Abstract
Aspergillus fumigatus is an important cause of life-threatening invasive fungal disease in patients with compromised immune systems. Resistance to itraconazole in A. fumigatus is closely linked to amino acid substitutions in Cyp51A that replace Gly54. In an effort to develop a new class of molecular diagnostic assay that can rapidly assess drug resistance, a multiplexed assay was established. This assay uses molecular beacons corresponding to the wild-type cyp51A gene and seven mutant alleles encoding either Arg54, Lys54, Val54, Trp54, or Glu54. Molecular beacon structure design and real-time PCR conditions were optimized to increase the assay specificity. The multiplex assay was applied to the analysis of chromosomal DNA samples from a collection of 48 A. fumigatus clinical and laboratory-derived isolates, most with reduced susceptibility to itraconazole. The cyp51A allelic identities for codon 54 were established for all of the strains tested, and mutations altering Gly54 in 23 strains were revealed. These mutations included G54W (n = 1), G54E (n = 12), G54K (n = 3), G54R (n = 3), and G54V (n = 4). Molecular beacon assay results were confirmed by DNA sequencing. Multiplex real-time PCR with molecular beacons is a powerful technique for allele differentiation and analysis of resistance mutations that is dynamic and suitable for rapid high-throughput assessment of drug resistance.
Aspergillus fumigatus is a common cause of invasive mold infections in humans resulting from an alteration of immune status due to AIDS, cancer, or solid organ or bone marrow transplantation. High rates of morbidity and mortality for patients with invasive aspergillosis, despite conventional antifungal therapy, are reported (14). Itraconazole, a triazole antifungal drug approved in 1992, has been widely used for treatment and prophylaxis of fungal infections (6, 41). Although newer triazole drugs, such as voriconazole, ravuconazole, and posaconazole, have recently become available (9, 36), Aspergillus infections are still commonly treated with itraconazole (8).
Triazole drugs are fungistatic, which can result in the development of secondary resistance in infecting strains. Itraconazole resistance in clinical isolates as well as in laboratory mutant strains has been reported (3, 5, 18, 24, 32). Triazole drugs bind to the active site of the fungal cytochome P450 14-α-sterol demethylase, which catalyzes the 14α demethylation of ergosterol precursors (22) and which is encoded by cyp51A in Aspergillus (29). Several amino acid substitutions in Cyp51A resulting in itraconazole resistance in A. fumigatus have been described (7, 26, 33). Yet, unlike the multiplicity of triazole resistance mutations found in Candida spp. (19, 48), itraconazole resistance mutations in the A. fumigatus cyp51A gene are tightly linked to amino acid substitutions at residue 54, corresponding to glycine (7, 26, 33). A. fumigatus strains with G54R, G54V, G54W, and G54E substitutions were detected in clinical isolates as well as in spontaneous and UV-induced itraconazole-resistant laboratory mutants (7, 26, 30, 33). The G54K amino acid change conferred cross-resistance to both itraconazole and posaconazole (26). The replacement of the wild-type chromosomal cyp51A allele by mutant allele bearing the G161A nucleotide change in codon 54 led to the acquisition of resistance to itraconazole de novo (7).
Early diagnosis of invasive aspergillosis by conventional procedures utilizing blood or bronchial fluid specimens is difficult. The use of cell wall components such as galactomannan (GM) as a indicator of disease is a major advancement (10, 23, 27), but it has some deficiencies as well (27, 31, 47).
Early detection of fungi in blood or bronchial alveolar lavage fluid, with a rapid assessment of drug susceptibility, could improve the survival of patients with invasive disease by accelerating the initiation of appropriate antifungal treatment while the fungal loads are still low. Genomic differences among fungi offer an alternative to culturing for detection and identification, and nucleic acid-based amplification assays for the detection of fungal nucleic acids may be the optimal diagnostic approach because they are more rapid and sensitive than current culture-based and biochemical methods (21, 34).
We have exploited the power of molecular beacon technology to develop a new multiplex real-time PCR assay suitable for rapid detection of itraconazole resistance mutations in codon 54 of A. fumigatus cyp51A. Molecular beacons are small, self-reporting, single-stranded nucleic acid hairpin probes that brightly fluoresce when bound to their targets (44). They are particularly well suited for allele discrimination (1, 43) and multiplex applications (37, 45). The molecular beacon-based assay developed in this study provides the basis for rapid identification of drug resistance in A. fumigatus associated with Cyp51A alterations, which can serve as a platform for a more comprehensive rapid diagnostic tool to more routinely assess invasive aspergillosis.
MATERIALS AND METHODS
Strains, culture conditions, and susceptibility testing.
Itraconazole-resistant RIT strains were obtained by mutagenesis of parental wild-type H11-20 strain 20 (33). Clinical isolates of A. fumigatus were obtained from D. Denning (Manchester University, Manchester, United Kingdom) (7). All the strains were grown and maintained in yeast extract-peptone-dextrose or Sabouraud dextrose agar media as previously described (33). Itraconazole (Janssen Pharmaceutica, Titusville, N.J.) was dissolved in dimethylformamide (Sigma-Aldrich Corp., St. Louis, Mo.) prior to addition to culture media. MICs of itraconazole were determined according to the NCCLS M38-P microdilution methodology, as previously described (33, 34).
Molecular beacon and primer design.
The sequence of A. fumigatus gene cyp51A reported by Mellado et al. (29) (GenBank accession number AF338659) was used for molecular beacon and primer design. Molecular beacons covering the locus of diversity corresponding to codon 54 of cyp51A were designed with Beacon Designer, version 2.12, software (PREMIER Biosoft Int., Palo Alto, Calif.). Molecular beacons were labeled with fluorophores 5-carboxyfluorescein (FAM), 6-carboxy-2′,4,4′,5′,7,7′-hexachlorofluorescein (HEX), carboxy-X-rhodamine (ROX), and Quasar 670 (Q670) at the 5′ end and with dabcyl at the 3′ end. Molecular beacons were purchased from Biosearch Technologies Inc. PCR primers were designed with Beacon Designer, version 2.12, software and Oligo, version 4.04, software (Molecular Biology Insights Inc., Cascade, Co.) and were purchased from Sigma-Genosys, Woodlands, Tex.). All molecular beacons and PCR primers used in the study are listed in Table 1.
TABLE 1.
