Abstract
Transcription factor TEA domain family transcription factor 4 (Tead4) is one of the key factors involved in the differentiation of the trophectoderm (TE) in murine embryos. However, knowledge on the roles of TEAD4 in preimplantation development during bovine embryos is currently limited. This study examined the transcript and protein expression patterns of TEAD4 and attempted to elucidate the functions of TEAD4 during bovine preimplantation development using RNA interference. TEAD4 mRNA was found to be upregulated between the 16-cell and morula stages, and nuclear localization of the TEAD4 protein was detected at the morula stage, as well as in subsequent developmental stages. TEAD4 downregulation did not affect embryonic development until the blastocyst stage, and TEAD4-downregulated embryos were capable of forming the TE under both 5% and 21% O2 conditions. Results of gene expression analysis showed that TEAD4 downregulation did not affect the expression levels of POU class 5 transcription factor 1 (OCT-4), NANOG, caudal-type homeobox 2 (CDX2), GATA binding protein 3 (GATA3), and interferon-tau (IFNT). In conclusion, TEAD4 might be dispensable for development until the blastocyst stage and TE differentiation in bovine embryos.
Keywords: Bovine embryo, Preimplantation development, RNA interference, TEAD4
The earliest differentiation event during mammalian embryonic development occurs between the morula and blastocyst stages. In this process, the outer cells of the morula differentiate into the trophectoderm (TE), a single layer of polarized epithelial cells surrounding the blastocoel that subsequently develops into the placenta. The inner cells of a morula differentiate into the inner cell mass (ICM), a population of pluripotent cells attached to the inside of the TE that give rise to the fetus and extra-embryonic tissues [1]. Successful segregation between ICM and TE and correct embryonic development depend on different transcriptional programs that comprise interaction among multiple genes. In mice, gene knockout studies showed that TEA domain family transcription factor 4 (Tead4) is essential for TE development and acts as a master regulator of the TE-specific transcriptional program [2, 3]. Tead4-deficient embryos fail to develop the TE and lack the expression of TE-specific factors such as caudal-type homeobox 2 (Cdx2) and GATA binding protein 3 (Gata3) [2,3,4,5]. Tead4 expression is detectable in both the ICM and TE lineages [2]. However, Tead4 upregulates TE-specific genes only in the outer cells [6]. In the inner cells, Hippo signaling modulates Tead4 function by preventing nuclear accumulation of a Tead4 coactivator, Yes-associated protein (Yap). In contrast, in the outer cells, Yap can accumulate in the nucleus and form a complex with Tead4 to activate TE-specific genes [7]. A recent study showed that Tead4-null embryos can form a blastocoel and express Cdx2 and Gata3 under low oxidative stress conditions [8]. The authors concluded that Tead4 is involved in maintaining energy homeostasis, which dramatically changes between the morula and blastocyst stages in murine embryos [8].
We recently performed CDX2 downregulation in bovine embryos and reported that CDX2 functions in TE development and in regulating the molecular mechanism of TE [9]. In addition, CDX2 regulates the expression of interferon-tau (IFNT), which is secreted by TE lineage cells and acts as a pregnancy recognition factor [10]. TEAD4 is also expected to control TE differentiation in bovine embryos, but little is known about the roles of TEAD4 during embryonic development and the differentiation of ICM and TE. We previously performed mRNA expression analysis of TEAD4 at the blastocyst and elongation stages, and results showed that TEAD4 is expressed both in the ICM and TE [11]. Another study reported that nuclear localization of TEAD4 is observed only in the TE and that TEAD4 protein expression occurs only in the cytoplasm in ICM cells [12]. However, limited information is available on the temporal and spatial expression patterns of TEAD4 in bovine embryos, particularly at the developmental stages prior to blastocyst formation.
In the present study, we characterized the mRNA and protein expression patterns of TEAD4 in bovine embryos until the hatched blastocyst stage. In addition, we performed TEAD4 knockdown and investigated its effects on the developmental competence and expression of genes speculated to be related in the differentiation of ICM and TE. The present study aims to investigate the roles of TEAD4 during the early development of bovine embryos.
Materials and Methods
Chemicals
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless otherwise stated.
