Abstract
Inflammasomes are multiprotein complexes whose primary function is to activate caspase-1, which allows the cleavage of pro-IL-1β and pro-IL-18 to their mature forms. The production of these cytokines has been shown to be critical for host defense as well as the maintenance of intestinal homeostasis and protection against pathologic intestinal inflammation. More recently, there has been growing evidence that inflammasomes are also capable of regulating the composition of the gut microbiota in mice models, which has significant implications for intestinal health and disease. Specifically, the absence of inflammasome components has been associated with pathologic alterations in the gut microbiota, or dysbiosis, that can result in increased susceptibility to colitis and tumorigenesis. In this review, evidence that inflammasome signaling is important for promoting a healthful microbiome and potential mechanisms by which inflammasomes modulate the gut microbiome will be presented. A better understanding of the function of inflammasomes in microbiome regulation may lead to the development of effective strategies for the prevention and treatment of diseases driven by dysbiosis.
Keywords: inflammasomes, IL-18, microbiome, colitis
I. Introduction
Alterations in the gut microbiota that are associated with the pathogenesis of multiple diseases, including inflammatory bowel disease, colorectal cancer, and metabolic disorders, have often been collectively termed dysbiosis. However, factors that contribute to the development of dysbiosis remain to be fully understood. There is increasing evidence that host immunity plays a significant role in shaping of the gut microbiome. In particular, innate immune receptors that participate in inflammasome assembly and activation have been implicated in regulating the composition of the gut microbiota, and deficiencies in these receptors have been associated with perturbations in the gut microbiome that are associated with susceptibility to systemic and intestinal diseases in mice models. In this review, the function of inflammasomes and the consequences of inflammasome deficiency as it pertains to perturbations in the microbiome and disease susceptibility will be discussed.
II. Inflammasomes: regulators of IL-1β and IL-18 production
A hallmark of canonical inflammasome signaling is the activation of caspase-1, whose primary function is to cleave biologically inactive pro-IL-1β and pro-IL-18 to their mature, active forms1. Caspase-1 activation also leads to a specific form of macrophage cell death that usually occurs during infection, known as pyroptosis, which enables the release of intracellular cytokines important for host defense2. Inflammasomes are multiprotein complexes typically consisting of a pattern recognition receptor (PRR) belonging to either the Nod-like receptor (NLR) or the absent in melanoma (AIM)-2 like receptors (ALR) family3. A common feature of these receptors that participate in inflammasome assembly is the presence of a pyrin (PYD) domain, which is capable of interacting with the pyrin domain of the inflammasome adaptor protein ASC upon receptor activation. ASC also contains a caspase recruitment domain (CARD), and thus, recruitment of ASC allows the binding of the CARD-domain containing procaspase-1 through homotypic protein interactions. As certain NLRs also contain CARD domains, procaspase-1 may directly be recruited through interactions with these NLRs without the requirement for ASC. Thusfar, there have been 5 bona fide inflammasomes identified as evidenced by formation of inflammasome complexes and/or caspase-1 activation upon recognition of ligand, which includes both microbial-derived pattern-associated molecular patterns (PAMPs) or endogenous tissue injury-related damage-associated molecular patterns (DAMPs). These are the NLR containing, NLRP1, NLRP3, and NLRC4 inflammasomes, the ALR containing AIM2 inflammasome, and the pyrin inflammasome1, 4–10. The upstream signals for these inflammasomes are relatively well-defined and can be both microbial- and/or host-derived. For example, the lethal toxin of Bacillus anthracis and flagellin are PAMPs that activate the NLRP1 and NLRC4 inflammasomes, respectively4, 11. The AIM2 inflammasome recognizes cytosolic double-stranded DNA released by either pathogens or fecal microbiota6–8, 12, but can also sense host DNA aberrantly located in the cytosol13. Although cyclic GMP-AMP synthase (cGAS) is also an intracellular DNA sensor, its function is distinct from that of AIM2 in that it induces Type I IFNs, which in turn, potentiates the activity of AIM214–16. The NLRP3 inflammasome is unique in that it responds to a diverse array of stimuli, both microbial and non-microbial, such as bacterial pore-forming toxins, particulate matter, oxidized mitochondrial DNA, and extracellular ATP17–22. Collectively, these various stimuli can be linked to cellular injury and changes in homeostasis, and therefore, it is likely that NLRP3 senses a common downstream signal such as changes in cytosolic potassium ion concentration21. Whether inflammasomes such as AIM2 or NLRP3 respond differently to endogenous versus microbial activators remains to be determined.
More recently, the NLR family member NLRP6 has been considered to be capable of forming an inflammasome based on data demonstrating colocalization of NLRP6 with ASC in a prototypical speckled pattern, and impaired IL-18 production and caspase-1 activation in the colons of NLRP6-deficient mice23–26. However, the precise signal leading to assembly of an “NLRP6 inflammasome” remains unknown, making it difficult to definitively prove that NLRP6 activation is directly linked to activation of caspase-1. Levy et al. reported that IL-18 production within the colon is regulated by microbial metabolites, in particular, taurine, which was sufficient to induce caspase-1 activation and IL-18 production in the colons of GF mice fed taurine in the drinking water26. Interestingly the ability of taurine to induce IL-18 was dependent on NLRP6, suggesting that taurine may be an upstream activator of NLRP626.