DNA oligonucleotides used in work
| Primer or molecular beacon | Sequencea | 5′ end modification | 3′ end modification | Purpose |
|---|---|---|---|---|
| Af210F | GTCTCTCATTCGTCCTTGTCCT | None | None | Sequencing PCR primer |
| Af709R | CGTTGAGAATAAACTCGTTCCC | None | None | Sequencing PCR primer |
| CYP51AS | TCATTGGGTCCCATTTCTG | None | None | Real-time PCR primer |
| CYP51AA | GCACGCAAAGAAGAACTTG | None | None | Real-time PCR primer |
| AAG-FAM | CGCGATCATCAGTTACAAGATTGATCCATCGCG | FAM | Dabcyl | AAG allele probe |
| GGG-FAM | CGCGATCATCAGTTACGGGATTGATCCATCGCG | FAM | Dabcyl | Wild-type allele probe |
| GAA-HEX | CGCGATCATCAGTTACGAAATTGATCCATCGCG | HEX | Dabcyl | GAA allele probe |
| AAG-HEX | CGCGATCATCAGTTACAAGATTGATCOATCGCG | HEX | Dabcyl | AAG allele probe |
| GAG-HEX | CGCGATCATCAGTTACGAGATTGATCCATCGCG | HEX | Dabcyl | GAG allele probe |
| AGG-HEX | CGCGATCATCAGTTACAGGATTGATCCATCGCG | HEX | Dabcyl | AGG allele probe |
| GTG-HEX | CGCGATCATCAGTTACGTGATTGATCCATCGCG | HEX | Dabcyl | GTG allele probe |
| TGG-HEX | CGCGATCATCAGTTACTGGATTGATCCATCGCG | HEX | Dabcyl | TGG allele probe |
| CGG-HEX | CGCGATCATCAGTTACCGGATTGATCCATCGCG | HEX | Dabcyl | CGG allele probe |
| TGG-ROX | CGCGATCATCAGTTACTGGATTGATCCATCGCG | ROX | Dabcyl | TGG allele probe |
| CGG-ROX | CGCGATCATCAGTTACCGGATTGATCCATCGCG | ROX | Dabcyl | CGG allele probe |
| GAA-Q670 | CGCGATCATCAGTTACGAAATTGATCCATCGCG | Q670 | Dabcyl | GAA allele probe |
| GTG-Q670 | CGCGATCATCAGTTACGTGATTGATCCATCGCG | Q670 | Dabcyl | GTG allele probe |
| GGG-T | AAAAGGATCAATCCCGTAACTGATGAAAA | None | None | Wild-type allele target |
| GAG-T | AAAAGGATCAATCTCGTAACTGATGAAAA | None | None | GAG allele target |
| AAG-T | AAAAGGATCAATCTTGTAACTGATGAAAA | None | None | AAG allele target |
| GAA-T | AAAAGGATCAATTTCGTAACTGATGAAAA | None | None | GAA allele target |
| AGG-T | AAAAGGATCAATCCTGTAACTGATGAAAA | None | None | AGG allele target |
| CGG-T | AAAAGGATCAATCCGGTAACTGATGAAAA | None | None | CGG allele target |
| TGG-T | AAAAGGATCAATCCAGTAACTGATGAAAA | None | None | TGG allele target |
| GTG-T | AAAAGGATCAATCACGTAACTGATGAAAA | None | None | GTG allele target |
Probe domains of molecular beacons and target domains of allele targets are underlined. Sequences for molecular beacon probe domains corresponding to codon 54 in A. fumigatus gene cyp51A and complement codons in allele targets are in boldface.
Thermal denaturation profile determination.
Single-stranded oligonucleotides used as DNA targets corresponding to different cyp51A alleles are listed in Table 1. Molecular beacon-target hybridization was investigated with the Stratagene Mx4000 multiplex quantitative PCR system. The “molecular beacon melting curve” option was chosen in the Mx4000 software for data monitoring and analysis. Each hybridization reaction mixture contained 1× Core PCR buffer (Stratagene, La Jolla, Calif.), 3 or 5 mM MgCl2, 100 pmol of individual oligonucleotides, and 5 pmol of molecular beacons. The test reaction mixtures were subjected to heating at 95°C for 3 min and cooling to 80°C, with subsequent cooling down to 25°C in 112 30-s steps with a temperature gradient of −0.5°C/step. Fluorescence output was measured at the end of each step. The final data of the molecular beacon melting curve experiment were converted to a “SYBR Green (with dissociation curve)” output. The values for melting temperatures (Tm) were calculated by Mx4000 software as temperature points corresponding to maximal values of the first derivative of the fluorescence output [−Rn′(T)].
DNA extraction, PCR amplification, and DNA sequencing.
A. fumigatus chromosomal DNA was extracted from cells grown for 24 h in Sabouraud dextrose agar media, as previously described (33). Chromosomal DNA of strain R7-1 was provided by P. Mann (26). Amplification of a 500-bp fragment of cyp51A was performed on an iCycler thermal cycler (Bio-Rad Laboratories, Hercules, Calif.). Each 100-μl PCR mixture contained 25 pmol of each of the Af210F and Af709R primers, 2.5 U of iTaq DNA polymerase (Bio-Rad Laboratories), 0.5 mM deoxynucleoside triphosphates, 50 mM KCl, 4 mM MgCl2, 20 mM Tris-HCl (pH 8.4), and about 100 ng of A. fumigatus chromosomal DNA. The cycling conditions were 1 cycle of 3 min at 95°C; 35 cycles of 30 s at 95°C, 30 s at 55°C, and 1 min at 72°C; and 1 cycle of 3 min at 72°C. PCR products were purified with the Montage PCR purification kit (Millipore). PCR products for sequencing were obtained and purified with the CEQ dye terminator cycle sequencing-Quick Start kit (Beckman Coulter, Inc., Fullerton, Calif.) according to the manufacturer recommendations on an iCycler thermal cycler. Af210F or Af709R primers were used for the sequencing reaction. The cycling conditions for sequencing PCR were 1 cycle of 3 min at 95°C and 30 cycles of 20 s at 96°C, 20 s at 50°C, and 1 min at 60°C. All DNA sequencing was performed on CEQ 8000 genetic analysis system (Beckman Coulter). CEQ 8000 genetic analysis system software (Beckman Coulter) was used for hardware control as well as for analysis of postrun sequencing results.
Real-time PCR.
Real-time PCR experiments were performed on a Stratagene Mx4000 multiplex quantitative PCR system with the “quantitative PCR (multiple standards)” setting. Reagents from the Brilliant QPCR core reagent kit were used for all reactions. Each 50-μl PCR mixture contained 1× Stratagene Core PCR buffer, 20 pmol of molecular beacons, 25 pmol of each of the cyp51AS and cyp51AA primers (Table 1), 2.5 U of Stratagene SureStart Taq DNA polymerase, 0.4 mM deoxynucleoside triphosphates, and 3 or 5 mM MgCl2. In multiplex PCR experiments 20 pmol of each molecular beacon (Table 1) was added to the reaction mixture. Amplification of a 70-bp fragment from A. fumigatus chromosomal DNA utilized 100 ng of DNA, while use of the 500-bp PCR fragment as a template required 10 pg of DNA (about 300 nmol) per reaction. PCRs were performed with the following parameters: 1 cycle of 10 min at 95°C and 45 cycles of 30 s at 95°C, 30 s at 61°C, and 30 s at 72°C. Annealing temperatures of 55 and 57°C were used when PCR experiments were performed in multiplex format. The filter gain setting of the Mx4000 system was changed to FAM-940 HEX-720 CY5-700 ROX-740 with the aim of equalization of the fluorescence signals from different molecular beacons. The fluorescence was measured three times during the annealing step.
Data processing.