In vitro fertilization (IVF) and embryo culture
Bovine oocytes were obtained by aspirating ovaries collected at a local slaughterhouse, as previously described [11]. Cumulus–oocyte complexes (COCs) were matured in a drop of IVMD-101 medium (Research Institution for the Functional Peptides, Yamagata, Japan) [13] and covered with mineral oil at 39ºC in a humidified atmosphere of 5% CO2 in air for 20 h. After in vitro maturation, COCs were transferred to a drop of IVF-100 medium (Research Institution for the Functional Peptides). Subsequently, frozen-thawed semen was centrifuged twice at 600 × g for 5 min in IVF-100 medium, and the spermatozoa were added to the COCs at a final concentration of 5 × 106 sperm per milliliter. COCs and spermatozoa were incubated at 39ºC in a humidified atmosphere of 5% CO2 in air for 6 h. After insemination, cumulus cells and excess spermatozoa were removed from presumptive zygotes by pipetting. Following microinjection of short interfering RNA (siRNA), the embryos were incubated in modified TALP (mTALP) medium [14] containing 1 mg/ml bovine serum albumin (BSA) at 39ºC in a humidified atmosphere of 5% CO2, 5% O2, and 90% N2, or of 5% CO2 in air. On Day 2 (IVF = Day 0), the embryos were transferred to mTALP medium supplemented with 3% (v/v) newborn calf serum (Thermo Fisher Scientific, Waltham, MA, USA) and cultured at 39ºC in a humidified atmosphere of 5% CO2, 5% O2, and 90% N2, or of 5% CO2 in air until Day 8. Embryonic developmental rates were assessed on Day 2 (2-cell ≤), Day 4 (16-cell ≤), Day 5 (32-cell ≤), Day 6 (morula ≤), Day 7 (blastocyst ≤), and Day 8 (blastocyst ≤ and hatching blastocyst ≤).
Design of siRNA and microinjection into embryos
The target site for TEAD4 was selected from the bovine sequence (GenBank accession no. XM_010827947.1). Specific siRNA was designed using the online software BLOCK-iTTM RNAi Designer (Thermo Fisher Scientific; http://rnaidesigner.thermofisher.com/rnaiexpress/). Both sense and antisense RNA sequences for siRNA were commercially synthesized (Table 1). After insemination, the denuded embryos were transferred to a drop of mTALP medium containing 1 mg/ml BSA for microinjection. Approximately 10 pl of 50 µM specific siRNA duplex was injected into the cytoplasm of each embryo using a FemtoJet (Eppendorf, Hamburg, Germany). Similarly, approximately 10 pl of 20 µM nonsilencing siRNA (AllStars Negative Control siRNA; Qiagen, Hilden, Germany) was injected as a control. The embryos were washed after microinjection and cultured as described above.
Table 1. Primers and siRNA sequences.
| Name | Nucleotide sequences (5ʹ–3ʹ)* | Annealing temperature | Fragment size | GenBank |
| (ºC) | (bp) | accession no. | ||
| TEAD4 | F- AAGTTCTGGGCAGACCTCAA | 60 | 249 | XM_010827947.1 |
| R- GTGCTTCAGCTTGTGGATGA | ||||
| CDX2 | F- GCCACCATGTACGTGAGCTACC | 55 | 140 | NM_001206299.1 |
| R- ACATGGTATCCGCCGTAGTCCGG | ||||
| GATA3 | F- ATGAAACCGAAACCCGATGG | 60 | 185 | NM_001076804 |
| R- TTCACAGCACTAGAGAGACC | ||||
| Oct-4 | F- GGTTCTCTTTGGAAAGGTGTTC | 52 | 314 | AF022987.1 |
| R- ACACTCGGACCACGTCTTTC | ||||
| NANOG | F- AATTCCCAGCAGCAAATCAC | 55 | 215 | DQ069776 |
| R- CCCTTCCCTCAAATTGACAC | ||||
| IFNT | F- GCAGTGCTTCAACCTCTTCC | 62 | 155 | AF238611.1 |
| R- TCCTTCCCATGTCAGAGTCC | ||||
| Histone H2A | F- AGGACGACTAGCCATGGACGTGTG | 60 | 209 | NM_174809 |
| R- CCACCACCAGCAATTGTAGCCTTG | ||||
| TEAD4 siRNA | S- GCUACGAGAAUGGCCACUATT | N/A | N/A | N/A |
| AS- UAGUGGCCAUUCUCGUAGCTT | N/A | N/A | N/A |
* F, forward; R, reverse; S, sense strand; AS, antisense strand.
