Abstract
Cytokinesis and cell division during pre-implantation embryonic development occur as an orchestrated spatiotemporal program. Cleavage, compaction, and blastulation in pre-implantation embryos are essential for successful implantation and pregnancy. Their alteration is associated with chromosomal imbalance and loss of developmental competence. In this study, we evaluated the time of cleavage and compaction as predictors for in vitro pre- and peri-implantation development and in utero implantation potential by time-lapse monitoring. Mouse 2-cell embryos were collected on 1.5 days post coitum (dpc) and were individually cultured to the outgrowth (OG) stage (7.5 dpc). Developmental stages were classified as 3-cell, 4-cell, 8-cell, morula, blastocyst, and OG. Cut-off times for successful blastocyst development were determined by receiver operating characteristic curve analysis. When cut-off times were set as 9 h for the third cleavage from the 2- to 4-cell stage, and 40 h for compaction from the 2-cell to morula stage, blastocyst and OG development rates, respectively, were significantly higher (P < 0.0001). Embryos were grouped according to the above cut-off time and transferred to the contralateral uterine horn on 3.5 dpc. Implantation rates in utero on 5.5 dpc were significantly higher in early third cleaved (≤ 9 h from 2- to 4-cell) and early compacted embryos (≤ 40 h from 2-cell to morula) than those in delayed embryos (P < 0.05). Therefore, the time of the third cleavage from 2- to the 4-cell stage and compaction from 2-cell to morula stage may be a useful morphokinetic parameter for predicting developmental potential, including successful implantation and pregnancy in human in vitro fertilization-embryo transfer programs.
Keywords: Cleavage, Compaction, Cut-off time, Implantation potential, Time-lapse monitoring
Mammalian embryos develop sequentially during pre-implantation embryogenesis, which starts with fertilization and ends with blastocyst implantation and coincides with dynamic changes in morphology and zygotic gene expression [1, 2]. Human in vitro fertilization-embryo transfer (IVF-ET) techniques are continuously being improved to increase implantation and pregnancy rates with better selection of transfer-competent embryos [3, 4]. Several studies have focused on identifying independent and useful predictors that can be used to select high-quality embryos in IVF laboratories [3,4,5,6,7,8,9,10]. A monitoring system to observe morphology during the course of in vitro pre-implantation embryonic development has been proposed [5,6,7,8,9,10,11]. Pre-implantation embryos with normal morphology may have chromosomal abnormalities, but can nonetheless reach the blastocyst stage. On the other hand, embryos with good morphology, which are chromosomally normal, may fail to differentiate into blastocysts [12]. To achieve a high implantation rate, transferable embryos are selected by considering various parameters beyond morphokinetics, such as the duration of oocyte activation [13], first cell division [14,15,16,17], pronuclear (PN) scoring [18], presence of evenly cleaved blastomeres [19], degree of fragmentation [20, 21], cleavage pattern [9], contraction pattern, and time to hatching [22, 23].
Cytokinesis and cell division during pre-implantation embryonic development occur as a spatiotemporally orchestrated program. [24, 25]. Generally, the time of the first cleavage division from the PN to 2-cell stage is highly variable between embryos within 22–30 h of insemination, whereas that of the second and third cleavages (from the 2- to 4-cell stage) is less variable, and can be detected 32–45 h after insemination [26]. Additionally, the compaction process involves functional changes, with expanding membrane channels serving as intracellular communication pathways [27]. The appearance of cleavage and compaction processes is the most important factor determining successful pre-implantation development [28, 29]. However, there is little information on human pre- and peri-implantation embryos in vivo owing to ethical restrictions. Several studies have shown that the first and second cleavage times are correlated with in vitro developmental competence and implantation in both humans and mice [9, 26, 30]. Many studies have found that transferring early cleaved embryos, rather than those exhibiting delayed development, yields higher implantation and pregnancy rates [31,32,33]. However, it is difficult to obtain conclusive evidence that implantation is dependent on the overall process of cleavage and compaction in pre-implantation development.
An in vitro outgrowth (OG) assay mimics implantation in the uterus, and enables experimental studies on implantation events and mechanisms. This assay has also revealed the relationship between metabolism based on morphokinetics of pre-implantation embryos and implantation potential [34], and has been used as an alternative tool to study the trophoblastic invasion and motility [34,35,36,37].
In this study, we showed that monitoring the cleavage and compaction times with a time-lapse imaging system was advantageous for predicting successful blastocyst development and implantation of mouse embryos in vitro and in utero. We found that the time of the third cleavage to the 4-cell stage and compaction to the morula stage was a useful morphokinetic parameter for predicting the potential of mouse pre-implantation embryos to develop into blastocysts and implant in utero. Our results provide important evidence that determining morphokinetic parameters with real-time monitoring could improve implantation and pregnancy rates in human IVF-ET programs.
Materials and Methods
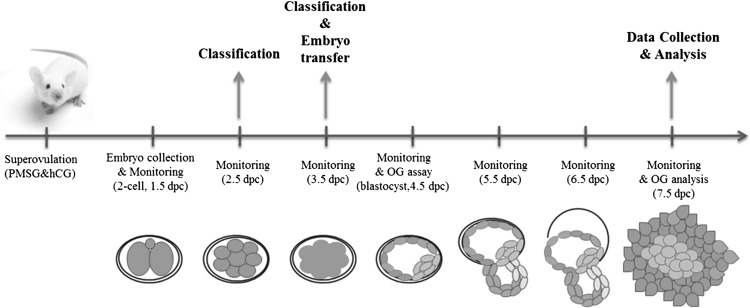
The overall scheme is presented in Fig. 1.
Fig. 1.
Schematic illustration of the experiment for investigating the relationship between cleavage and compaction timings and developmental competence. Mouse 2-cell embryos were cultured to blastocyst and outgrowth stages, and embryonic development was monitored for 7 days (from 1.5 to 7.5 dpc). Data were collected at the end of monitoring (7.5 dpc). Classification, embryo transfer, and outgrowth assay were performed. dpc, days post coitum.
Animal care and embryo collection
Inbred ICR (Institute of Cancer Research) female mice (6–8 weeks old) were induced to superovulate by intraperitoneal injection of 5 IU serum gonadotropin from a pregnant mare (Sigma, St. Louis, MO, USA) followed by injection with 5 IU human chorionic gonadotropin (hCG; Sigma) 46 h later. Superovulated female mice were mated with fertile male mice and euthanized by cervical dislocation 46 h after hCG injection. The day of vaginal plugging was designated as 0.5 days post-coitum (dpc).
Experimental animal protocols were approved by the Eulji University Institutional Animal Care and Use Committee (EUIACUC 12-19).