Fluorescence signals coming from Mx4000 system during PCR amplification were monitored with Mx4000 software in real time. At the end of each run the amplification plot data were converted to graphic format and stored as image files or exported into Microsoft Office Excel and stored as spreadsheet files. For multiplex PCRs, the final results of PCR amplifications were converted from a “quantitative PCR (multiple standards)” type of experiment to the “quantitative plate read” type of experiment. Total changes in fluorescence for individual fluorophores (Rpost − Rpre) were taken as values for analysis. Results were converted to graphic or numerical format and stored as image or spreadsheet files.
RESULTS
Itraconazole resistance has been tightly linked to cyp51A mutations in the codon for Gly54, resulting in five different amino substitutions (G54K, G54V, G54R, G54E, and G54W) (7, 26, 33), and such linkage provides an ideal opportunity to develop a nucleic acid-based diagnostic for A. fumigatus that can rapidly assess the drug resistance status of an infecting organism. Since multiple mutations at codon 54 confer resistance, including single and multiple nucleotide changes, it was important that the nucleic acid-based diagnostic assay possess intrinsic allele specificity and the feasibility of simultaneously detecting a range of mutations in multiplex format. Molecular beacon technology was chosen because it is a superior platform relative to other self-reporting probes for allele discrimination and multiplex assay development (44, 45).
Design and validation of molecular beacon to wild-type Gly54.
A molecular beacon complementary to the cyp51A wild-type allele was synthesized with a 21-nucleotide probe target sequence (hairpin loop) domain and a 6-nucleotide stem domain with the 5′ end of the beacon labeled with FAM as the fluorophore and the 3′ end modified with a dabcyl quencher. The sequence of the probe domain was identical to the sequence of cyp51A starting from nucleotide position 150 and ending at position 170. Codon GGG, corresponding to wild-type Gly54, occupied nucleotide positions 11 to 13 of the beacon probe domain. The beacon was tested with oligonucleotides representing the wild-type allele and seven known cyp51A itraconazole resistance alleles (Table 1). The molecular beacon showed appropriate thermal behavior and hybridization with DNA targets. The efficiency of molecular beacon annealing to different targets varied depending on target nucleotide content and divalent cation concentration, with experimentally derived Tm values for each of the eight beacon-target hybrids shown in Table 2. The stability of intermolecular hybrids of GGG-FAM molecular beacons and artificial oligonucleotide targets decreased in the order of GGG-T > GTG-T > AGG-T > GAG-T > TGG-T > CGG-T > AAG-T > GAA-T. As expected, hybrids with double mismatches possessed the lowest stability. Among noncomplement targets, the GGG-FAM beacon formed the most stable single-mismatched hybrid with GTG-T. The temperature interval between Tm of the most stable mismatched beacon-target hybrid and Tm of the complement beacon-target hybrid represented the condition allowing specific allele-discriminative binding of the molecular beacon or window of discrimination. As shown in Table 2, the window of discrimination for GGG-FAM molecular beacon was within the temperature ranges of 61.7 to 65.2°C under conditions of 3 mM Mg2+ and 63.7 to 67.2°C at 5 mM Mg2+.
TABLE 2.
Tms for CYP51A molecular beacons
|
Tma at Mg concn of:
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beacon | 3 mM for target:
|
5 mM for target:
|
||||||||||||||
| GGG-T | GAA-T | AAG-T | GAG-T | AGG-T | GTG-T | TGG-T | CGG-T | GGG-T | GAA-T | AAG-T | GAG-T | AGG-T | GTG-T | TGG-T | CGG-T | |
| AAG-FAM | 48.7 | 44.7 | 62.2 | 52.7 | 53.2 | 48.2 | 50.7 | 49.7 | 49.7 | 45.2 | 63.7 | 54.2 | 54.7 | 54.2 | 52.2 | 52.2 |
| GGG-FAM | 65.2 | 53.7 | 54.2 | 58.7 | 59.7 | 61.7 | 58.2 | 55.7 | 67.2 | 55.2 | 55.7 | 61.2 | 61.7 | 63.7 | 61.2 | 59.7 |
| GAA-HEX | 45.7 | 61.2 | 41.7 | 50.2 | 35.2 | 45.2 | 41.7 | 36.2 | 46.7 | 63.7 | 47.7 | 52.7 | 41.2 | 46.7 | 42.2 | 41.7 |
| AAG-HEX | 46.2 | 39.7 | 61.2 | 52.2 | 51.7 | 49.7 | 48.2 | 47.2 | 48.7 | 44.7 | 63.2 | 53.2 | 53.2 | 51.2 | 50.7 | 49.7 |
| GAG-HEX | 55.7 | 55.7 | 56.7 | 63.2 | 50.2 | 57.7 | 50.7 | 50.7 | 57.2 | 56.2 | 57.2 | 64.2 | 51.7 | 58.7 | 52.7 | 52.2 |
| AGG-HEX | 54.7 | 42.2 | 55.2 | 49.7 | 61.7 | 49.7 | 56.2 | 58.2 | 56.7 | 44.7 | 57.2 | 51.7 | 63.7 | 52.2 | 58.2 | 59.7 |
| GTG-HEX | 54.7 | 47.7 | 48.7 | 54.7 | 49.7 | 63.7 | 51.7 | 49.7 | 56.2 | 48.7 | 51.2 | 57.2 | 50.7 | 65.2 | 53.7 | 52.2 |
| TGG-HEX | 56.2 | 47.2 | 53.7 | 52.7 | 58.7 | 53.7 | 63.7 | 60.2 | 57.7 | 48.2 | 54.7 | 53.2 | 59.7 | 55.2 | 65.2 | 61.7 |
| CGG-HEX | 54.7 | 40.7 | 47.7 | 48.2 | 54.2 | 50.2 | 54.7 | 65.2 | 55.2 | 42.7 | 49.2 | 50.2 | 55.2 | 51.7 | 57.2 | 66.2 |
| TGG-ROX | 52.7 | 39.2 | 49.7 | 46.2 | 56.2 | 48.2 | 62.7 | 58.2 | 54.2 | 39.7 | 51.2 | 49.2 | 57.7 | 50.2 | 63.7 | 59.7 |
| CGG-ROX | 52.7 | 40.7 | 47.7 | 46.7 | 53.7 | 49.7 | 53.2 | 64.2 | 54.2 | 42.2 | 48.7 | 49.7 | 54.7 | 51.2 | 55.2 | 65.7 |
| GAA-Q670 | 42.2 | 58.2 | 42.7 | 48.7 | 39.7 | 42.7 | 39.2 | 41.2 | 46.7 | 61.7 | 42.7 | 51.2 | 40.2 | 48.2 | 44.7 | 43.2 |
| GTG-Q670 | 44.7 | 46.2 | 47.7 | 55.2 | 42.7 | 61.2 | 49.7 | 48.2 | 49.7 | 48.2 | 50.2 | 56.2 | 49.2 | 64.2 | 51.7 | 50.2 |
Tms of complement molecular beacon-target hybrids are in boldface.
Real-time PCR evaluation of wild-type molecular beacon.