Determination of the relative abundance of gene transcripts
Total RNA was isolated from denuded metaphase II oocytes (confirmed based on the presence of one polar body), 1-cell, 2-cell, 4-cell, 8-cell, 16-cell, morula, blastocyst, expanded blastocyst, and hatched blastocyst-stage embryos. Oocytes and embryos at the appropriate developmental stage were treated with 0.1% (w/v) protease in phosphate-buffered saline (PBS) containing 1% (w/v) polyvinyl pyrrolidone for 5 min to remove the zona pellucida. Pools of 20 oocytes, 10 embryos from the 1-cell to the 8-cell stage, or 5 embryos from the other stages were added to 5 µl of lysis buffer [0.8% (v/v) Igepal (MP Biomedicals, LLC, Santa Ana, CA, USA), 5 mM dithiothreitol (Thermo Fisher Scientific) and 1 U/µl RNasin (Promega, Madison, WI, USA)], snap-frozen in liquid nitrogen, and subsequently stored at −80ºC. For validation of siRNA, one expanded blastocyst per sample was collected on Day 7, and RNA was extracted using the same method. RNA samples were heated at 80ºC for 5 min and then subjected to reverse transcription using the QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacturer’s instructions. The reaction mixture was then diluted with RNase free water to obtain a final volume of 31 µl. Real-time quantitative PCR was performed using StepOneTM (Thermo Fisher Scientific), and products were detected using SYBR Green included in the QuantiTect SYBR Green PCR Master Mix Kit (Qiagen). The amplification program was as follows: preincubation at 95ºC for 15 min to activate the HotStarTaq DNA Polymerase (Qiagen), followed by 45 cycles of denaturation at 94ºC for 15 sec, primer-specific annealing (Table 1) for 30 sec, and elongation at 72ºC for 30 sec. After completion of the last cycle, a melting curve was generated by starting fluorescence acquisition at 60ºC and recording measurements in 0.3ºC increments up to 95ºC. A final quantification step was performed using StepOneTM quantification software (Thermo Fisher Scientific). Expression levels of the target gene in each run were normalized to the internal standard Histone H2A.
Immunofluorescent staining and assessment of cell numbers
Embryos at the appropriate developmental stages were fixed in 4% (w/v) paraformaldehyde in PBS (Wako Pure Chemical Industries, Osaka, Japan) for 20 min at room temperature, and then washed twice for 10 min each in PBS containing 0.1% (v/v) Triton X-100 (TXPBS). Samples were subsequently permeabilized in 0.3% (v/v) Triton X-100 in PBS for 60 min, and washed twice in TXPBS for 10 min. Blocking was performed by incubation in 0.5% (w/v) BSA and 1% (w/v) skimmed milk in TXPBS for 90 min. Embryos were incubated with anti-TEAD4 primary antibody (1:1500; ab58310; Abcam, Cambridge, UK) in PBS supplemented with 0.5% (w/v) BSA and 0.05% (v/v) Triton X-100 at room temperature for 2 h, or with anti-CDX2 primary antibody (1:300; MU392A-UC; BioGenex, Fremont, CA, USA) in PBS supplemented with 0.5% (w/v) BSA and 0.05% (v/v) Triton X-100 at 37ºC overnight. Next, the embryos were washed four times for 15 min each in TXPBS. The embryos were then incubated with Alexa Fluor 488-conjugated goat anti-mouse secondary antibody (A-11029, Thermo Fisher Scientific) at a dilution of 1:400 at room temperature for 1 h in PBS containing 0.5% (w/v) BSA and 0.05% (v/v) Triton X-100. The embryos were washed four times in TXPBS for 20 min and subsequently mounted onto slides in a drop of VECTASHIELD Mounting Medium with 4’,6-diamidino-2-phenyndol (DAPI; Vector Laboratories, Burlingame, CA, USA). Fluorescent images were obtained using an inverted fluorescence microscope and a digital camera system (ECLIPSE Ti-U and DS-Fi2-L3; Nikon, Tokyo, Japan). Cell counting was performed using ImageJ (https://imagej.nih.gov/ij/).