Embryo collection and time-lapse monitoring of embryo culture to OG stage
Mouse 2-cell embryos were collected from oviducts on 1.5 dpc and cultured in pre-warmed Quinn’s Advantage blastocyst medium (SAGE/Origio, Måløv, Denmark) containing serum protein substitute (SAGE/Origio) in an incubator (Sanyo, Osaka, Japan) at 37°C and 5% CO2. The culture medium was replaced with Dulbecco’s modified Eagle medium (Gibco/Life Technologies, Carlsbad, CA, USA) containing 10% fetal bovine serum on 4.5 dpc for culture to the OG stage. Embryogenesis from the 2-cell to the OG stage on 7.5 dpc was observed using a Primo Vision system (Vitrolife, Gothenburg, Sweden) (Fig. 2). The digital camera for the time-lapse microscope was set to acquire a single image every 30 min for 6 days. Stages of embryo development were classified as 3-cell (3C), 4-cell (4C), 8-cell (8C), compacted morula (Mo), early blastocyst (EB), late blastocyst (LB), hatching blastocyst (HB), hatched blastocyst (Hed BL), and OG.
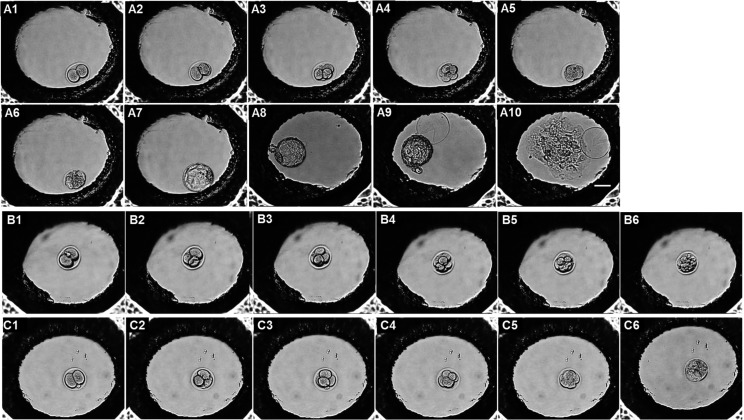
Fig. 2.
Time-lapse monitoring of embryos during development. Mouse 2-cell embryos from 1.5 dpc were cultured to the outgrowth (OG) stage (7.5 dpc) in a Primo Vision monitoring system, with images captured every 30 min. (A1–10) The developmental stages were 2-cell, 3-cell, 4-cell, 8-cell, morula, early blastocyst, late blastocyst, hatching blastocyst, hatched blastocyst, and OG. (B, C) Abnormal patterns of embryo development. (B1–6) Embryo arrested at the cleavage stage. (C1–6) Embryo that failed to undergo OG expansion. dpc, days post coitum.
Grouping analysis based on the time of each development stage
Mouse 2-cell embryos were collected on 1.5 dpc and were individually cultured and monitored up to the OG stage (7.5 dpc) by time-lapse imaging. Embryos were classified into success and failure groups based on their development to the blastocyst and OG stages on 4.5 and 7.5 dpc, respectively. Mean cumulative time and time interval between cell divisions was compared between embryos of the success and failure groups, whether or not they developed to the blastocyst and OG stages. Blastocyst development and OG rates were compared with respect to cut-off times determined by the receiver operating characteristic (ROC) curve analysis.
ROC curve analysis
ROC curves were generated using the ROCR library of the R programming language. Linear models were generated using the lm function of R. Area under the ROC curve (AUC) was estimated to assess the ability of sensitivity and specificity (%) to predict development to blastocyst and OG stages; that is, a greater AUC signified superior predictive capacity of the model.
Quantitative analysis of mRNA expression in embryos
We collected cleavage stage embryos (3C, 4C, 8C, and Mo) for quantitative real-time (RT)-PCR analysis. Total mRNA (10 μl) was isolated from five embryos, using an mRNA Direct kit (Dynal Biotech ASA, Oslo, Norway), and 80 μl cDNA was synthesized from 2 μl total mRNA using the PrimeScript 1st strand cDNA Synthesis kit (Takara Bio, Otsu, Japan). A fraction of the reverse-transcribed product (2 μl) was used directly for quantitative RT-PCR, which was carried out in a final reaction volume of 20 μl with SYBR Green (Applied Biosystems, Foster City, CA, USA) and primers amplifying cytokinesis-related genes such as the actin-binding protein anillin (Anln) and Ras homolog gene family member A (RhoA) and pluripotency markers such as octamer-binding transcription factor (Oct)-4 and caudal type homeobox (Cdx)-2 (Table 1). The internal standard for normalization was 16SrRNA. Expression levels were determined using a modified version of the 2−ΔΔCt method [38]. Experiments were performed at least three times.
Table 1. List of primers used for quantiative RT-PCR and expected product sizes.
| Gene | GenBank accession number | Primer sequences (5'–3') | Product size (bp) |
| Anln | NM_028390.3 | F: TGGGGCTGAGCAGATGGTCG | 274 |
| R: TCCGGGACTGGCCATAACTGAAGA | |||
| RhoA | NM_016802.4 | F: CATTGACAGCCCTGATAGTT | 120 |
| R: TCGTCATTCCGAAGGTCCTT | |||
| Oct-4 | NM_013633.3 | F: TCAGGTTGGACTGGGCCTAGT | 100 |
| R: GGAGGTTCCCTCTGAGTTGCTT | |||
| Cdx-2 | NM_007673.3 | F: CGCAGAACTTTGTCAGTCCTCCGCAGTACC | 254 |
| R: GTATTCGGCGGGGCTGCTGTAGCCCATAGC | |||
| 16S rRNA | XM_003688749.3 | F: AGATGATGCAGCCGCGC | 163 |
| R: GCTACCAGGGCCTTTGAGATGGA |
Analysis of in utero implantation after embryo transfer
Implantation rates after embryo transfer were assessed within the cut-off times described above. Embryos were divided into early and delayed groups according to the third cleavage at 9 h from 2C as the 3C and 4C groups, and into another group according to compaction at 40 h from 2C as the 8C and Mo groups. Embryos were transferred on 3.5 dpc into the contralateral uterine horn of pseudopregnant female mice obtained by mating with a vasectomized male. After embryo transfer 2 days later on 5.5 dpc, implantation sites in the uterine horns of pregnant mice were detected by intravenous injection of Chicago Blue dye. Clear blue bands in utero were considered sites of successful implantation.
Statistical analysis
All experiments were performed at least in triplicate. The statistical significance of two-group comparisons was evaluated by the Student’s t test or Fisher’s exact test, whereas for multiple comparisons, one-way analysis of variance or a χ2 test was performed. P values < 0.05 were considered statistically significant.