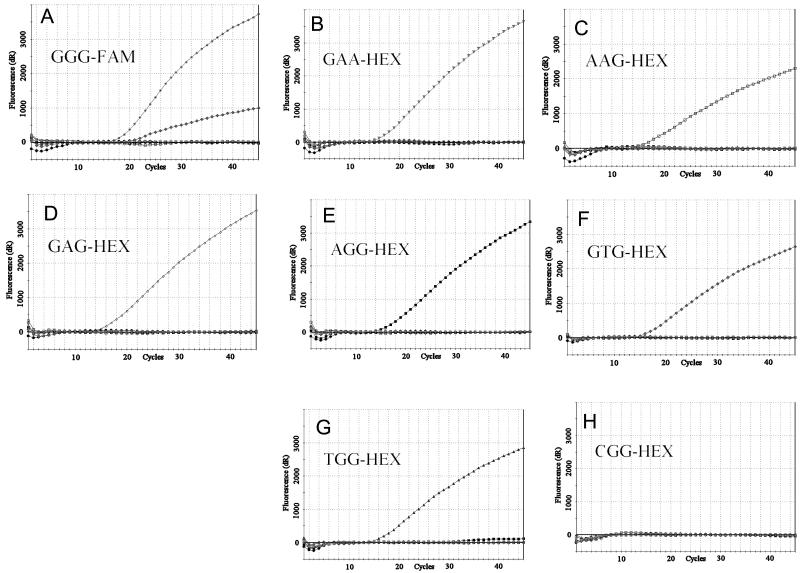
DNA templates for real-time PCR evaluation of molecular beacons were obtained by amplification of 0.5-kb PCR fragments of cyp51A from chromosomal DNA of A. fumigatus strains H11-20 (wild-type GGG allele), RIT12 (GAG), RIT15 (AAG), RIT18 (GAA), RIT51 (AGG), R7-1 (TGG), and Br181 (GTG) (Table 3). The wild-type GGG-FAM molecular beacon was tested in real-time PCR experiments against the above cyp51A alleles. At an annealing temperature of 61°C and 3 mM Mg2+, the GGG-FAM molecular beacon formed a stable hybrid with the wild-type DNA target, which was detected by observing a high level of fluorescence that increased during the amplification process (Fig. 1A). These experimental conditions also provided superior discrimination for all six tested mutant alleles, except for the GTG allele, in which nonspecific hybridization was detected at about 27% of that for the GGG complement allele (Fig. 1A). The GGG-FAM beacon possessed some affinity for the GTG allele, which was also observed by temperature profiling (Table 2). Increasing the annealing temperature for real-time PCR improved the discrimination characteristics of GGG-FAM beacon for the GTG allele, although there was a reduction in total fluorescence for the wild-type allele.
TABLE 3.
A. fumigatus strains analyzed by multiplex molecular beacon real-time PCR
| Strain | MIC (μg/ml) | 54th codon of cyp51A | Amino acid |
|---|---|---|---|
| H11-20 | 0.5 | GGG | Gly |
| R7-1 | >16a | TGG | Trp |
| RIT7 | >16 | GGG | Gly |
| RIT12 | >16 | GAG | Glu |
| RIT14 | >16 | GGG | Gly |
| RIT15 | >16 | AAG | Lys |
| RIT18 | >16 | GAA | Glu |
| RIT19b | >16 | AGG | Arg |
| RIT27 | >16 | GGG | Gly |
| RIT30A | >16 | GGG | Gly |
| RIT31 | >16 | GGG | Gly |
| RIT32 | >16 | GAA | Glu |
| RIT33 | >16 | GGG | Gly |
| RIT34 | >16 | GAG | Glu |
| RIT35 | >16 | GGG | Gly |
| RIT37 | >16 | GGG | Gly |
| RIT38 | >16 | AAG | Lys |
| RIT39 | >16 | GAA | Glu |
| RIT40 | >16 | GAG | Glu |
| RIT41 | >16 | AGG | Arg |
| RIT42 | >16 | GGG | Gly |
| RIT43 | >16 | GAA | Glu |
| RIT44 | >16 | GGG | Gly |
| RIT45 | >16 | GAA | Glu |
| RIT46 | >16 | GGG | Gly |
| RIT47 | >16 | GGG | Gly |
| RIT48 | >16 | GGG | Gly |
| RIT50A | >16 | GGG | Gly |
| RIT51 | >16 | AGG | Arg |
| RIT52 | >16 | GGG | Gly |
| RIT53 | >16 | GAA | Glu |
| RIT54 | >16 | GGG | Gly |
| RIT55 | >16 | AGG | Arg |
| RIT56 | >16 | GGG | Gly |
| Af41 | 0.25 | GGG | Gly |
| Af72 | >16 | GAG | Glu |
| Af90 | >16 | GGG | Gly |
| Br128 | >16 | GGG | Gly |
| Br130 | >16 | GAG | Glu |
| Br181 | 8.0 | GTG | Val |
| F/5211 | 0.5 | GGG | Gly |
| F/6919 | >16 | GGG | Gly |
| F/7075 | >16 | GAG | Glu |
| F/7763 | 1.0 | GGG | Gly |
| SO/3626 | 1.0 | GGG | Gly |
| SO/3827A | >16 | GTG | Val |
| SO/3827B | >16 | GTG | Val |
| SO/3829 | >16 | GTG | Val |
Data are from Mann et al. (26).
Additional mutation T164A resulting in an I55N amino acid change was revealed in the RIT19 strain by sequencing.
FIG. 1.
cyp51A allele discrimination by molecular beacons. Real-time PCR was used to evaluate the specificity of eight different molecular beacons designed to distinguish the wild-type allele and seven mutant alleles. Each panel shows an individual molecular beacon and its relevant Gly54 allele recognition sequence. Real-time PCRs were performed with separate 500-bp templates corresponding to the GGG (*), GAA (▾), AAG (□), GAG (⋄), AGG (▪), GTG (♦), and TGG (▴) cyp51A alleles and a blank (no DNA; •) with the indicated specific molecular beacons. Each panel shows a composite representation of eight separate template reactions with the same molecular beacon.
Design and evaluation of molecular beacons to Gly54 mutant sequences.
After the suitability of a wild-type GGG-FAM molecular beacon for target discrimination was confirmed in real-time PCR experiments, seven molecular beacons targeted to all known Gly54 codon mutations were synthesized. The mutant beacons comprised single- or double-nucleotide substitutions identical to that found in the Gly54 GGG codon of cyp51A of itraconazole-resistant A. fumigatus strains. The cyp51A alleles considered in beacon design were the GAG (7, 26, 33), GAA (33), AAG (33), AGG (7, 26, 33), TGG (26), GTG (7), and CGG (P. Mann, personal communication) alleles. The compositions of all mutant beacons were identical to that of the GGG-FAM beacon except for allele-specific nucleotide substitutions in the probe sequence (Table 1). All seven beacons were labeled with HEX at the 5′ end. The temperature profiles for eight DNA targets were determined, and Tm values are listed in Table 2. Most molecular beacons recognizing mutant sequences had thermal profiles different from the GGG-FAM beacon profile. In general, they formed less-stable intramolecular hybrids with mismatched DNA targets and possessed lower Tms. For beacons GAA-HEX, AAG-HEX, and AGG-HEX, temperatures for duplex formation were so low that the corresponding window of discrimination did not overlap with that of the wild-type probe GGG-FAM, which would pose a problem for multiplex applications. However, an overall increase in probe-target hybrid stability and a concomitant increase in Tm values were achieved by increasing the concentration of Mg2+ from 3 to 5 mM in the reaction mixture. Under this condition, all molecular beacons (mutant and wild type) could be run in a real-time PCR experiment at the same annealing temperature of 61°C and showed excellent discrimination against both wild-type and mutant alleles (Fig. 1). Nonspecific hybridization was not observed for molecular beacons recognizing mutant sequences under these conditions.