NLRP12 is another member that is also believed to be capable of functioning as part of an inflammasome under certain conditions based on early data demonstrating caspase-1 activation in transfection experiments in vitro using caspase-1, ASC, and NLRP12 expression plasmids27. In addition, Nlrp12−/− bone marrow-derived macrophages exhibited decreased caspase-1 activation and impaired secretion of both IL-1β and IL-18 in response to the pathogen Yersinia28. It is important to note that the activities of NLRP12 and NLRP6 are not necessarily limited to IL-1β or IL-18 production, respectively. For example, both have been suggested to negatively regulate NFκB and MAPK activation in response to microbial ligands29–31. In addition, NLRP6 has been shown to bind viral RNA via the RNA helicase Dhx5 and is capable of regulating Type I/II IFN responses to mediate resistance to viral infection in a caspase-1-independent process32.
III. Inflammasomes: regulators of intestinal homeostasis
As proinflammatory cytokines, IL-1β and IL-18 have important functions in host defense. For example, IL-18 promotes Th1 responses by inducing IFNγ production33, and IL-1β induces neutrophil influx, activates both myeloid cells and lymphocytes, and promotes Th17 differentiation34. Although elevated levels of IL-1β and IL-18 have been observed in patients with inflammatory bowel disease (IBD), mouse models of chemically-induced colitis using dextran sulfate sodium (DSS) have suggested that inflammasomes are important for maintaining intestinal homeostasis and reducing susceptibility to DSS-induced morbidity and mortality. More specifically, mice deficient in NLRP1, NLRP3, NLRP6, NLRP12, AIM2, or ASC have greater weight loss, significantly greater induction of proinflammatory mediators and increased inflammation and epithelial damage within the colon12, 24, 25, 30, 31, 35–39(Table I). Similarly, susceptibility to colitis associated tumorigenesis using the (azoxymethane) AOM/DSS model in which mice are administered the carcinogen AOM followed by multiple rounds of colitis-inducing DSS, was also increased12, 25, 30, 31, 35, 36, 39, 40. With the exception of NLRP12, which was shown to negatively regulate NFκB and MAPK signaling to limit colitis and tumorigenesis, a common mechanism by which NLRP3, NLRP6 and AIM2 reduces colitis susceptibility is through the production of IL-18, which is important for tissue repair, epithelial restitution and consequently resolution of inflammation24, 25, 41, 42. Although IL-1β levels were also observed to be low in Nlrp1−/− and Nlrp3−/− mice, IL-1β levels were markedly elevated in Nlrp6−/− and AIM2−/− mice, suggesting that IL-1β is not necessarily protective in this model and that the production of IL-1β and IL-18 are not coupled25, 35, 38. Importantly, administration of recombinant IL-18 into caspase-1−/−, Nlrp1−/− or Nlrp3−/− mice ameliorated colitis35, 37, 42. Furthermore, IL-18−/− mice also developed more severe colitis after DSS treatment41. However, unlike mice fully deficient in IL-18, mice in which IL-18 is conditionally knocked out in either the epithelial or hematopoietic compartments resulted actually in protection against DSS-induced colitis, suggesting that concerted IL-18 production by different cell types may affect disease outcomes. Indeed, bone marrow chimera experiments have yielded conflicting results regarding the relative importance of inflammasome signaling in the epithelial versus hematopoietic compartment. Non-hematopoietic cells are the predominant sources of IL-18 production and studies by Elinav et al., have suggested that non-hematopoietic production of IL-18 is associated with protection against DSS-induced colitis24, 25. However, IL-18 production by inflammatory monocytes reduced DSSinduced mortality in Nlrp6−/− mice43. Conflicting results were also observed with bone marrow chimera experiments to determine the relative importance of NLRP3 signaling in epithelial versus hematopoietic cells with one study demonstrating the importance of intact NLRP3 signaling in bone marrow-derived cells to reduce colitis susceptibility, but non-hematopoietic cells in another36, 37. These inconsistencies highlight the complexity of inflammasome signaling and may also reflect differences in experimental setup as well as other contributory factors such as facility-dependent differences in the gut microbiota.
Table 1.