Results
Differences in mean times of each developmental stage are related to the potential for development to the blastocyst stage
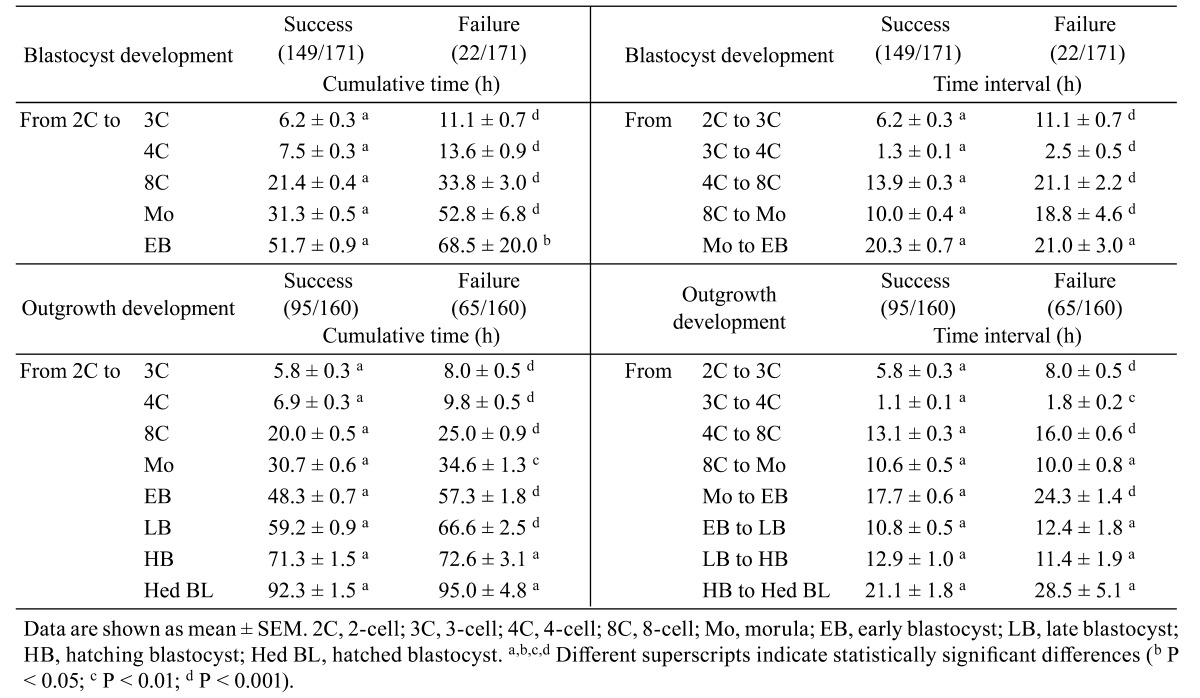
Mouse 2-cell embryos were cultured to the OG stage for 6 days (from 1.5 to 7.5 dpc), and developmental stages were classified as 3C, 4C, 8C, Mo, EB, LB, HB, Hed BL, and OG, as shown in Fig. 2. Cultured embryos developed to blastocyst (87.1%, 149/171) and OG (53.2%, 91/171) stages. Mean cumulative times of each stage were shorter in embryos of the group with successful blastocyst development as compared to that in the failure group (Table 2 ). Mean time intervals between cell divisions to the blastocyst stage, except for Mo and EB, were significantly shorter than those in the failure group (P < 0.001).
Differences in mean times of each developmental stage are related to the potential for development to the OG stage
The mean cumulative times and time intervals to reach the OG stage on 7.5 dpc from each developmental stage were compared between embryos of the success and failure groups (Table 2). Mean cumulative times for each developmental stage to the blastocyst stage were significantly shorter than those for embryos in the failure group (P < 0.01). On the other hand, mean cumulative times after hatching were similar between the two groups (P > 0.05). The mean time intervals between 2C to 3C, 3C to 4C, 4C to 8C, and Mo to EB were significantly shorter than those in the group that failed to reach OG stage (P < 0.01).
Table 2. Mean times for each developmental stage with respect to developmental potential to blastocyst and outgrowth stages.
Sensitivity and specificity of developmental potential to reach blastocyst stage determined based on the time intervals of each development stage
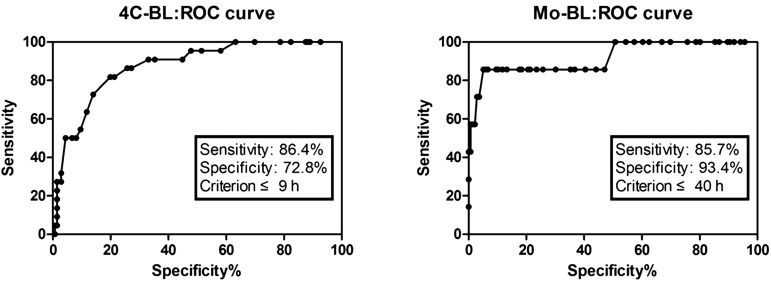
The cut-off time for the highest sensitivity and specificity at each developmental stage was set according to the ROC analysis. When the cut-off time was set as 9 h from 2C to 4C, sensitivity and specificity of blastocyst development were 86.4% and 72.8%, respectively. A cut-off time of 40 h from 2C to Mo showed the highest sensitivity and specificity of blastocyst development, 85.7% and 93.4%, respectively (Fig. 3).
Fig. 3.
ROC curve analysis of sensitivity and specificity of developmental potential to the blastocyst stage according to cut-off times from 2- to 4-cell and morula stages. Cut-off values corresponding to true-positive and false-negative rates are shown on the curve. To predict blastocyst development: AUCROC for sensitivity score = 86.4; AUCROC for specificity score = 72.8 within 9 h from 2- to 4-cell stage; AUCROC for sensitivity score = 85.7; AUCROC for specificity score = 93.4 within 40 h from 2-cell to morula stage.