Multiplex real-time PCR detection of Gly54 mutations.
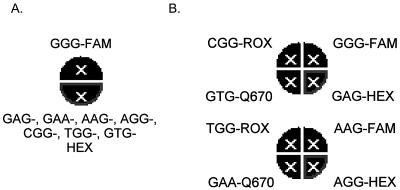
Once target-specific hybridization with complement DNA target was validated for each beacon, we investigated the possibility of combining them in a multiplex real-time PCR format. A single-reaction real-time PCR assay with eight different molecular beacons (one wild type beacon labeled with FAM and seven mutant beacons labeled with HEX) and a 0.5-kb template corresponding to each cyp51A allele was performed. The reaction conditions included a 5 mM Mg2+ concentration and an annealing temperature of 55°C. In spite of such a low temperature at the annealing step, only specific hybridization was observed for molecular beacons in multiplex real-time PCRs. HEX fluorescence (mutant beacons) was observed only when one or more of the mutant templates were present, while the FAM signal from the GGG-FAM molecular beacon was detected only when the wild-type allele was present. The layout of the single-tube multiplex PCR format is shown in Fig. 2A. In this format, the fluorescence signal indicated the presence of either wild-type or mutant sequences corresponding to itraconazole susceptibility or resistance, respectively. The advantage of this format is that, in a single assay, mutant (resistant) and wild-type (susceptible) strains can be easily distinguished. However, this format cannot be used to identify specific mutations of Gly54 because each mutant beacon was labeled with the same fluorophore.
FIG. 2.
Multiplex assay formats. (A) Graphic output of the Stratagene Mx4000 software for the multiplex single-tube assay. The top semicircle is highlighted when the FAM signal is observed, reporting the presence of the wild-type cyp51A allele. The bottom semicircle is highlighted when the HEX signal is observed, reporting the presence of any of seven mutant cyp51A alleles. (B) Graphic output from the Stratagene Mx4000 software for the multiplex double-tube assay. Each quadrant of the eight total sectors in the two circles represents an allele-specific molecular beacon labeled with HEX, ROX, Q670, or FAM, as indicated. The top right sector of the upper circle is highlighted when the FAM signal is observed, reporting the presence of the wild-type cyp51A allele. All other sectors are highlighted when the HEX, ROX, Q670, or FAM signal is observed, reporting the presence of a specific mutant cyp51A allele.
To expand the applicability of the multiplex assay to distinguish separate alleles, the molecular beacons were labeled with different fluorophores: FAM, HEX, ROX, or the CY5 analog Q670 (Table 1). This configuration required two PCRs to assess all seven mutations, since only four colors could be distinguished in a single reaction. The first PCR mixture contained molecular beacons with high Tms against complement DNA targets: GGG-FAM, GAG-HEX, GTG-Q670, and CGG-ROX. The second PCR mixture contained molecular beacons with somewhat lower Tms: AAG-FAM, AGG-HEX, GAA-Q670, and TGG-ROX. The system specificity was further optimized by adjusting the Mg2+ concentration up to 3 mM for the first reaction and to 5 mM for the second reaction. A uniform annealing temperature of 57°C was used for both reactions. The layout of this double-tube multiplex PCR format is shown at Fig. 2B. The assay system was tested by adding individual 0.5-kb cyp51A templates to each of two reaction mixtures. Under these conditions, specific hybridization with complement cyp51A alleles was observed for all molecular beacons except the GGG-FAM wild-type beacon, which exhibited some level of nonspecific hybridization with the GTG allele. To avoid any possible false-positive results coming from nonspecific hybridization of the wild-type GGG-FAM beacon, the threshold fluorescence level was adjusted to values close to those obtained for complement beacon-target pairs. This multiplex format allowed all seven specific cyp51A alleles bearing mutations in the Gly54 codon, along with the wild-type allele, to be distinguished in a real-time assay.
Application of an allele-specific panel.
The multiplex real-time PCR assay was applied to the analysis of a collection of 48 A. fumigatus isolates comprising itraconazole-susceptible (n = 1; parental strain) and -resistant (n = 33) laboratory strains obtained during previous in vitro studies (26, 33) and itraconazole-susceptible (n = 4) and -resistant (n = 11) clinical isolates (Table 3). Chromosomal DNA from each strain was isolated and used as a template in both single-tube and double-tube formats. The assay in the single-tube format revealed only the presence of wild-type or mutant cyp51A alleles, while the system in the double-tube format specified the particular Gly54 mutations. Out of 48 total strains tested, 23 isolates bearing mutant cyp51A alleles with GAG, GAA, AAG, AGG, TGG, or GTG at codon 54 were found. All 23 strains with G54 mutations exhibited elevated levels of itraconazole resistance. Of the 14 clinical isolates, 7 were found to have resistance mutations (Table 3). In all cases but one, the presence of a resistant allele detected by the assay correlated with drug susceptibility testing. The single negative result came from the sample of RIT19 DNA, where a secondary mutation in codon 55 of cyp51A was revealed. For this reason, none of the eight molecular beacons (mutant and wild type) could hybridize specifically with the RIT19 DNA. All strains were subjected to DNA sequencing to validate the real-time determination.
DISCUSSION
The multiplex PCR assay for itraconazole resistance presented in this study provides a foundation for rapid analysis of target site drug resistance mutations that has the potential to extend molecular diagnostics beyond pathogen identification to include simultaneous evaluation of drug susceptibility. This approach represents a natural progression from earlier studies utilizing individual LightCycler probes for rapid identification of triazole resistance mutations in Aspergillus and Candida spp. (15). Microbial drug resistance remains a complicating factor for treatment of fungal infections. While fluconazole resistance in yeasts has remained largely constant (39, 42), resistance to newer triazole drugs (38) and the echinocandins (4) poses new challenges, since in some cases novel molecular mechanisms contribute to resistance (12). Mutations in cyp51A conferring resistance to itraconazole, especially at the Gly54 codon, are well established (7, 26, 33) and represent suitable targets for molecular analysis that have been exploited in this study. The relevance of cyp51A mutations for clinically observed itraconazole resistance needs more thorough investigation, but it is significant, as demonstrated by the fact that 7 of 14 clinical isolates with reduced itraconazole susceptibility were found to contain Gly54 mutations (Table 3). The strong linkage between target site mutations and phenotypic in vitro resistance provided the primary rationale for focusing on these mutations as a surrogate marker for triazole resistance. In fact, of the 48 examined laboratory and clinical isolates for which itraconazole MICs were elevated, 23 were found to contain mutations in codon 54 of the cyp51A gene (Table 3).