Increased susceptibility to inflammation-related disorders in inflammasome-deficient mice is associated with changes in IL-1b, IL-18, and microbiome composition. NASH, non-alcoholic steatohepatitis; NAFLD, non-alcoholic fatty liver disease
| Mice Strain |
Phenotype | Phenotype Transmissible ? |
IL-1β level s |
IL-18 level s |
Microbiome Changes |
|---|---|---|---|---|---|
| Nlrp1−/− | ↑ DSS-induced colitis35 |
Yes | ↓ | ↓ | Not identified |
| ↑ colitis- associated tumors35 |
|||||
| Nlrp3−/− | ↑ DSS-induced colitis36,37 |
Yes | ↓ | ↓ | ↑Enterobacteriaceae48 |
| ↑ colitis- associated tumors36,40 |
|||||
| Nlrp6−/− | ↑ DSS- colitis24,25 |
Yes | ↑ | ↓ | ↑Prevotellaceae, Prevotella, TM724 |
| ↑ colitis- associated tumors25, 39,49 |
|||||
| Nlrp12−/− | ↑ DSS-induced colitis30,31 |
Yes | ↑ | ↓ | Not identified |
| ↑ colitis- associated tumors30,31 |
|||||
| Aim2−/− | ↑ DSS-induced colitis12,38 |
Yes | ↑ | ↓ | ↑ Akkermansia, Anaeroplasma47 |
| ↑ Enterobacteriaceae38 | |||||
| ↑ colitis- associated tumors47 |
↑ Prevotella, Bacteroides12 |
||||
| ↓ Prevotella, Anaerostipes, and Paraprevotella47 |
|||||
| Asc−/− | ↑ DSS-induced colitis |
Yes | ↓ | ↓ | ↑Prevotellaceae, Prevotella,TM724 |
| ↑ colitis- associated tumors |
↑Porphyromonadaceae46 | ||||
| ↑NASH/NAFL D |
|||||
|
Caspase1 −/− |
↑ DSS-induced colitis42 |
Not tested | ↓ | ↓ | Not identified |
| ↑ colitis- associated tumors40 |
Despite the redundant activities of the different inflammasomes, it is also not clear why colitis susceptibility as a result of deficiency in any one inflammasome is not compensated by other functional inflammasomes. One possibility is that there are innate immune receptor-specific activities beyond IL-18 production that contribute to intestinal homeostasis. For example, NLRP6 also regulates goblet cell function through autophagy pathways as well as promotes Muc2 secretion in response to bacterial signals, both of which are important for maintaining an intact mucus layer that provides an effective barrier against bacterial-driven inflammation44, 45. As each of the inflammasomes respond to different upstream signals, the presence or absence of these signals and the magnitude of response may also dictate the relative contributions of the different inflammasomes during inflammation.
IV. Inflammasome deficiency is associated with disease-promoting dybiosis
Reduction in the severity of DSS-induced colitis with antibiotic treatment provided the first evidence that the gut microbiota contributes significantly to the phenotype of inflammasome-deficient mice24, 35. Additional evidence was provided by cohousing wildtype (WT) with inflammasome-deficient mice or crossfostering WT or inflammasome-deficient mice with mothers of the opposite genotype, which demonstrated the transmissibility of colitis through transfer of microbiota. In a seminal study by Elinav et al., when WT mice were either cohoused for 4 weeks with Asc−/− mice or crossfostered at birth with Asc−/− mothers, they developed more severe colitis comparable to that of singly housed mice, which strongly suggested that Asc−/− mice harbored colitogenic bacteria that can be transferred into WT mice to increase colitis susceptibility. Cohousing WT with susceptible caspase-1-deficient mice revealed similar results, with cohoused WT mice developing more severe colitis than singly housed, non-cohoused WT mice. With additional cohousing experiments between WT and different NLR members associated with inflammasome activity, it was determined that only Nlrp6−/− mice was capable of transmitting increased colitis susceptibility to WT mice24. On the other hand, WT mice cohoused with AIM2-, NLRP12-, or NLRC4-deficient mice did not develop worse colitis symptoms than singly housed WT mice. Based on these experiments, it was concluded that inflammasome-deficient mice and specifically, NLRP6–deficient mice, harbored an altered microbiome characterized by the enrichment of potentially colitogenic bacteria that can be transferred to WT mice upon cohousing or crossfostering. Indeed, conventionalized germfree (GF) Nlrp6−/− mice developed a microbiome that resembled that of specific-pathogen free (SPF) Nlrp6−/− mice and distinct from that of conventionalized GF WT26.
To identify potentially colitogenic bacteria that were increased in abundance in SPF Nlrp6−/− mice, 16S rRNA sequencing was performed on the fecal microbiota, and Prevotella, a member of the family Prevotellaceae as well as the phylum TM7 were identified to be significantly associated with Nlrp6−/−, Asc−/−, caspase1−/−, IL-18−/− and cohoused WT mice that developed severe DSS-induced colitis. However, whether the accumulation of these bacterial populations are truly regulated by inflammasome activity or merely reflect colony-specific changes remain to be determined. Nonetheless, antibiotic treatment of Nlrp6−/− mice leading to reduction in the relative abundance of Prevotellaceae resulted in amelioration of colitis symptoms although this result suggests only a correlation and not a direct causal relationship between Prevotellaceae abundance and colitis susceptibility24.