Prediction of potential for developing to the blastocyst stage within 9 and 40 h of the set cut-off time
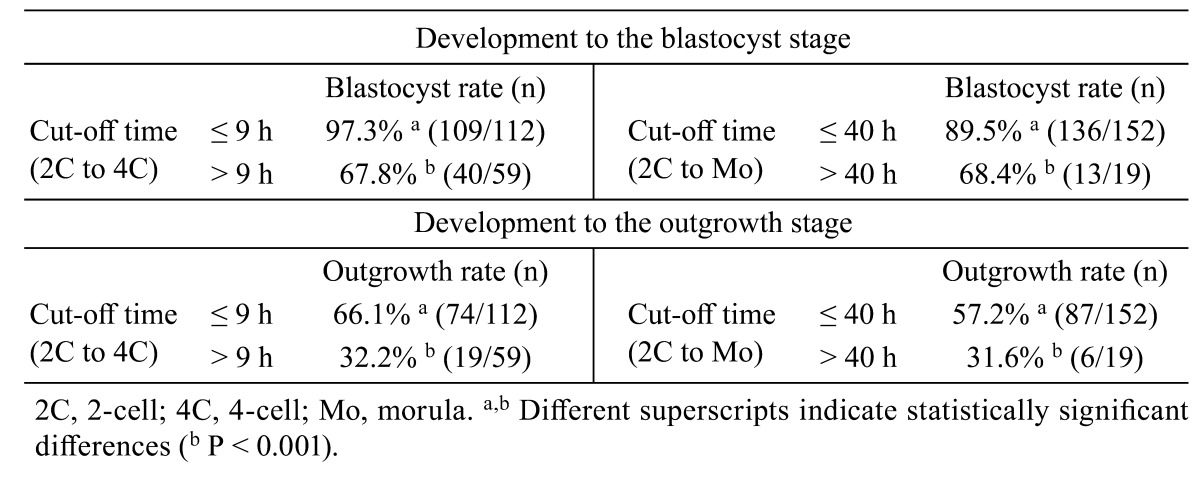
All monitored embryos were grouped according to the above cut-off times of 9 h and 40 h. We found that 97.3% (109/112) of embryos taking ≤ 9 h from 2C to 4C developed into blastocysts, whereas only 67.8% (40/59) of those taking > 9 h from 2C to 4C successfully developed (P < 0.001). In addition, 89.5% (136/152) of embryos taking ≤ 40 h from 2C to Mo developed to blastocysts whereas 68.4% (13/19) of those taking > 40 h successfully developed (P < 0.001, Table 3). When applying the above cut-off times, OG rates were significantly higher in early cleaved and compacted embryos than in delayed embryos (Table 3).
Table 3. Developmental potential to the blastocyst and outgrowth stages according to cut-off times from 2-to 4-cell and morula stages.
Gene expression within the cut-off times for cleavage and compaction events
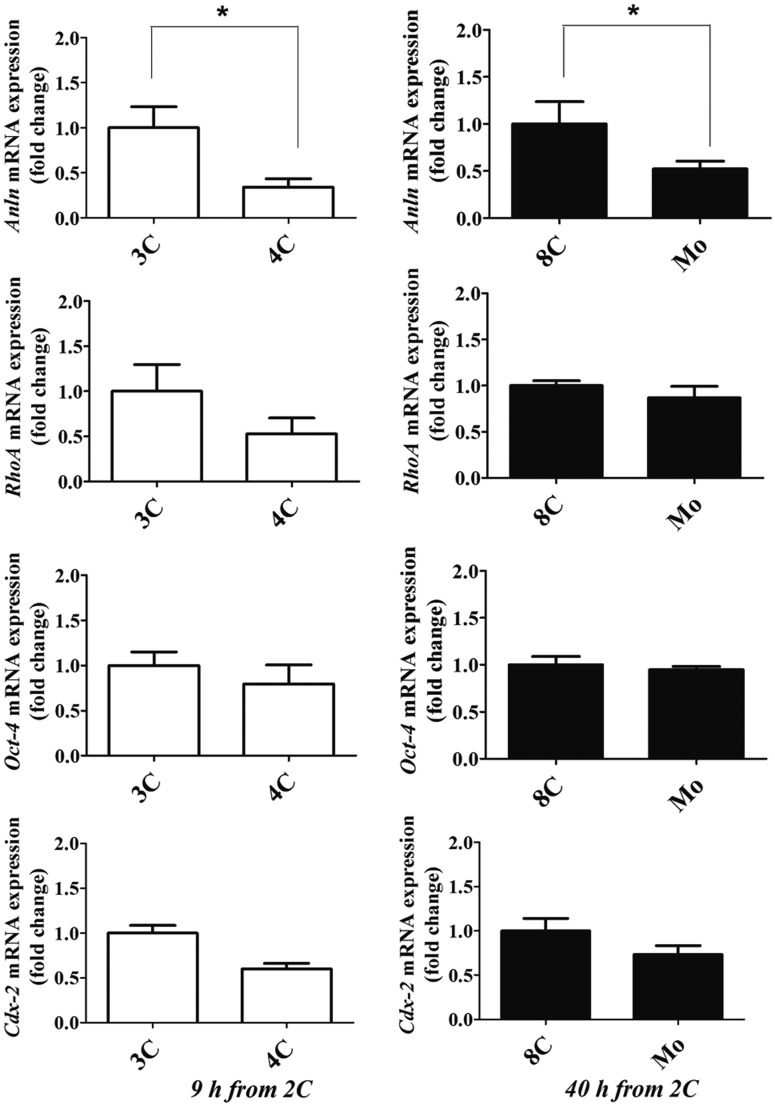
The mRNAs levels of the cytokinesis-related genes Anln and RhoA, the pluripotency marker Oct-4, and the trophoblast marker Cdx-2 were determined by quantitative RT-PCR. Embryos were divided into two groups according to the cut-off times of 9 h and 40 h from 2C to 4C and Mo, respectively (Fig. 4). Embryos taking ≤ 9 h from 2C to 4C showed lower Anln expression than those taking > 9 h from 2C to 4C. In addition, embryos that underwent compaction within 40 h from 2C to Mo expressed a lower level of Anln mRNA than those that compacted after > 40 h from 2C to Mo (P < 0.01, Fig. 5). There were no differences in other genes among groups.
Fig. 4.
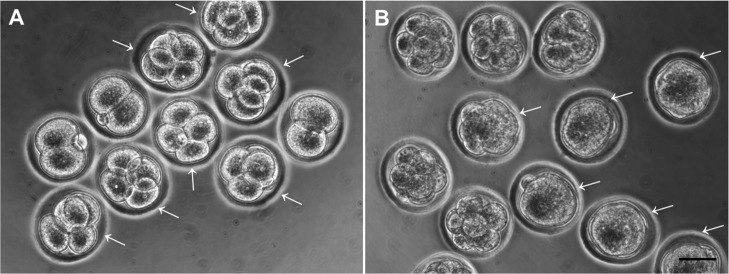
Embryos at each time point of cleavage and compaction events. Mouse 2-cell embryos were cultured and classified as early-developed or delayed embryos at 9 h from 2-cell (A) and 40 h from 2-cell (B). Arrows indicate early cleaved or compacted embryos. Scale bars = 50 µm.
Fig. 5.