The development of fast, accurate, and sensitive diagnostic assays for the identification of invasive aspergillosis remains an important goal to overcome current deficiencies associated with standard microbiological identification. Rapid clinical diagnosis and aggressive preemptive therapy can limit morbidity and morality associated with invasive fungal disease. Yet, most clinical laboratories still rely on culture-based technology with phenotypic end points that can take several days for positive identification and even longer to determine drug susceptibility. In addition to causing time delays, these techniques often lack adequate sensitivity and specificity, and organisms such as Aspergillus are difficult to culture from blood. This often means that empirical therapy must begin in the absence of positive pathogen identification. To circumvent this problem, non-culture-based techniques are emerging rapidly and include tests such as the enzyme-linked immunosorbent assay for the GM antigen and (1→3)-β-d-glucan (BDG) (13), as well as real-time PCR-based assays for Aspergillus-specific DNA (17). Nucleic acid-based diagnostics provide rapid and sensitive results, reducing the time needed for diagnostic work-up to a few hours, and rapid pathogen identification can be achieved with high fidelity from both culture and primary specimens (17, 37). Real-time PCR assays have been reported to have a sensitivity higher than those of the GM and BDG tests (11), although such a determination is controversial since conclusive studies are lacking.
PCR-based amplification of highly conserved rRNA genes and intergenic sequences is the most reliable approach for identification of fungi (20). The fidelity of these assays has improved markedly with the emergence of real-time self-reporting nucleic acid probes, which can be used to detect one to five organisms per milliliter of blood (16), and include LightCycler (16), TaqMan (2), and molecular beacon (35, 37) probes. Real-time PCR with high-fidelity self-reporting probes enables both PCR amplification and detection to be performed in a sealed tube, which reduces the possibility of contamination and allows product formation to be continuously monitored and validated. The fidelity of authentic target recognition by self-reporting real-time probes is critical to a clinical microbiology laboratory because PCR amplification has the potential to amplify small amounts of target DNA from contaminating organisms and even human DNA. Furthermore, real-time probes are quantitative and have a large dynamic range, exceeding 1 million times that of the starting target.
Molecular beacons were selected in this study because they can accurately distinguish allelic differences in a DNA sequence by detecting single nucleotide differences (28). This property is derived from inherent energetic properties of the hairpin structure that make mismatched probe-target hybrids less thermodynamically stable than hybrids between corresponding linear probes, resulting in a wider temperature range for discrimination between perfect matches and single-nucleotide changes (1). Molecular beacon technology has been successfully applied to mutational analysis of bacteria and viruses for single-nucleotide polymorphism genotyping, allele differentiation, qualitative microorganism identification, and quantitative gene expression and viral-load assays (25, 35, 37, 40, 46). The versatility of such a high-fidelity probe system for nucleic acid-based detection now extends beyond simple pathogen detection and includes a rapid assessment of drug resistance where specific changes in the DNA are linked to resistance.
The ultimate goal of actively discriminating between cyp51A alleles differing in a single nucleotide was achieved by optimizing molecular beacon design and real-time PCR conditions. Since mutations conferring resistance to itraconazole in A. fumigatus are clustered at a single locus corresponding to codon 54 of the cyp51A gene, the analysis was restricted by surrounding this locus. The probe domains of synthesized molecular beacons reproduced sequences of eight known cyp51A alleles (the wild-type allele and seven mutant alleles) starting from nucleotide position 150 and ending at 170. The probe length of 21 nucleotides and a stem sequence of 6 nucleotides provided sufficient specificity for detection of point mutations.
Application of molecular beacons in individual PCRs showed specificity suitable for allele differentiation (Fig. 1); only a minor nonspecific hybridization was noted for the GGG-FAM wild-type beacon with respect to the GTG allele. Importantly, no nonspecific interactions were observed for the beacons recognizing mutant sequences. Such high discriminating power is especially important for prospective application of the assay for analysis of total-DNA samples from potentially mixed A. fumigatus populations where a small amount of mutant cyp51A DNA could be accurately detected in a background of wild-type DNA.
A single multiplex reaction assay that combines numerous probes and that is capable of identifying multiple pathogens is an efficient and cost-effective approach for a clinical microbiology laboratory. Such assays require that probes representing different targets be reliably resolved in the same reaction vessel. Multiplexing in its simplest format required a single tube and included all eight (wild-type plus seven mutant) molecular beacons. The wild-type beacon was labeled with FAM, while all mutant beacons were labeled with HEX. The assay was robust and detected either the wild-type sequence or a mutant sequence. Combining all eight allele-specific molecular beacons in a single multiplex PCR assay made possible the differentiation of DNA from wild-type and itraconazole-resistant strains with mutations in cyp51A. Although the assay was allele specific, it was designed to give a wild-type (FAM fluorescence) or mutant (HEX fluorescence) output, and it could not be used to distinguish which mutant allele was present in the template. This assay format is powerful because many probes can be included in a single reaction with a common set of PCR primers, and it accurately determines resistance if a mutant beacon signal is detected. For clinical laboratories, such an output may be sufficient.
The multiplex format utilizing individually labeled molecular beacons made possible genotyping of specific cyp51A itraconazole resistance mutations. The Mx4000 real-time system capabilities were restricted to analysis of no more than four fluorophores per tube, so the assay of eight beacons labeled by FAM, HEX, ROX, and Q670 was split between two PCR tubes. In both multiplex formats, the PCR amplification of individual wild-type or mutant DNA target produced distinct and robust fluorescence signals. No false-negative or false-positive results due to hybridization or PCR failure and no nonspecific hybridization were ever observed. The only case in which no fluorescence signal was obtained was for a DNA sample of the RIT19 strain, which had two missense mutations G161A and T163A in codons 54 and 55 of cyp51A gene. This result provided evidence of specificity not only with respect to known mutations at codon 54 but also to other potential mutations at the cyp51A locus covered by molecular beacons.
It is important to recognize that, although the multiplex assay developed in this work focused on resistance-associated mutations in codon 54 of cyp51A, in fact, the platform is dynamic and can be expanded to include mutations, once identified, at other loci. These include the recently described Met220 mutations described by Mellado and colleagues (30). In principle, a broader platform that includes a comprehensive inventory of target site mutations, along with a quantitative assessment of drug efflux transporter gene expression levels, could then accurately identify most, if not all, resistant strains in a rapid assay. Thus, the overall robustness and ultimate clinical value of the assay will depend on the characterization of resistance alleles and the prevalence of certain mutations or mutation “hot spots.”
In conclusion, we have developed a multiplex molecular beacon real-time PCR assay that permits simple, rapid, and reliable diagnostics of the itraconazole resistance mutations in the A. fumigatus cyp51A gene. Thus, the notion of identifying an invasive pathogen such as Aspergillus from a primary specimen and simultaneously assessing its drug susceptibility status is entirely feasible and has important implications for therapeutic management of patients.