In addition to colitis, Asc−/−, caspase1−/−, Nlrp3−/−, or Nlrp6−/− mice are also more susceptible to the development of non-alcoholic fatty liver disease (NAFLD) than WT mice when mice are fed a methionine-choline-deficient diet46. This phenotype was transmissible to WT mice upon cohousing, suggesting that the severity of NAFLD is, in part, dependent on the microbiome. Comparison of the microbiomes of WT mice that were singly-housed or cohoused with Asc−/− mice confirmed the increased abundance of Prevotellaceae in WT mice cohoused with Asc−/− mice although this was not maintained upon change in diet. Rather, there was a significant expansion in bacterial members belonging to the family Porphyromonadaceae in WT mice cohoused with Asc−/− mice compared to singly housed WT mice after placement on a methioninecholine- deficient diet to induce NAFLD. These experiments suggest only a correlation between abundance of Porphyromonodaceae and therefore, additional experiments involving GF monoassociation studies will be helpful to determine a causal relationship between Prevotellaceae and Porphyomondaceae and the severity of colitis and NAFLD, respectively.
Elinav et al. previously reported that cohousing WT mice with Aim2−/− or Nlrp12−/− mice did not result in exacerbated DSS-induced colitis24. However, Hu et al. demonstrated amelioration of DSS-induced colitis in Aim2−/− mice after cohousing with WT mice, suggesting that WT mice may harbor protective bacteria populations that can be transferred to Aim2−/− mice although the possibility that the cohousing that was performed in this study was insufficient to permit transfer of potentially colitogenic bacteria found in Aim2−/− mice38. Regardless, GF mice colonized with the microbiota of Aim2−/− mice developed more severe colitis than GF WT mice colonized with WT microibota suggesting that the microbiome associated with AIM2-deficiency can confer increased colitis susceptibility. Similarly, in a separate study by Man et al., AIM2 deficiency was associated with increased susceptibility to colitis-associated tumorigenesis, which decreased after cohousing with WT mice47. Based on a survey of the abundance of various bacterial populations by qPCR, Hu et al. did not observe an enrichment of Prevotellaceae or TM7 in Aim2−/− mice unlike in Nlrp6−/− mice. However, there were significantly increased levels of bacteria belonging to the family Enterobacteriaceae and, in particular, E. coli. In the study by Man et al., a more comprehensive survey by 16S rRNA sequencing was performed on mice feces and demonstrated instead relatively increased abundances of Akkermansia muciniphila and Anaeroplasma species, but decreased abundances of Prevotella, Anaerostipes, and Paraprevotella species in single housed Aim2−/− mice compared to that of WT, which was reversed upon cohousing with WT mice, suggesting that the enrichment or depletion of these bacterial populations can modulate tumor susceptibility. This study did not identify Enterobacteriaceae, which may be related to facility-dependent differences in microbiota. However, this also raises questions on whether the observed differences in microbiota can be directly attributed to AIM2 deficiency as it is unclear whether the microbiome analysis was performed on Aim2−/− and WT littermates. Additional experiments looking at heterozygote matings and conventionalization of GF WT and Aim2−/− mice will help resolve this issue.
Although less well-characterized, the microbiomes of NLRP1 or NLRP3-deficient mice have also been suggested to be different from that of WT mice. Terminal restriction fragment polymorphism analysis of the fecal microbiota of WT mice derived from littermates of Nlrp3−/− mice showed significant differences in the microbiome composition compared to that of WT mice48. Specifically, they identified several bacterial genera that were differentially abundance between the two genotypes, including the enrichment of Enterobacteriaceae in Nlrp3−/− mice. However, which of these differentially abundance bacterial populations are responsible for the disease phenotype remains unknown. There is also data suggesting that NLRP1-deficient mice have increased susceptibility to colitis, which is improved with antibiotics and can be transferred to WT mice upon cohousing, but it remains to be determined as well whether NLRP1 truly regulates the gut microbiota.