Gene expression at each time point of cleavage and compaction events. Mouse 2-cell embryos were cultured and classified as 3- or 4-cell at 9 h from 2-cell, and 8-cell and morula at 40 h from 2-cell. mRNA expression of Anln, RhoA, Oct-4, and Cdx-2 was compared between early and delayed embryos within each cut-off time. Data are shown as mean ± SEM. * P < 0.05.
Enhanced implantation rates in utero based on cut-off times of third cleavage and compaction events
Embryos were divided into two groups (from 2C to 3C or 4C at 9 h) and two other groups (from 2C to 8C or Mo at 40 h) based on ROC curve analysis. On 3.5 dpc, embryos from each group were transferred to the contralateral uterine horn of pseudopregnant females. Implantation sites were detected by intravenous injection of Chicago blue dye on 5.5 dpc. Implantation rates were higher for transferred embryos with a cut-off time of ≤ 9 h from 2C to 4C than for those with a cut-off time of > 9 h (78.6 ± 8.7% vs. 42.9 ± 14.0%, P < 0.01). In addition, implantation rates were higher for transferred embryos with a cut-off time of ≤ 40 h from 2C to Mo compared to those with a cut-off time of > 40 h (54.2 ± 9.3% vs. 33.3 ± 6.3%, P < 0.01) (Table 4).
Table 4. Implantation rates in utero after embryo transfer.
| Group of transferred embryos | 9 h from 2C |
40 h from 2C |
||
| 3C | 4C | 8C | Mo | |
| No. of transferred embryos | 42 | 42 | 48 | 48 |
| No. of implanted embryos (mean number) | 18 (2.6 ± 0.8) a | 33 (4.7 ± 0.5) b | 16 (2.0 ± 0.4) a | 26 (3.3 ± 0.6) b |
| Implantation rate | 42.9 ± 14.0% a | 78.6 ± 8.7% b | 33.3 ± 6.3% a | 54.2 ± 9.3% b |
Data are shown as mean ± SEM. 2C, 2-cell; 3C, 3-cell; 4C, 4-cell; 8C. 8-cell; Mo, morula. Embryos were classified as 3C and 4C at 9 h from 2C, respectively, and as 8C and Mo at 40 h from 2C, then transferred to the contralateral uterine horn on 3.5 dpc. a,b Different superscripts indicate statistical differences (b P < 0.05).
Discussion
Embryo selection based on morphological grading and biochemical markers is not always associated with higher implantation rates. High-quality blastocysts lead to high pregnancy and implantation rates, but some still do not implant into the uterus; this is related to embryonic biochemical factors such as O2 consumption [39, 40], pyruvate and glucose uptake [34, 41], lactate production [42], the secretion of factors such as platelet-activating factor and insulin-like growth factor [43], and the activity of the enzymes involved in acid and carbohydrate metabolism pathways [44, 45]. However, measuring these parameters typically requires special instruments and training, limiting their practical application. Therefore, more efficient methods for assessing embryo viability and development competence based on alternative criteria are needed.
Some researchers have proposed PN morphological scoring to select viable embryos for transfer in human assisted reproductive technology (ART) programs. PN morphology has a significant effect on embryo developmental potential in vitro, which therefore affects day 3 and 5 morphological scores and the ability of these embryos to develop to the blastocyst stage [18, 46]. However, one study reported that PN scoring is not correlated with implantation and pregnancy rates [47]. As such, the question of which markers are useful for selecting good-quality embryos remains controversial.
Early cleavage of pre-implantation embryos has been suggested as an additional criterion for embryo selection, as it has been found to be correlated with implantation potential and enhanced viability compared to embryos that exhibit delayed cleavage [5, 15, 48, 49]. The time of the first cleavage alters gene expression associated with implantation potential, and slowly cleaving embryos exhibit reduced viability, chromosomal abnormalities, and decreased oxygen consumption at the blastocyst stage, thereby lowering hatching and pregnancy rates [50, 51]. A higher mitochondrial DNA copy number is also associated with a higher rate of zygotic cleavage in humans and pigs [52, 53]. In addition, chromosomal imbalances, such as aneuploidy, frequently occur in delayed-cleaved embryos that comprise unequal blastomeres [19, 54]; the first cleavage from PN to 2C may be affected by intrinsic factors within the oocyte, the sperm, or both [26].
Compaction is an important event during pre-implantation embryo development, and embryos with apparently normal cleavage do not always reach morula and blastocyst stages at the appropriate time. When embryos are compacted at the 4-cell stage, they may miss the chance to develop further, which requires differentiation into the inner cell mass and trophectoderm [12]. Partial compaction is also a negative indicator of developmental competence and pregnancy rate [55]. Embryos that undergo compaction and cavitation become distinguishable and reflect embryo commitment to the next stage of development [56]. It has been reported that embryo quality on day 2 may be linked to the ability to undergo compaction on day 3 [57].
Here, we compared the mean times of cleavage and compaction between embryos that successfully reached the blastocyst stage and those that failed to do so. Embryo with shorter mean cleavage and compaction times showed increased development and higher outgrowth rates compared to embryos with longer times. These results demonstrate that embryos with early third cleavage and compaction events had higher potential for development to the blastocyst stage and consequent implantation in vitro and in utero.
ROC curve analysis is a well-established method for identifying precise diagnostic points. Sensitivity and specificity are key parameters used to determine the accuracy and efficiency of diagnostic tests [58, 59]. In the context of the present study, sensitivity was defined as the proportion of embryos that developed successfully to the blastocyst and OG stages, whereas specificity was defined as the proportion of embryos that failed to reach these stages. We predicted blastocyst developmental competence and implantation potential based on ROC analysis. Embryos that underwent third cleavage division and compaction within the cut-off time had the highest sensitivity (86.4%) and specificity (93.4%) for blastocyst development. Although we predicted a high rate of OG development, there was no meaningful data in the ROC analysis; that is, several cut-off times were unsatisfactory for establishing OG potential. Therefore, there is still room for improvement in this regard; further studies with larger sample sizes are needed to optimize specificity and sensitivity.
With the exception of Anln gene expression, the expression of other genes examined in the different embryo groups was similar regardless of cut-off time. Anln is implicated in cytoskeletal dynamics during cellularization and cytokinesis and interacts with RhoA, which regulates cytoskeletal dynamics, cell cycle progression, and cell transformation [60]. Anln, which is a maternal transcript, is highly expressed at the zygote stage, with mRNA expression declining during development to blastocyst stage [9, 61]. Oct-4 is a key marker of the pluripotency regulatory network involved in the self-renewal of undifferentiated embryonic stem cells via a reciprocal interaction with Cdx-2 [62]. We also analyzed other pluripotency genes such as Nanog, Sox-2, and c-Myc, and found that these genes showed similar expression patterns regardless of cut-off time (data not shown). In this study, only the Anln level differed between embryos with early and delayed development due to the shortened cleavage interval. Cleavage and compaction times may be related to cytokinesis-related gene expression in pre-implantation embryos [24]. We did not compare the mRNA expression in early and delayed groups when they reached to same stage. Further investigation will be required to analyze mRNA expression in blastocysts from early and delayed groups.