Acknowledgments
We acknowledge the following for providing A. fumigatus strains or chromosomal DNA: Gustavo H. Goldman (University of São Paulo, Ribeirao Preto, Brazil), David W. Denning and Andrew Sharp (Hope Hospital, Manchester, United Kingdom), and Paul Mann and Paul McNicholas (Schering Plough Research Institute, Kenilworth, N.J.). We also thank David Denning for his advice and critical review of the manuscript.
This work was supported by NIH/NIAID grant AI055767 to D.S.P.
REFERENCES
- 1.Bonnet, G., S. Tyagi, A. Libchaber, and F. R. Kramer. 1999. Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc. Natl. Acad. Sci. USA 96:6171-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandt, M. E., A. A. Padhye, L. W. Mayer, and B. P. Holloway. 1998. Utility of random amplified polymorphic DNA PCR and TaqMan automated detection in molecular identification of Aspergillus fumigatus. J. Clin. Microbiol. 36:2057-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dannaoui, E., E. Borel, M. F. Monier, M. A. Piens, S. Picot, and F. Persat. 2001. Acquired itraconazole resistance in Aspergillus fumigatus. J. Antimicrob. Chemother. 47:333-340. [DOI] [PubMed] [Google Scholar]
- 4.Denning, D. 2003. Echinocandin antifungal drugs. Lancet 362:1142-1151. [DOI] [PubMed] [Google Scholar]
- 5.Denning, D. W., S. A. Radford, K. L. Oakley, L. Hall, E. M. Johnson, and D. W. Warnock. 1997. Correlation between in-vitro susceptibility testing to itraconazole and in-vivo outcome of Aspergillus fumigatus infection. J. Antimicrob. Chemother. 40:401-414. [DOI] [PubMed] [Google Scholar]
- 6.Denning, D. W., R. M. Tucker, L. H. Hanson, and D. A. Stevens. 1989. Treatment of invasive aspergillosis with itraconazole. Am. J. Med. 86:791-800. [DOI] [PubMed] [Google Scholar]
- 7.Diaz-Guerra, T. M., E. Mellado, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2003. A point mutation in the 14α-sterol demethylase gene cyp51A contributes to itraconazole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 47:1120-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feldmesser, M. 2003. New and emerging antifungal agents: impact on respiratory infections. Am. J. Respir. Med. 2:371-383. [DOI] [PubMed] [Google Scholar]
- 9.Gupta, A. K., and E. Tomas. 2003. New antifungal agents. Dermatol. Clin. 21:565-576. [DOI] [PubMed] [Google Scholar]
- 10.Herbrecht, R., V. Letscher-Bru, C. Oprea, B. Lioure, J. Waller, F. Campos, O. Villard, K. L. Liu, S. Natarajan-Ame, P. Lutz, P. Dufour, J. P. Bergerat, and E. Candolfi. 2002. Aspergillus galactomannan detection in the diagnosis of invasive aspergillosis in cancer patients. J. Clin. Oncol. 20:1898-1906. [DOI] [PubMed] [Google Scholar]
- 11.Kami, M., T. Fukui, S. Ogawa, Y. Kazuyama, U. Machida, Y. Tanaka, Y. Kanda, T. Kashima, Y. Yamazaki, T. Hamaki, S. Mori, H. Akiyama, Y. Mutou, H. Sakamaki, K. Osumi, S. Kimura, and H. Hirai. 2001. Use of real-time PCR on blood samples for diagnosis of invasive aspergillosis. Clin. Infect. Dis. 33:1504-1512. [DOI] [PubMed] [Google Scholar]
- 12.Kartsonis, N. A., J. Nielsen, and C. M. Douglas. 2003. Caspofungin: the first in a new class of antifungal agents. Drug Resist. Updates 6:197-218. [DOI] [PubMed] [Google Scholar]
- 13.Kawazu, M., Y. Kanda, Y. Nannya, K. Aoki, M. Kurokawa, S. Chiba, T. Motokura, H. Hirai, and S. Ogawa. 2004. Prospective comparison of the diagnostic potential of real-time PCR, double-sandwich enzyme-linked immunosorbent assay for galactomannan, and a (13)-β-d-glucan test in weekly screening for invasive aspergillosis in patients with hematological disorders. J. Clin. Microbiol. 42:2733-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin, S. J., J. Schranz, and S. M. Teutsch. 2001. Aspergillosis case-fatality rate: systematic review of the literature. Clin. Infect. Dis. 32:358-366. [DOI] [PubMed] [Google Scholar]
- 15.Loeffler, J., L. Hagmeyer, H. Hebart, N. Henke, U. Schumacher, and H. Einsele. 2000. Rapid detection of point mutations by fluorescence resonance energy transfer and probe melting curves in Candida species. Clin. Chem. 46:631-635. [PubMed] [Google Scholar]
- 16.Loeffler, J., N. Henke, H. Hebart, D. Schmidt, L. Hagmeyer, U. Schumacher, and H. Einsele. 2000. Quantification of fungal DNA by using fluorescence resonance energy transfer and the light cycler system. J. Clin. Microbiol. 38:586-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loeffler, J., K. Kloepfer, H. Hebart, L. Najvar, J. R. Graybill, W. R. Kirkpatrick, T. F. Patterson, K. Dietz, R. Bialek, and H. Einsele. 2002. Polymerase chain reaction detection of aspergillus DNA in experimental models of invasive aspergillosis. J. Infect. Dis. 185:1203-1206. [DOI] [PubMed] [Google Scholar]
- 18.Loeffler, J., and D. A. Stevens. 2003. Antifungal drug resistance. Clin. Infect. Dis. 36:S31-S41. [DOI] [PubMed] [Google Scholar]
- 19.Loffler, J., S. L. Kelly, H. Hebart, U. Schumacher, C. Lass-Florl, and H. Einsele. 1997. Molecular analysis of cyp51 from fluconazole-resistant Candida albicans strains. FEMS Microbiol. Lett. 151:263-268. [DOI] [PubMed] [Google Scholar]
- 20.Lott, T. J., R. J. Kuykendall, and E. Reiss. 1993. Nucleotide sequence analysis of the 5.8S rDNA and adjacent ITS2 region of Candida albicans and related species. Yeast 9:1199-1206. [DOI] [PubMed] [Google Scholar]
- 21.Luo, G., and T. G. Mitchell. 2002. Rapid identification of pathogenic fungi directly from cultures by using multiplex PCR. J. Clin. Microbiol. 40:2860-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lupetti, A., R. Danesi, M. Campa, M. Del Tacca, and S. Kelly. 2002. Molecular basis of resistance to azole antifungals. Trends Mol. Med. 8:76-81. [DOI] [PubMed] [Google Scholar]
- 23.Maertens, J., J. Verhaegen, K. Lagrou, J. Van Eldere, and M. Boogaerts. 2001. Screening for circulating galactomannan as a noninvasive diagnostic tool for invasive aspergillosis in prolonged neutropenic patients and stem cell transplantation recipients: a prospective validation. Blood 97:1604-1610. [DOI] [PubMed] [Google Scholar]
- 24.Manavathu, E. K., O. C. Abraham, and P. H. Chandrasekar. 2001. Isolation and in vitro susceptibility to amphotericin B, itraconazole and posaconazole of voriconazole-resistant laboratory isolates of Aspergillus fumigatus. Clin. Microbiol. Infect. 7:130-137. [DOI] [PubMed] [Google Scholar]