How bacterial populations that accumulate in the gut of inflammasome-deficient mice promote disease remains to be fully elucidated, but may be related to their ability to upregulate the production of proinflammatory mediators such as CCL5, IL-6 and TNF-α. Elinav et al. reported that their naive Nlrp6−/− mice exhibited low, subclinical levels of intestinal inflammation associated with elevated levels of CCL5, a chemokine that induces the recruitment of immune cells24. The increased production of CCL5 in Nlrp6−/− mice was attributed to host responses to colitogenic bacteria residing in Nlrp6−/− mice since CCL5 levels were upregulated in WT mice upon cohousing with Nlrp6−/− mice. Despite the increased relative abundance of Prevotellaceae in CCL5−/− mice upon cohousing with Nlrp6−/− mice, cohoused CCL5−/− mice were resistant to exacerbated DSS-induced colitis, suggesting that CCL5 contributes to worsening of DSS-induced colitis upon transfer of colitogenic bacteria. Similarly, increased susceptibility to colitis-associated tumors in WT mice cohoused with Nlrp6−/− mice was abrogated in cohoused CCL5−/− mice49. In addition to CCL5, the production of IL-6 by the epithelium also contributed to increased colitis-associated tumorigenesis49. In the case of NAFLD, susceptibility to liver inflammation and steatosis was mediated by TLR signaling pathways since Myd88/TRIF-doubly deficient mice cohoused with Asc−/− mice had decreased severity of liver disease compared to WT mice cohoused with Asc−/− mice. Although live bacteria were not observed in the liver, levels of TLR4 and TLR9 agonists were significantly increased in the portal circulation of WT mice cohoused with Asc−/− mice and fed the methionine-choline-deficient diet, suggesting that PAMPs from proinflammatory bacteria enriched in the gut of Asc−/− mice can contribute to systemic disease46. It is also important to note that the colitogenic effects of bacteria that are capable of promoting inflammatory cytokine production may only be realized in the context of a breached epithelial barrier together with defective repair mechanisms as inflammasome-deficient mice do not develop frank spontaneous colitis. In addition, inflammatory cytokines including TNFα and IL-6, which can be induced by bacterial stimulation of PRRs, are still important for epithelial repair and homeostasis50, 51, but are detrimental when produced beyond a certain threshold in the context of chronic, sustained inflammation. Altogether, these results suggest that deficiencies in inflammasome signaling result in perturbations in the gut microbiota that lead to the accumulation of bacteria capable of exacerbating pro-inflammatory responses in the appropriate context that predispose to inflammation-related diseases including colitis, tumorigenesis, and metabolic syndrome.
V. Mechanisms of inflammasome regulation of the microbiota: the IL-18/AMP axis
The mechanism by which inflammasome signaling influences the composition of the gut microbiota remains unclear. However, a common theme that has emerged is a predominant role for IL-18 in modulating the microbiome (Figure 1). The increased susceptibility of colitis in Asc−/−, Nlrp6−/−, Nlrp1−/−, Nlrp3−/−, Aim2−/− or caspase-1−/− mice that consistently exhibit decreased levels of IL-18 as opposed to IL-1β. IL-18−/− mice also have increased severity to DSS-induced colitis and colitis-associated tumorigenesis compared to WT mice41. Elinav et al. also determined that WT mice cohoused with IL-18−/− mice, but not IL-1β−/− or IL-1R−/− mice developed increased sensitivity to DSS colitis, suggesting that similar to deficiencies in other inflammasome components, IL-18-deficiency results in alterations in the gut microbiota that can be transmitted to WT mice and affect colitis severity24.
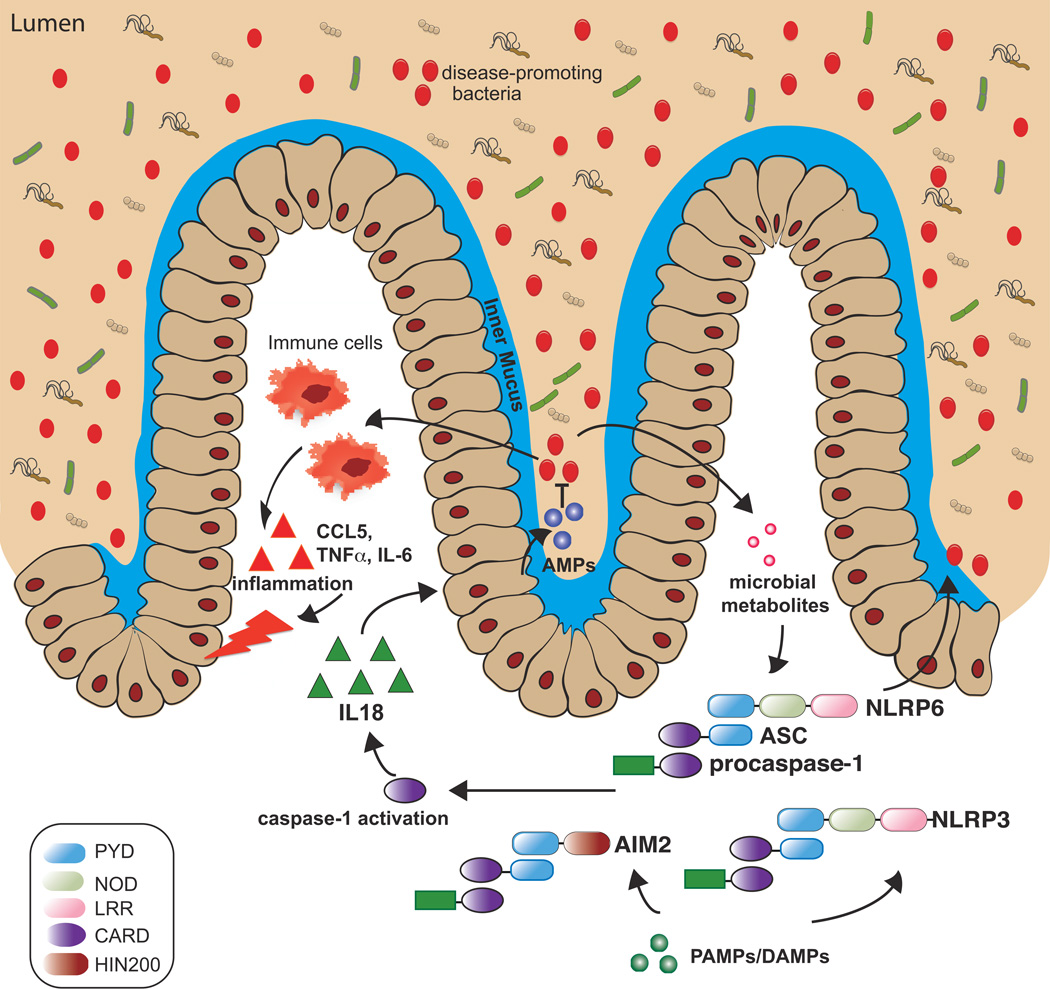
Figure 1. Inflammasome activity affects the composition of the microbiome in part via IL-18 signaling.