In embryo transfer experiments, we obtained the highest implantation rate (78.6 ± 8.7%) in embryos with a cut-off time of ≤ 9 h from 2C to 4C. We also found that Mo at ≤ 40 h from 2C had higher implantation potential in utero compared to the potential of embryos at 8C. Our results suggest that embryos with third cleavage at 9 h and compaction at 40 h not only have higher probability of developing into blastocysts, but also higher implantation potential. It is well-known that embryos within appropriate times of the third cleavage and compaction exhibit better developmental competence than those that undergo late cleavage and delayed compaction. Indeed, transferrable 4-cell embryos with uniform blastomeres were manually selected 2 days after insemination in human IVF-ET programs [63,64,65].
In conclusion, our study showed that the cumulative time from 2C to 4C and from 2C to Mo predicts blastocyst development and implantation potential in utero. Specifically, embryos exhibiting short third cleavage intervals had a higher probability of blastocyst formation, OG, and implantation in utero. In addition, the compaction time was related to further embryonic developmental competence and implantation potential. Our results provide evidence that analyzing morphokinetic parameters by real-time monitoring may improve the efficacy of selection of transferrable embryos with high implantation potential in human IVF-ET programs.
Conflict of interest
The authors declare that they have no conflicts of interest.
Acknowledgments
This project was financially supported by a Korea Healthcare Technology R&D Project grant from the Ministry of Health and Welfare (A120043).
References
- 1.Niakan KK, Han J, Pedersen RA, Simon C, Pera RA. Human pre-implantation embryo development. Development 2012; 139: 829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zernicka-Goetz M, Morris SA, Bruce AW. Making a firm decision: multifaceted regulation of cell fate in the early mouse embryo. Nat Rev Genet 2009; 10: 467–477. [DOI] [PubMed] [Google Scholar]
- 3.Yang Z, Zhang J, Salem SA, Liu X, Kuang Y, Salem RD, Liu J. Selection of competent blastocysts for transfer by combining time-lapse monitoring and array CGH testing for patients undergoing preimplantation genetic screening: a prospective study with sibling oocytes. BMC Med Genomics 2014; 7: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mansour R, Ishihara O, Adamson GD, Dyer S, de Mouzon J, Nygren KG, Sullivan E, Zegers-Hochschild F. International Committee for Monitoring Assisted Reproductive Technologies world report: Assisted Reproductive Technology 2006. Hum Reprod 2014; 29: 1536–1551. [DOI] [PubMed] [Google Scholar]
- 5.Nakahara T, Iwase A, Goto M, Harata T, Suzuki M, Ienaga M, Kobayashi H, Takikawa S, Manabe S, Kikkawa F, Ando H. Evaluation of the safety of time-lapse observations for human embryos. J Assist Reprod Genet 2010; 27: 93–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pribenszky C, Mátyás S, Kovács P, Losonczi E, Zádori J, Vajta G. Pregnancy achieved by transfer of a single blastocyst selected by time-lapse monitoring. Reprod Biomed Online 2010; 21: 533–536. [DOI] [PubMed] [Google Scholar]
- 7.Pribenszky C, Losonczi E, Molnár M, Lang Z, Mátyás S, Rajczy K, Molnár K, Kovács P, Nagy P, Conceicao J, Vajta G. Prediction of in-vitro developmental competence of early cleavage-stage mouse embryos with compact time-lapse equipment. Reprod Biomed Online 2010; 20: 371–379. [DOI] [PubMed] [Google Scholar]
- 8.Meseguer M, Rubio I, Cruz M, Basile N, Marcos J, Requena A. Embryo incubation and selection in a time-lapse monitoring system improves pregnancy outcome compared with a standard incubator: a retrospective cohort study. Fertil Steril 2012; 98: 1481–1489.e10. [DOI] [PubMed] [Google Scholar]
- 9.Wong CC, Loewke KE, Bossert NL, Behr B, De Jonge CJ, Baer TM, Reijo Pera RA. Non-invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nat Biotechnol 2010; 28: 1115–1121. [DOI] [PubMed] [Google Scholar]
- 10.Lundin K, Ahlström A. Quality control and standardization of embryo morphology scoring and viability markers. Reprod Biomed Online 2015; 31: 459–471. [DOI] [PubMed] [Google Scholar]
- 11.Castelló D, Motato Y, Basile N, Remohí J, Espejo-Catena M, Meseguer M. How much have we learned from time-lapse in clinical IVF? Mol Hum Reprod 2016; 22: 719–727. [DOI] [PubMed] [Google Scholar]
- 12.Alikani M, Calderon G, Tomkin G, Garrisi J, Kokot M, Cohen J. Cleavage anomalies in early human embryos and survival after prolonged culture in-vitro. Hum Reprod 2000; 15: 2634–2643. [DOI] [PubMed] [Google Scholar]
- 13.Nagy ZP, Janssenswillen C, Janssens R, De Vos A, Staessen C, Van de Velde H, Van Steirteghem AC. Timing of oocyte activation, pronucleus formation and cleavage in humans after intracytoplasmic sperm injection (ICSI) with testicular spermatozoa and after ICSI or in-vitro fertilization on sibling oocytes with ejaculated spermatozoa. Hum Reprod 1998; 13: 1606–1612. [DOI] [PubMed] [Google Scholar]
- 14.Shoukir Y, Campana A, Farley T, Sakkas D. Early cleavage of in-vitro fertilized human embryos to the 2-cell stage: a novel indicator of embryo quality and viability. Hum Reprod 1997; 12: 1531–1536. [DOI] [PubMed] [Google Scholar]
- 15.Giorgetti C, Hans E, Terriou P, Salzmann J, Barry B, Chabert-Orsini V, Chinchole JM, Franquebalme JP, Glowaczower E, Sitri MC, Thibault MC, Roulier R. Early cleavage: an additional predictor of high implantation rate following elective single embryo transfer. Reprod Biomed Online 2007; 14: 85–91. [DOI] [PubMed] [Google Scholar]