Statistical analysis
Percentage data for embryonic development were subjected to an arcsine transformation. Transformed values were analyzed using one-way analysis of variance (ANOVA). Data for temporal TEAD4 gene expression and the mRNA expression levels of TEAD4 at the morula stage and IFNT were analyzed using the Kruskal-Wallis test, followed by multiple pairwise comparisons using Scheffé’s method. The mRNA expression levels of TEAD4 at the expanded blastocyst stage and of CDX2, GATA3, POU class 5 transcription factor 1 (OCT-4), and NANOG were analyzed using one-way ANOVA followed by multiple pairwise comparisons using the Tukey-Kramer method. A P value < 0.05 was considered statistically significant.
Results
Temporal and spatial expression of TEAD4 mRNA and protein in early embryos
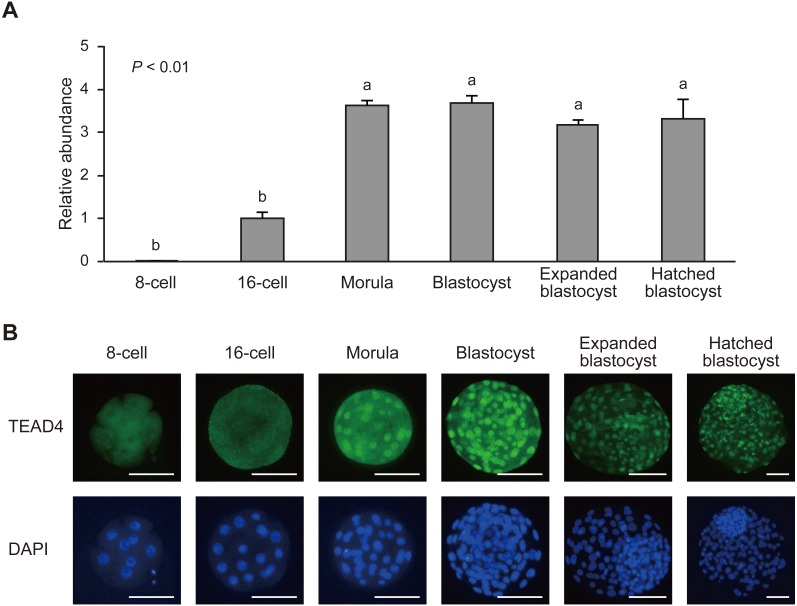
Metaphase II stage oocytes and in vitro-fertilized embryos were collected at different developmental stages from the 1-cell stage to hatched blastocyst, and analyzed expression levels of TEAD4 mRNA and the expression pattern of TEAD4 protein using real-time quantitative PCR and immunofluorescent staining, respectively (Fig. 1). TEAD4 mRNA showed very late threshold cycles until the 8-cell stage, indicating low starting quantities (Fig. 1A, data from the metaphase II stage to the 4-cell stage not shown). TEAD4 expression slightly increased (P = 0.09) from the 8-cell to the 16-cell stage and exhibited approximately fourfold upregulation from the 16-cell to the morula stage. After the morula stage, TEAD4 was consistently and strongly expressed until the hatched blastocyst stage.
Fig. 1.
TEAD4 mRNA and protein profiles during the bovine preimplantation development. (A) Relative abundance (mean ± SEM; n = 6) of TEAD4 transcripts in 8-cell, 16-cell, morula, blastocyst, expanded blastocyst, and hatched blastocyst. a, b Different characters indicate a significant difference (P < 0.01). (B) Representative images of TEAD4 protein expression. Embryos were labeled with TEAD4 antibody (green) and DAPI for DNA (blue). Scale bars, 100 µm.
To characterize TEAD4 protein expression dynamics, we stained bovine embryos from the 8-cell stage to the hatched blastocyst stage, using TEAD4-specific antibody (Fig. 1B). A diffuse TEAD4 signal was consistently observed in the cytoplasm until the 16-cell stage. A nuclear TEAD4 signal was detected in the morula stage. After the blastocyst stage, strong nuclear TEAD4 signals were observed in almost all blastomeres.