Activation of PRRs that participate in inflammasome assembly such as NLRP6, NLRP3, and AIM2 by their respective signals results in caspase-1 activation followed by production of mature, biologically active IL-18. IL-18, in turn, upregulates the production of antimicrobial peptide (AMP), which can affect the relative abundance of bacterial populations through its bacteriocidal activity. In the absence of inflammasome signaling, dysbiosis develops with the accumulation of disease-promoting bacteria (red dots) that can upregulate inflammatory responses such as the production of CCL5, IL-6 and TNFα that can cause inflammatory disease.
Besides inducing Th1 responses through the upregulation of IFNγ, IL-18 also upregulates the production of anti-microbial peptides (AMPs), which mediate bacterial clearance26. AMPs are expressed by the epithelium consistent with high expression levels of IL-1824, 25. Decreased levels of specific AMPs have been observed in Asc−/−, caspase-1−/−, AIM2−/− or Nlrp6−/− mice compared to WT mice, and administration of recombinant IL-18 restored levels of specific AMPs26,38. Notably, Nlrp6−/−, Asc−/− or IL-18−/− mice had decreased levels of the AMPS, Ang1, Ang4, Relnβ, and Itln1, while Aim2−/− had decreased production of Reg3γ and Reg3β, and β-defensin212, 26, 38. In addition, colon crypt secretions obtained from Nlrp3−/− mice ex vivo had decreased bactericidal activity against E. coli in vitro compared to that from WT mice48. That AMP production may contribute to the abundance of certain bacterial populations in inflammasome-deficient mice was further suggested by experiments in which the injection of Ang4 into Asc−/− mice resulted in changes in overall diversity and community structure of the gut microbiome although the resultant microbiome remained significantly different from WT mice26. In addition, injection of IL-18 into Aim2−/− mice reduced the relative abundance of E. coli, which was significantly increased in Aim2−/− mice compared to WT mice. Although AMPs can exhibit species-selective bacteriocidal activity52, it is not clear whether impaired AMPs entirely explains changes in abundance of specific bacterial populations, and it possible that IL-18 regulates other processes than the microbiome. Also of note, the fecal microbiome of IL-18−/− mice was different from that of Asc−/− or Nlrp6−/− mice suggesting that additional mechanisms independent of IL-18 may contribute to microbiome changes in Asc−/− or Nlrp6−/− mice. For example, regulation of mucus secretion by NLRP6 may theoretically affect the abundance of bacteria that can utilize mucin oligosaccharides for nutrition.
VI. Conclusion
Significant advances have been made in understanding the influence of the immune system on the composition of the gut microbiota. Studies involving the natural colonization of GF WT and mutant mice have provided the strongest evidence that inflammasome signaling, in particular, through NLRP6, can dictate the composition of the gut microbiota with respect to certain bacterial populations. Additional studies looking at littermate mice and GF mice will help determine more rigorously whether other inflammasome components truly affect microbiome composition that include not only bacteria, but also viruses and fungi. Similarly, whether inflammasome deficiency results in changes in specific microbial populations that directly promote or cause disease also needs to be further clarified. Unlike AIM2 that has been shown to directly bind to cytosolic double-stranded DNA, there is no conclusive evidence as yet that inflammasome-associated PRRs interact directly with their activating microbial or endogenous molecules. A better understanding of the precise signals that activate each of the inflammasomes to regulate microbiome composition would be important for developing therapeutic strategies to prevent the development of dysbiosis and disease.
Highlights.
Inflammasomes regulate the production of IL-1β and IL-18.
Inflammasome deficiency results in increased susceptibility to colitis in mice.
Impaired inflammasome activity and IL-18 production are associated with dysbiosis.
IL-18-mediated antimicrobial peptide production may regulate the gut microbiome.