- 16.Sakkas D, Percival G, DArcy Y, Sharif K, Afnan M. Assessment of early cleaving in vitro fertilized human embryos at the 2-cell stage before transfer improves embryo selection. Fertil Steril 2001; 76: 1150–1156. [DOI] [PubMed] [Google Scholar]
- 17.Fenwick J, Platteau P, Murdoch AP, Herbert M. Time from insemination to first cleavage predicts developmental competence of human preimplantation embryos in vitro. Hum Reprod 2002; 17: 407–412. [DOI] [PubMed] [Google Scholar]
- 18.Scott L. Pronuclear scoring as a predictor of embryo development. Reprod Biomed Online 2003; 6: 201–214. [DOI] [PubMed] [Google Scholar]
- 19.Hardarson T, Hanson C, Sjögren A, Lundin K. Human embryos with unevenly sized blastomeres have lower pregnancy and implantation rates: indications for aneuploidy and multinucleation. Hum Reprod 2001; 16: 313–318. [DOI] [PubMed] [Google Scholar]
- 20.Rubio I, Galán A, Larreategui Z, Ayerdi F, Bellver J, Herrero J, Meseguer M. Clinical validation of embryo culture and selection by morphokinetic analysis: a randomized, controlled trial of the EmbryoScope. Fertil Steril 2014; 102: 1287–1294.e5. [DOI] [PubMed] [Google Scholar]
- 21.Van Royen E, Mangelschots K, De Neubourg D, Valkenburg M, Van de Meerssche M, Ryckaert G, Eestermans W, Gerris J. Characterization of a top quality embryo, a step towards single-embryo transfer. Hum Reprod 1999; 14: 2345–2349. [DOI] [PubMed] [Google Scholar]
- 22.Niimura S. Time-lapse videomicrographic analyses of contractions in mouse blastocysts. J Reprod Dev 2003; 49: 413–423. [DOI] [PubMed] [Google Scholar]
- 23.Niimura S, Ogata T, Okimura A, Sato T, Uchiyama Y, Seta T, Nakagawa H, Nakagawa K, Tamura Y. Time-lapse videomicrographic observations of blastocyst hatching in cattle. J Reprod Dev 2010; 56: 649–654. [DOI] [PubMed] [Google Scholar]
- 24.Zernicka-Goetz M. Cleavage pattern and emerging asymmetry of the mouse embryo. Nat Rev Mol Cell Biol 2005; 6: 919–928. [DOI] [PubMed] [Google Scholar]
- 25.Sakkas D. Cleavage in the preimplantation embryo: it is all about being in the right place at the right time! Mol Hum Reprod 2016; 22: 679–680. [DOI] [PubMed] [Google Scholar]
- 26.Lechniak D, Pers-Kamczyc E, Pawlak P. Timing of the first zygotic cleavage as a marker of developmental potential of mammalian embryos. Reprod Biol 2008; 8: 23–42. [DOI] [PubMed] [Google Scholar]
- 27.Ma M, Zhou L, Guo X, Lv Z, Yu Y, Ding C, Zhang P, Bi Y, Xie J, Wang L, Lin M, Zhou Z, Huo R, Sha J, Zhou Q. Decreased cofilin1 expression is important for compaction during early mouse embryo development. Biochim Biophys Acta 2009; 1793: 1804–1810. [DOI] [PubMed] [Google Scholar]
- 28.Pratt HP, Ziomek CA, Reeve WJ, Johnson MH. Compaction of the mouse embryo: an analysis of its components. J Embryol Exp Morphol 1982; 70: 113–132. [PubMed] [Google Scholar]
- 29.Kojima Y, Tam OH, Tam PP. Timing of developmental events in the early mouse embryo. Semin Cell Dev Biol 2014; 34: 65–75. [DOI] [PubMed] [Google Scholar]
- 30.Mio Y, Maeda K. Time-lapse cinematography of dynamic changes occurring during in vitro development of human embryos. Am J Obstet Gynecol 2008; 199: 660.e1–5. [DOI] [PubMed] [Google Scholar]
- 31.Tsai YC, Chung MT, Sung YH, Tsai TF, Tsai YT, Lin LY. Clinical value of early cleavage embryo. Int J Gynaecol Obstet 2002; 76: 293–297. [DOI] [PubMed] [Google Scholar]
- 32.Isiklar A, Mercan R, Balaban B, Alatas C, Aksoy S, Urman B. Early cleavage of human embryos to the two-cell stage. A simple, effective indicator of implantation and pregnancy in intracytoplasmic sperm injection. J Reprod Med 2002; 47: 540–544. [PubMed] [Google Scholar]
- 33.Sakkas D, Shoukir Y, Chardonnens D, Bianchi PG, Campana A. Early cleavage of human embryos to the two-cell stage after intracytoplasmic sperm injection as an indicator of embryo viability. Hum Reprod 1998; 13: 182–187. [DOI] [PubMed] [Google Scholar]
- 34.Lee YS, Thouas GA, Gardner DK. Developmental kinetics of cleavage stage mouse embryos are related to their subsequent carbohydrate and amino acid utilization at the blastocyst stage. Hum Reprod 2015; 30: 543–552. [DOI] [PubMed] [Google Scholar]
- 35.González IM, Martin PM, Burdsal C, Sloan JL, Mager S, Harris T, Sutherland AE. Leucine and arginine regulate trophoblast motility through mTOR-dependent and independent pathways in the preimplantation mouse embryo. Dev Biol 2012; 361: 286–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gangloff YG, Mueller M, Dann SG, Svoboda P, Sticker M, Spetz JF, Um SH, Brown EJ, Cereghini S, Thomas G, Kozma SC. Disruption of the mouse mTOR gene leads to early postimplantation lethality and prohibits embryonic stem cell development. Mol Cell Biol 2004; 24: 9508–9516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakayama T, Fujiwara H, Maeda M, Inoue T, Yoshioka S, Mori T, Fujii S. Human peripheral blood mononuclear cells (PBMC) in early pregnancy promote embryo invasion in vitro: HCG enhances the effects of PBMC. Hum Reprod 2002; 17: 207–212. [DOI] [PubMed] [Google Scholar]
- 38.Jeong YJ, Choi HW, Shin HS, Cui XS, Kim NH, Gerton GL, Jun JH. Optimization of real time RT-PCR methods for the analysis of gene expression in mouse eggs and preimplantation embryos. Mol Reprod Dev 2005; 71: 284–289. [DOI] [PubMed] [Google Scholar]
- 39.Wale PL, Gardner DK. Time-lapse analysis of mouse embryo development in oxygen gradients. Reprod Biomed Online 2010; 21: 402–410. [DOI] [PubMed] [Google Scholar]
- 40.Tejera A, Herrero J, Viloria T, Romero JL, Gamiz P, Meseguer M. Time-dependent O2 consumption patterns determined optimal time ranges for selecting viable human embryos. Fertil Steril 2012; 98: 849–857.e1−3. [DOI] [PubMed] [Google Scholar]
- 41.Gardner DK, Wale PL, Collins R, Lane M. Glucose consumption of single post-compaction human embryos is predictive of embryo sex and live birth outcome. Hum Reprod 2011; 26: 1981–1986. [DOI] [PubMed] [Google Scholar]