Effect of siRNA injection on TEAD4 expression
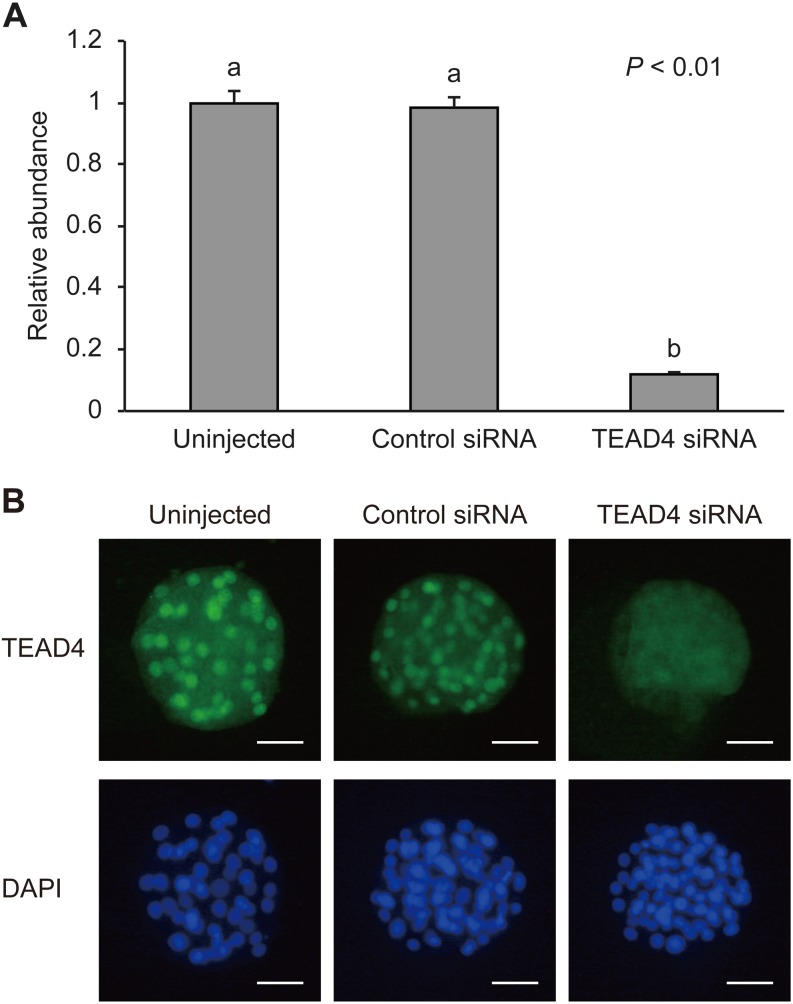
To determine the efficacy of TEAD4 downregulation using RNA interference, we examined the mRNA and protein expression levels of TEAD4 in uninjected, control siRNA-injected, and TEAD4-specific siRNA (TEAD4 siRNA)-injected morula-stage embryos on Day 5 (Fig. 2). The relative abundance of TEAD4 mRNA in the embryos injected with TEAD4 siRNA was significantly lower (P < 0.01) than that in uninjected and control siRNA-injected embryos (Fig. 2A). Protein expression of TEAD4 was evaluated via immunofluorescent staining. Nuclear signals from TEAD4 siRNA-injected embryos were difficult to detect at the morula stage (Fig. 2B).
Fig. 2.
Validation of TEAD4 siRNA at the morula stage on Day 5. (A) Relative abundance (mean ± SEM) of TEAD4 transcripts in uninjected (n = 6), control siRNA-injected (n = 5), or TEAD4 siRNA-injected embryos (n = 6). a, b Different characters indicate a significant difference (P < 0.01). (B) Representative images of TEAD4 immunofluorescent staining of morula-stage uninjected, control siRNA-injected, or TEAD4 siRNA-injected embryos. Embryos were labeled with TEAD4 antibody (green) and DAPI for DNA (blue). Scale bars, 50 µm.