- 25.Manganelli, R., E. Dubnau, S. Tyagi, F. R. Kramer, and I. Smith. 1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol. Microbiol. 31:715-724. [DOI] [PubMed] [Google Scholar]
- 26.Mann, P. A., R. M. Parmegiani, S.-Q. Wei, C. A. Mendrick, X. Li, D. Loebenberg, B. DiDomenico, R. S. Hare, S. S. Walker, and P. M. McNicholas. 2003. Mutations in Aspergillus fumigatus resulting in reduced susceptibility to posaconazole appear to be restricted to a single amino acid in the cytochrome P450 14α-demethylase. Antimicrob. Agents Chemother. 47:577-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marr, K. A., S. A. Balajee, L. McLaughlin, M. Tabouret, C. Bentsen, and T. J. Walsh. 2004. Detection of galactomannan antigenemia by enzyme immunoassay for the diagnosis of invasive aspergillosis: variables that affect performance. J. Infect. Dis. 190:641-649. [DOI] [PubMed] [Google Scholar]
- 28.Marras, S. A., F. R. Kramer, and S. Tyagi. 1999. Multiplex detection of single-nucleotide variations using molecular beacons. Genet. Anal. 14:151-156. [DOI] [PubMed] [Google Scholar]
- 29.Mellado, E., T. M. Diaz-Guerra, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2001. Identification of two different 14-α sterol demethylase-related genes (cyp51A and cyp51B) in Aspergillus fumigatus and other Aspergillus species. J. Clin. Microbiol. 39:2431-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mellado, E., G. Garcia-Effron, L. Alcazar-Fuoli, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2004. Substitutions at methionine 220 in the 14α-sterol demethylase (Cyp51A) of Aspergillus fumigatus are responsible for resistance in vitro to azole antifungal drugs. Antimicrob. Agents Chemother. 48:2747-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mennink-Kersten, M. A., J. P. Donnelly, and P. E. Verweij. 2004. Detection of circulating galactomannan for the diagnosis and management of invasive aspergillosis. Lancet Infect. Dis. 4:349-357. [DOI] [PubMed] [Google Scholar]
- 32.Moore, C. B., N. Sayers, J. Mosquera, J. Slaven, and D. W. Denning. 2000. Antifungal drug resistance in Aspergillus. J. Infect. 41:203-220. [DOI] [PubMed] [Google Scholar]
- 33.Nascimento, A. M., G. H. Goldman, S. Park, S. A. E. Marras, G. Delmas, U. Oza, K. Lolans, M. N. Dudley, P. A. Mann, and D. S. Perlin. 2003. Multiple resistance mechanisms among Aspergillus fumigatus mutants with high-level resistance to itraconazole. Antimicrob. Agents Chemother. 47:1719-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.National Committee for Clinical Laboratory Standards. 1998. Reference method for broth dilution antifungal susceptibility testing of conidium-forming filamentous fungi. Proposed standard M38-P. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 35.Park, S., M. Wong, S. A. E. Marras, E. W. Cross, T. E. Kiehn, V. Chaturvedi, S. Tyagi, and D. S. Perlin. 2000. Rapid identification of Candida dubliniensis using a species-specific molecular beacon. J. Clin. Microbiol. 38:2829-2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearson, M. M., P. D. Rogers, J. D. Cleary, and S. W. Chapman. 2003. Voriconazole: a new triazole antifungal agent. Ann. Pharmacother. 37:420-432. [DOI] [PubMed] [Google Scholar]
- 37.Perlin, D. S., and S. Park. 2001. Rapid identification of fungal pathogens: molecular approaches for a new millennium. Rev. Med. Microbiol. 12:S13-S20. [Google Scholar]
- 38.Pfaller, M. A., D. J. Diekema, S. A. Messer, L. Boyken, and R. J. Hollis. 2003. Activities of fluconazole and voriconazole against 1,586 recent clinical isolates of Candida species determined by broth microdilution, disk diffusion, and Etest methods: report from the ARTEMIS Global Antifungal Susceptibility Program, 2001. J. Clin. Microbiol. 41:1440-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfaller, M. A., S. A. Messer, R. J. Hollis, R. N. Jones, G. V. Doern, M. E. Brandt, and R. A. Hajjeh. 1999. Trends in species distribution and susceptibility to fluconazole among blood stream isolates of Candida species in the United States. Diagn. Microbiol. Infect. Dis. 33:217-222. [DOI] [PubMed] [Google Scholar]
- 40.Piatek, A. S., A. Telenti, M. R. Murray, H. El-Hajj, W. R. Jacobs, Jr., F. R. Kramer, and D. Alland. 2000. Genotypic analysis of Mycobacterium tuberculosis in two distinct populations using molecular beacons: implications for rapid susceptibility testing. Antimicrob. Agents Chemother. 44:103-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pierard, G. E., J. E. Arrese, and C. Pierard-Franchimont. 2000. Itraconazole. Expert Opin. Pharmacother. 1:287-304. [DOI] [PubMed] [Google Scholar]
- 42.Sanglard, D., and F. C. Odds. 2002. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect. Dis. 2:73-85. [DOI] [PubMed] [Google Scholar]
- 43.Turenne, C. Y., S. E. Sanche, D. J. Hoban, J. A. Karlowsky, and A. M. Kabani. 1999. Rapid identification of fungi by using the ITS2 genetic region and an automated fluorescent capillary electrophoresis system. J. Clin. Microbiol. 37:1846-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tyagi, S., D. P. Bratu, and F. R. Kramer. 1998. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 16:49-53. [DOI] [PubMed] [Google Scholar]
- 45.Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14:303-308. [DOI] [PubMed] [Google Scholar]
- 46.Vet, J. A., A. R. Majithia, S. A. Marras, S. Tyagi, S. Dube, B. J. Poiesz, and F. R. Kramer. 1999. Multiplex detection of four pathogenic retroviruses using molecular beacons. Proc. Natl. Acad. Sci. USA 96:6394-6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Viscoli, C., M. Machetti, P. Cappellano, B. Bucci, P. Bruzzi, M. T. Van Lint, and A. Bacigalupo. 2004. False-positive galactomannan platelia Aspergillus test results for patients receiving piperacillin-tazobactam. Clin. Infect. Dis. 38:913-916. [DOI] [PubMed] [Google Scholar]
- 48.White, T. C., S. Holleman, F. Dy, L. F. Mirels, and D. A. Stevens. 2002. Resistance mechanisms in clinical isolates of Candida albicans. Antimicrob. Agents Chemother. 46:1704-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]