Acknowledgments
The author apologizes for any work not cited due to space constraints. GYC has been supported by the National Institutes of Health (NIH) grant R01CA166879, R21CA205636, and R21CA19144 as well as an American Cancer Society Research Scholar Grant.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 2.Fernandes-Alnemri T, Wu J, Yu JW, Datta P, Miller B, Jankowski W, et al. The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 2007;14:1590–1604. doi: 10.1038/sj.cdd.4402194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broz P, Dixit VM. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol. 2016;16:407–420. doi: 10.1038/nri.2016.58. [DOI] [PubMed] [Google Scholar]
- 4.Franchi L, Amer A, Body-Malapel M, Kanneganti TD, Ozoren N, Jagirdar R, et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat Immunol. 2006;7:576–582. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- 5.Kanneganti TD, Ozoren N, Body-Malapel M, Amer A, Park JH, Franchi L, et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006;440:233–236. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- 6.Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 7.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu JW, Wu J, Zhang Z, Datta P, Ibrahimi I, Taniguchi S, et al. Cryopyrin and pyrin activate caspase-1, but not NF-kappaB, via ASC oligomerization. Cell Death Differ. 2006;13:236–249. doi: 10.1038/sj.cdd.4401734. [DOI] [PubMed] [Google Scholar]
- 10.Xu H, Yang J, Gao W, Li L, Li P, Zhang L, et al. Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature. 2014;513:237–241. doi: 10.1038/nature13449. [DOI] [PubMed] [Google Scholar]
- 11.Boyden ED, Dietrich WF. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat Genet. 2006;38:240–244. doi: 10.1038/ng1724. [DOI] [PubMed] [Google Scholar]
- 12.Ratsimandresy RA, Indramohan M, Dorfleutner A, Stehlik C. The AIM2 inflammasome is a central regulator of intestinal homeostasis through the IL-18/IL-22/STAT3 pathway. Cell Mol Immunol. 2016 doi: 10.1038/cmi.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jakobs C, Perner S, Hornung V. AIM2 Drives Joint Inflammation in a Self-DNA Triggered Model of Chronic Polyarthritis. PLoS One. 2015;10:e0131702. doi: 10.1371/journal.pone.0131702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Man SM, Karki R, Malireddi RK, Neale G, Vogel P, Yamamoto M, et al. The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat Immunol. 2015;16:467–475. doi: 10.1038/ni.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Storek KM, Gertsvolf NA, Ohlson MB, Monack DM. cGAS and Ifi204 cooperate to produce type I IFNs in response to Francisella infection. J Immunol. 2015;194:3236–3245. doi: 10.4049/jimmunol.1402764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mariathasan S, Weiss DS, Newton K, McBride J, O'Rourke K, Roose-Girma M, et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 18.Martinon F, Petrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 19.Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dostert C, Petrilli V, Van Bruggen R, Steele C, Mossman BT, Tschopp J. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science. 2008;320:674–677. doi: 10.1126/science.1156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munoz-Planillo R, Kuffa P, Martinez-Colon G, Smith BL, Rajendiran TM, Nunez G. K(+) efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity. 2013;38:1142–1153. doi: 10.1016/j.immuni.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimada K, Crother TR, Karlin J, Dagvadorj J, Chiba N, Chen S, et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012;36:401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grenier JM, Wang L, Manji GA, Huang WJ, Al-Garawi A, Kelly R, et al. Functional screening of five PYPAF family members identifies PYPAF5 as a novel regulator of NF-kappaB and caspase-1. FEBS Lett. 2002;530:73–78. doi: 10.1016/s0014-5793(02)03416-6. [DOI] [PubMed] [Google Scholar]
- 24.Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen GY, Liu M, Wang F, Bertin J, Nunez G. A functional role for Nlrp6 in intestinal inflammation and tumorigenesis. J Immunol. 2011;186:7187–7194. doi: 10.4049/jimmunol.1100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levy M, Thaiss CA, Zeevi D, Dohnalova L, Zilberman-Schapira G, Mahdi JA, et al. Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling. Cell. 2015;163:1428–1443. doi: 10.1016/j.cell.2015.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Manji GA, Grenier JM, Al-Garawi A, Merriam S, Lora JM, et al. PYPAF7, a novel PYRIN-containing Apaf1-like protein that regulates activation of NF-kappa B and caspase-1-dependent cytokine processing. J Biol Chem. 2002;277:29874–29880. doi: 10.1074/jbc.M203915200. [DOI] [PubMed] [Google Scholar]