Effect of TEAD4 downregulation on the development of bovine embryos
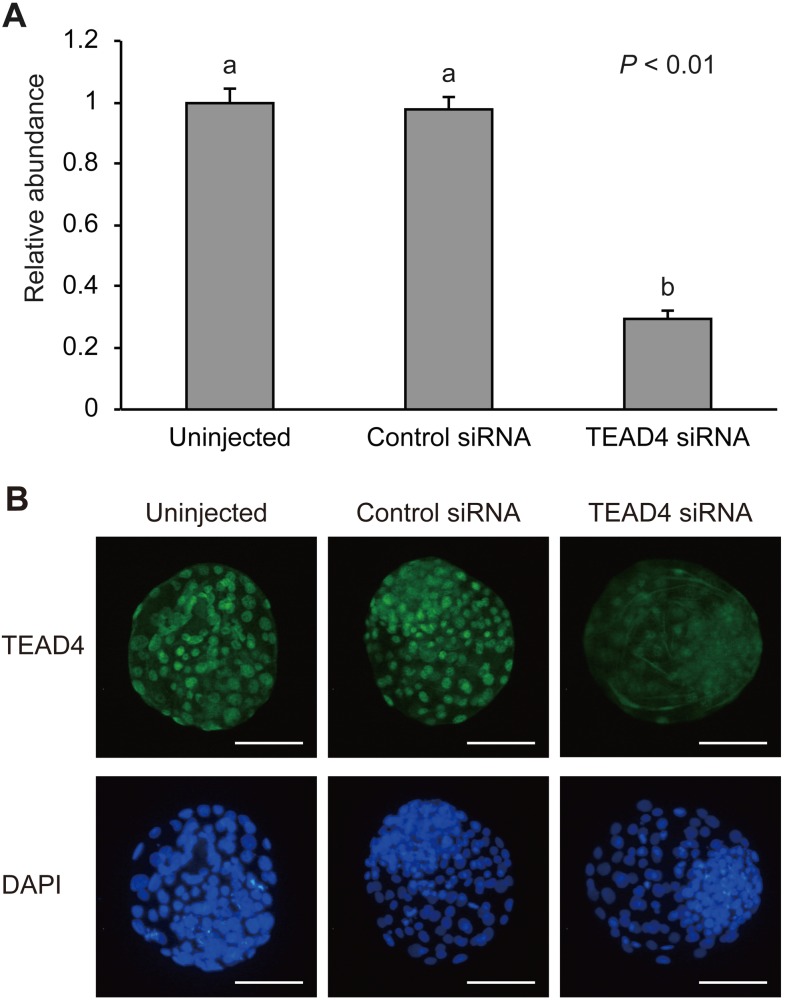
We evaluated the in vitro developmental competence of TEAD4 siRNA-injected embryos (Table 2). There was no significant difference in the rate of development during all the developmental stages until Day 8 among the experimental groups. The embryos with TEAD4 downregulation could form blastocoels, expand themselves, and hatch from the zona pellucida. To determine whether siRNA injection can maintain TEAD4 downrgregulation beyond the morula stage on Day 5, we evaluated TEAD4 expression at the expanded blastocyst stage on Day 7 (Fig. 3). The relative abundance of TEAD4 mRNA in the embryos injected with TEAD4 siRNA was significantly lower (P < 0.01) than that in the uninjected and control siRNA-injected embryos (Fig. 3A). Although TEAD4 signals were observed in some nuclei of TEAD4 siRNA-injected embryos, the signals were weaker compared to those in uninjected and control siRNA-injected embryos (Fig. 3B). Therefore, we concluded that the blastocyst formation could have given rise under the TEAD4-downregulated condition in bovine embryos.
Table 2. Effect of TEAD4 siRNA injection on in vitro development of bovine embryos*.
| Treatment | Number of embryos cultured | No. (%)† of embryos developed to |
||||||
| Day 2 | Day 4 | Day 5 | Day 6 | Day 7 | Day 8 |
|||
| 2-cell ≤ | 16-cell ≤ | 32-cell ≤ | Morula ≤ | Blastocyst ≤ | Blastocyst ≤ | Hatching blastocyst ≤ | ||
| Uninjected | 230 | 196 (85.2) | 153 (66.5) | 138 (60.0) | 135 (58.7) | 137 (59.6) | 134 (58.3) | 63 (27.4) |
| Control siRNA | 248 | 213 (85.9) | 159 (64.1) | 140 (56.5) | 138 (55.6) | 137 (55.2) | 135 (54.4) | 59 (23.8) |
| TEAD4 siRNA | 241 | 192 (79.7) | 143 (59.3) | 122 (50.6) | 115 (47.7) | 114 (47.3) | 124 (51.5) | 50 (20.7) |
* Experiments were replicated six times. † Percentages of the number of embryos cultured.