- 42.Gott AL, Hardy K, Winston RM, Leese HJ. Non-invasive measurement of pyruvate and glucose uptake and lactate production by single human preimplantation embryos. Hum Reprod 1990; 5: 104–108. [DOI] [PubMed] [Google Scholar]
- 43.Kawamura K, Chen Y, Shu Y, Cheng Y, Qiao J, Behr B, Pera RA, Hsueh AJ. Promotion of human early embryonic development and blastocyst outgrowth in vitro using autocrine/paracrine growth factors. PLoS ONE 2012; 7: e49328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leese HJ. Metabolism of the preimplantation embryo: 40 years on. Reproduction 2012; 143: 417–427. [DOI] [PubMed] [Google Scholar]
- 45.Sfontouris IA, Lainas GT, Sakkas D, Zorzovilis IZ, Petsas GK, Lainas TG. Non-invasive metabolomic analysis using a commercial NIR instrument for embryo selection. J Hum Reprod Sci 2013; 6: 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Borini A, Lagalla C, Cattoli M, Sereni E, Sciajno R, Flamigni C, Coticchio G. Predictive factors for embryo implantation potential. Reprod Biomed Online 2005; 10: 653–668. [DOI] [PubMed] [Google Scholar]
- 47.Berger DS, Zapantis A, Merhi Z, Younger J, Polotsky AJ, Jindal SK. Embryo quality but not pronuclear score is associated with clinical pregnancy following IVF. J Assist Reprod Genet 2014; 31: 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lundin K, Bergh C, Hardarson T. Early embryo cleavage is a strong indicator of embryo quality in human IVF. Hum Reprod 2001; 16: 2652–2657. [DOI] [PubMed] [Google Scholar]
- 49.Salumets A, Hydén-Granskog C, Mäkinen S, Suikkari AM, Tiitinen A, Tuuri T. Early cleavage predicts the viability of human embryos in elective single embryo transfer procedures. Hum Reprod 2003; 18: 821–825. [DOI] [PubMed] [Google Scholar]
- 50.Sugimura S, Akai T, Hashiyada Y, Somfai T, Inaba Y, Hirayama M, Yamanouchi T, Matsuda H, Kobayashi S, Aikawa Y, Ohtake M, Kobayashi E, Konishi K, Imai K. Promising system for selecting healthy in vitro-fertilized embryos in cattle. PLoS ONE 2012; 7: e36627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sugimura S, Akai T, Hashiyada Y, Aikawa Y, Ohtake M, Matsuda H, Kobayashi S, Kobayashi E, Konishi K, Imai K. Effect of embryo density on in vitro development and gene expression in bovine in vitro-fertilized embryos cultured in a microwell system. J Reprod Dev 2013; 59: 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murakoshi Y, Sueoka K, Takahashi K, Sato S, Sakurai T, Tajima H, Yoshimura Y. Embryo developmental capability and pregnancy outcome are related to the mitochondrial DNA copy number and ooplasmic volume. J Assist Reprod Genet 2013; 30: 1367–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.El Shourbagy SH, Spikings EC, Freitas M, St John JC. Mitochondria directly influence fertilisation outcome in the pig. Reproduction 2006; 131: 233–245. [DOI] [PubMed] [Google Scholar]
- 54.Yadav BR, King WA, Betteridge KJ. Relationships between the completion of first cleavage and the chromosomal complement, sex, and developmental rates of bovine embryos generated in vitro. Mol Reprod Dev 1993; 36: 434–439. [DOI] [PubMed] [Google Scholar]
- 55.Tao J, Tamis R, Fink K, Williams B, Nelson-White T, Craig R. The neglected morula/compact stage embryo transfer. Hum Reprod 2002; 17: 1513–1518. [DOI] [PubMed] [Google Scholar]
- 56.Skiadas CC, Jackson KV, Racowsky C. Early compaction on day 3 may be associated with increased implantation potential. Fertil Steril 2006; 86: 1386–1391. [DOI] [PubMed] [Google Scholar]
- 57.Le Cruguel S, Ferré-LHôtellier V, Morinière C, Lemerle S, Reynier P, Descamps P, May-Panloup P. Early compaction at day 3 may be a useful additional criterion for embryo transfer. J Assist Reprod Genet 2013; 30: 683–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seli E, Bruce C, Botros L, Henson M, Roos P, Judge K, Hardarson T, Ahlström A, Harrison P, Henman M, Go K, Acevedo N, Siques J, Tucker M, Sakkas D. Receiver operating characteristic (ROC) analysis of day 5 morphology grading and metabolomic Viability Score on predicting implantation outcome. J Assist Reprod Genet 2011; 28: 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mallol A, Piqué L, Santaló J, Ibáñez E. Morphokinetics of cloned mouse embryos treated with epigenetic drugs and blastocyst prediction. Reproduction 2016; 151: 203–214. [DOI] [PubMed] [Google Scholar]
- 60.Piekny AJ, Glotzer M. Anillin is a scaffold protein that links RhoA, actin, and myosin during cytokinesis. Curr Biol 2008; 18: 30–36. [DOI] [PubMed] [Google Scholar]
- 61.Sharif B, Fadero T, Maddox AS. Anillin localization suggests distinct mechanisms of division plane specification in mouse oogenic meiosis I and II. Gene Expr Patterns 2015; 17: 98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu G, Schöler HR. Role of Oct4 in the early embryo development. Cell Regen (Lond) 2014; 3: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ziebe S, Petersen K, Lindenberg S, Andersen AG, Gabrielsen A, Andersen AN. Embryo morphology or cleavage stage: how to select the best embryos for transfer after in-vitro fertilization. Hum Reprod 1997; 12: 1545–1549. [DOI] [PubMed] [Google Scholar]
- 64.Kirkegaard K, Sundvall L, Erlandsen M, Hindkjær JJ, Knudsen UB, Ingerslev HJ. Timing of human preimplantation embryonic development is confounded by embryo origin. Hum Reprod 2016; 31: 324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ahlstrom A, Park H, Bergh C, Selleskog U, Lundin K. Conventional morphology performs better than morphokinetics for prediction of live birth after day 2 transfer. Reprod Biomed Online 2016; 33: 61–70. [DOI] [PubMed] [Google Scholar]