- 28.Vladimer GI, Weng D, Paquette SW, Vanaja SK, Rathinam VA, Aune MH, et al. The NLRP12 inflammasome recognizes Yersinia pestis. Immunity. 2012;37:96–107. doi: 10.1016/j.immuni.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anand PK, Malireddi RK, Lukens JR, Vogel P, Bertin J, Lamkanfi M, et al. NLRP6 negatively regulates innate immunity and host defence against bacterial pathogens. Nature. 2012;488:389–393. doi: 10.1038/nature11250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaki MH, Vogel P, Malireddi RK, Body-Malapel M, Anand PK, Bertin J, et al. The NOD-like receptor NLRP12 attenuates colon inflammation and tumorigenesis. Cancer Cell. 2011;20:649–660. doi: 10.1016/j.ccr.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allen IC, Wilson JE, Schneider M, Lich JD, Roberts RA, Arthur JC, et al. NLRP12 suppresses colon inflammation and tumorigenesis through the negative regulation of noncanonical NF-kappaB signaling. Immunity. 2012;36:742–754. doi: 10.1016/j.immuni.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang P, Zhu S, Yang L, Cui S, Pan W, Jackson R, et al. Nlrp6 regulates intestinal antiviral innate immunity. Science. 2015;350:826–830. doi: 10.1126/science.aab3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novick D, Kim S, Kaplanski G, Dinarello CA. Interleukin-18, more than a Th1 cytokine. Semin Immunol. 2013;25:439–448. doi: 10.1016/j.smim.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 34.Joosten LA, Netea MG, Dinarello CA. Interleukin-1beta in innate inflammation, autophagy and immunity. Semin Immunol. 2013;25:416–424. doi: 10.1016/j.smim.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 35.Williams TM, Leeth RA, Rothschild DE, Coutermarsh-Ott SL, McDaniel DK, Simmons AE, et al. The NLRP1 inflammasome attenuates colitis and colitis-associated tumorigenesis. J Immunol. 2015;194:3369–3380. doi: 10.4049/jimmunol.1402098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allen IC, TeKippe EM, Woodford RM, Uronis JM, Holl EK, Rogers AB, et al. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J Exp Med. 2010;207:1045–1056. doi: 10.1084/jem.20100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zaki MH, Boyd KL, Vogel P, Kastan MB, Lamkanfi M, Kanneganti TD. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity. 2010;32:379–391. doi: 10.1016/j.immuni.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu S, Peng L, Kwak YT, Tekippe EM, Pasare C, Malter JS, et al. The DNA Sensor AIM2 Maintains Intestinal Homeostasis via Regulation of Epithelial Antimicrobial Host Defense. Cell Rep. 2015;13:1922–1936. doi: 10.1016/j.celrep.2015.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Normand S, Delanoye-Crespin A, Bressenot A, Huot L, Grandjean T, Peyrin-Biroulet L, et al. Nod-like receptor pyrin domain-containing protein 6 (NLRP6) controls epithelial self-renewal and colorectal carcinogenesis upon injury. Proc Natl Acad Sci U S A. 2011;108:9601–9606. doi: 10.1073/pnas.1100981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zaki MH, Vogel P, Body-Malapel M, Lamkanfi M, Kanneganti TD. IL-18 production downstream of the Nlrp3 inflammasome confers protection against colorectal tumor formation. J Immunol. 2010;185:4912–4920. doi: 10.4049/jimmunol.1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salcedo R, Worschech A, Cardone M, Jones Y, Gyulai Z, Dai RM, et al. MyD88-mediated signaling prevents development of adenocarcinomas of the colon: role of interleukin 18. J Exp Med. 2010;207:1625–1636. doi: 10.1084/jem.20100199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dupaul-Chicoine J, Yeretssian G, Doiron K, Bergstrom KS, McIntire CR, LeBlanc PM, et al. Control of intestinal homeostasis, colitis, and colitis-associated colorectal cancer by the inflammatory caspases. Immunity. 2010;32:367–378. doi: 10.1016/j.immuni.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 43.Seregin SS, Golovchenko N, Schaf B, Chen J, Eaton KA, Chen GY. NLRP6 function in inflammatory monocytes reduces susceptibility to chemically induced intestinal injury. Mucosal Immunol. 2016 doi: 10.1038/mi.2016.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wlodarska M, Thaiss CA, Nowarski R, Henao-Mejia J, Zhang JP, Brown EM, et al. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell. 2014;156:1045–1059. doi: 10.1016/j.cell.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Birchenough GM, Nystrom EE, Johansson ME, Hansson GC. A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science. 2016;352:1535–1542. doi: 10.1126/science.aaf7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Man SM, Zhu Q, Zhu L, Liu Z, Karki R, Malik A, et al. Critical Role for the DNA Sensor AIM2 in Stem Cell Proliferation and Cancer. Cell. 2015;162:45–58. doi: 10.1016/j.cell.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirota SA, Ng J, Lueng A, Khajah M, Parhar K, Li Y, et al. NLRP3 inflammasome plays a key role in the regulation of intestinal homeostasis. Inflamm Bowel Dis. 2011;17:1359–1372. doi: 10.1002/ibd.21478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu B, Elinav E, Huber S, Strowig T, Hao L, Hafemann A, et al. Microbiota-induced activation of epithelial IL-6 signaling links inflammasome-driven inflammation with transmissible cancer. Proc Natl Acad Sci U S A. 2013;110:9862–9867. doi: 10.1073/pnas.1307575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 51.Zhan Y, Chen PJ, Sadler WD, Wang F, Poe S, Nunez G, et al. Gut microbiota protects against gastrointestinal tumorigenesis caused by epithelial injury. Cancer Res. 2013;73:7199–7210. doi: 10.1158/0008-5472.CAN-13-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat Immunol. 2003;4:269–273. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]