Fig. 3.
Validation of TEAD4 siRNA at the expanded blastocyst stage on Day 7. (A) Relative abundance (mean ± SEM; n = 12) of TEAD4 transcripts in uninjected, control siRNA-injected, or TEAD4 siRNA-injected embryos. a, b Different characters indicate a significant difference (P < 0.01). (B) Representative images of TEAD4 immunofluorescent staining of expanded blastocyst-stage uninjected, control siRNA-injected, or TEAD4 siRNA-injected embryos. Embryos were labeled with TEAD4 antibody (green) and DAPI for DNA (blue). Scale bars, 100 µm.
Next, to evaluate the effect of TEAD4 downregulation on cell composition of ICM and TE cell numbers, we performed immunostaining against CDX2, which is exclusively expressed in TE cells, and subsequently counted total cell number and CDX2-positive cell number. ICM cell number was calculated by subtracting the counts of CDX2-positive cells from the total cell count. However, there was no significant difference in ICM/TE ratios among uninjected (0.43, n = 24), control siRNA-injected (0.37, n =20), and TEAD4 siRNA-injected embryos (0.40, n = 21).
Quantitative PCR gene expression analysis
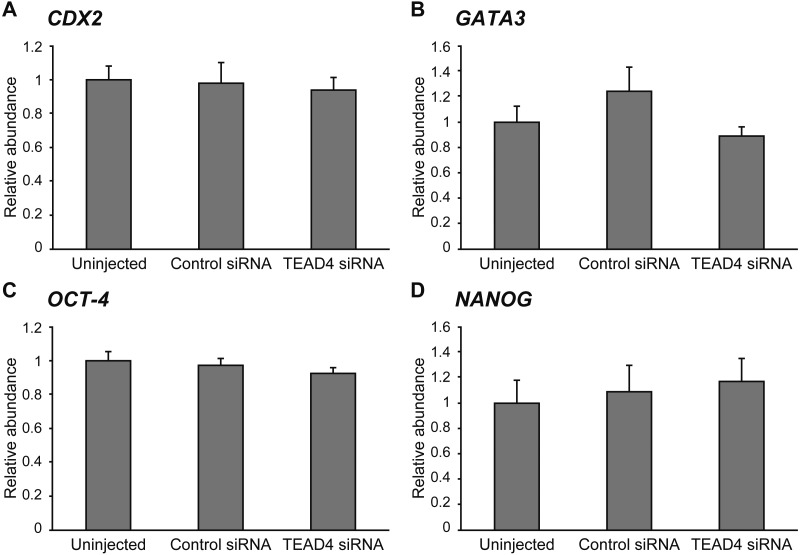
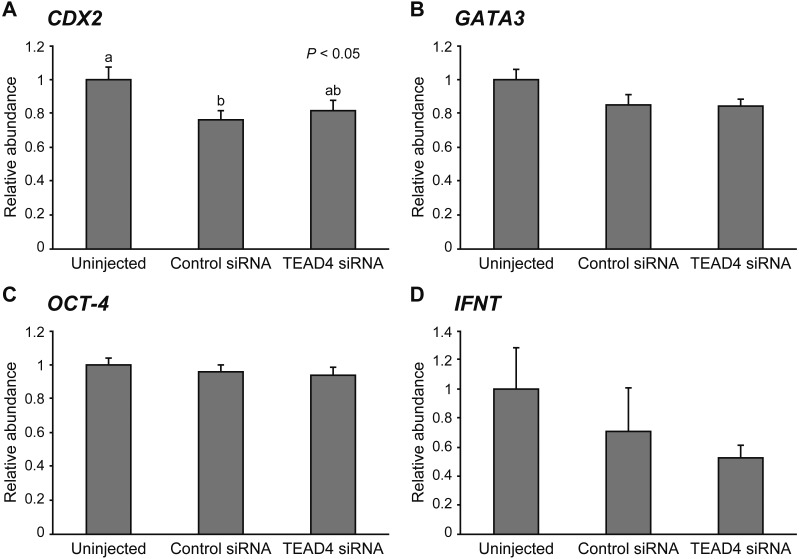
To clarify the effect of TEAD4 downregulation on gene expression, we evaluated the mRNA levels of CDX2, GATA3, OCT-4, and NANOG at the morula stage (Fig. 4) and of CDX2, GATA3, OCT-4, and IFNT at the expanded blastocyst stage (Fig. 5). There was no significant difference in all transcript levels of the genes among the experimental groups at the morula stage on Day 5. At the expanded blastocyst stage on Day 7, the relative abundances of CDX2, GATA3, OCT-4, and IFNT in TEAD4-downregulated embryos remained unchanged compared to the uninjected and control siRNA-injected embryos. Although CDX2 mRNA level in the control siRNA-injected embryos was significantly lower (P < 0.05) than that in the uninjected embryos, no significant difference was observed between control siRNA-injected embryos and TEAD4 siRNA-injected embryos.
