstert.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 37.Velthut-Meikas A, Simm J, Tuuri T, Tapanainen JS, Metsis M, Salumets A. Research resource: small RNA-seq of human granulosa cells reveals miRNAs in FSHR and aromatase genes. Mol Endocrinol. 2013;27(7):1128–41. doi: 10.1210/me.2013-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kishimoto T. Cell-cycle control during meiotic maturation. Curr Opin Cell Biol. 2003;15(6):654–63. doi: 10.1016/j.ceb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 39.Stitzel ML, Seydoux G. Regulation of the oocyte-to-zygote transition. Science. 2007;316(5823):407–8. doi: 10.1126/science.1138236. [DOI] [PubMed] [Google Scholar]
- 40.Schier AF. The maternal-zygotic transition: death and birth of RNAs. Science. 2007;316(5823):406–7. doi: 10.1126/science.1140693. [DOI] [PubMed] [Google Scholar]
- 41.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312(5770):75–9. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 42.Whitaker M. Calcium at fertilization and in early development. Physiol Rev. 2006;86(1):25–88. doi: 10.1152/physrev.00023.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Machaty Z. Signal transduction in mammalian oocytes during fertilization. Cell Tissue Res. 2016;363(1):169–83. doi: 10.1007/s00441-015-2291-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schatten H, Sun QY. New insights into the role of centrosomes in mammalian fertilization and implications for ART. Reproduction. 2011;142(6):793–801. doi: 10.1530/REP-11-0261. [DOI] [PubMed] [Google Scholar]
- 45.Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. 2011;13(9):1016–23. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev : MMBR. 2011;75(1):50–83. doi: 10.1128/MMBR.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Odile ML, Heloise C, Julia M, Robert B, Patrick C. MAPK/ERK activity is required for the successful progression of mitosis in sea urchin embryos. Dev Biol. 2016 doi: 10.1016/j.ydbio.2016.11.018. [DOI] [PubMed] [Google Scholar]
- 48.Feng R, Sang Q, Zhu Y, Fu W, Liu M, Xu Y, et al. MiRNA-320 in the human follicular fluid is associated with embryo quality in vivo and affects mouse embryonic development in vitro. Sci Rep. 2015;5:8689. doi: 10.1038/srep08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kropp J, Khatib H. Characterization of microRNA in bovine in vitro culture media associated with embryo quality and development. J Dairy Sci. 2015;98(9):6552–63. doi: 10.3168/jds.2015-9510. [DOI] [PubMed] [Google Scholar]
- 50.Armstrong DA, Green BB, Seigne JD, Schned AR, Marsit CJ. MicroRNA molecular profiling from matched tumor and bio-fluids in bladder cancer. Mol Cancer. 2015;14:194. doi: 10.1186/s12943-015-0466-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cha DJ, Franklin JL, Dou Y, Liu Q, Higginbotham JN, Demory Beckler M, et al. KRAS-dependent sorting of miRNA to exosomes. eLife. 2015;4:e07197. doi: 10.7554/eLife.07197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guzman N, Agarwal K, Asthagiri D, Yu L, Saji M, Ringel MD, et al. Breast cancer-specific miR signature unique to extracellular vesicles includes “microRNA-like” tRNA fragments. Mol Cancer Res : MCR. 2015;13(5):891–901. doi: 10.1158/1541-7786.MCR-14-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Joshi GK, Deitz-McElyea S, Liyanage T, Lawrence K, Mali S, Sardar R, et al. Label-free nanoplasmonic-based short noncoding RNA sensing at attomolar concentrations allows for quantitative and highly specific assay of MicroRNA-10b in biological fluids and circulating exosomes. ACS Nano. 2015;9(11):11075–89. doi: 10.1021/acsnano.5b04527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Melman YF, Shah R, Danielson K, Xiao J, Simonson B, Barth A, et al. Circulating MicroRNA-30d is associated with response to cardiac resynchronization therapy in heart failure and regulates cardiomyocyte apoptosis: a translational pilot study. Circulation. 2015;131(25):2202–16. doi: 10.1161/CIRCULATIONAHA.114.013220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 146 kb)
(XLSX 49 kb)
(XLSX 13 kb)
(XLSX 13 kb)
(XLSX 20 kb)
(XLSX 18 kb)
(XLSX 14 kb)
(XLSX 20 kb)
(XLSX 11 kb)