Fig. 4.
Relative abundance (mean ± SEM) of (A) CDX2, (B) GATA3, (C) OCT-4, and (D) NANOG transcripts at the morula stage in uninjected (n = 6), control siRNA-injected (n = 5), and TEAD4 siRNA-injected embryos (n = 6).
Fig. 5.
Relative abundance (mean ± SEM) of (A) CDX2, (B) GATA3, (C) OCT-4, and (D) IFNT transcripts at the expanded blastocyst stage in uninjected (n = 12), control siRNA-injected (n = 12), and TEAD4 siRNA-injected embryos (n = 12). a, b Different characters indicate a significant difference (P < 0.05).
Developmental rates under the high O2 condition
Tead4-deficient murine embryos successfully formed the TE, that is, form the blastocyst cavity under the low O2 (5%) cultural condition. However, the embryos failed to develop to blastocyst in atmospheric O2 (21%) [8]. For the cultural condition in cows, embryos are generally cultured at 5% O2. In the present study, TEAD4-downregulated embryos that were able to form the blastocyst cavity were cultured at 5% O2. Therefore, we evaluated the developmental competence of TEAD4-downregulated embryos cultured in 21% O2 (Table 3). However, no significant difference in the rate of development during all the developmental stages was observed among the experimental groups.
Table 3. Effect of TEAD4 siRNA injection on in vitro development under 21% O2 conditions*.
| Treatment | Number of embryos cultured | No. (%)† of embryos developed to |
||||||
| Day 2 | Day 4 | Day 5 | Day 6 | Day 7 | Day 8 |
|||
| 2-cell ≤ | 16-cell ≤ | 32-cell ≤ | Morula ≤ | Blastocyst ≤ | Blastocyst ≤ | Hatching blastocyst ≤ | ||
| Uninjected | 228 | 198 (86.8) | 149 (65.4) | 106 (46.5) | 99 (43.4) | 93 (40.8) | 84 (36.8) | 12 (5.3) |
| Control siRNA | 212 | 179 (84.4) | 131 (61.8) | 106 (50.0) | 99 (46.7) | 102 (48.1) | 94 (44.3) | 5 (2.4) |
| TEAD4 siRNA | 217 | 196 (90.3) | 143 (65.9) | 117 (53.9) | 104 (47.9) | 88 (40.6) | 93 (42.9) | 9 (4.1) |
* Experiments were replicated six times. † Percentages of the number of embryos cultured.
Discussion
In the present study, we analyzed TEAD4 expression patterns during early embryonic development. To elucidate the mechanisms underlying ICM/TE differentiation, we downregulated TEAD4 expression in bovine embryos using RNA interference. We attempted to analyze TEAD4 mRNA expression dynamics using real-time PCR and observed very late threshold cycles until the 8-cell stage. However, Histone H2A expression, which was used as the internal standard, also showed very late threshold cycles until the 4-cell stage. Although the results obtained until the 4-cell stage were insufficient to evaluate TEAD4 mRNA dynamics during the earlier developmental stages, results of this study showed that TEAD4 transcripts are weakly expressed at the 8-cell and 16-cell stages but become upregulated starting from the morula stage. In the present study, nuclear TEAD4 expression was detected beginning at the morula stage and was observed in almost all cells, including the ICM, in blastocoel-formed embryos. In murine embryos, Tead4 mRNA was not detected in oocytes and 1-cell-stage embryos [3]. Tead4 is expressed prior to Cdx2, which is expressed from the 8-cell stage and is regulated by Tead4. Tead4 expression begins at the 2-cell stage and continues until the 4-cell stage, after which expression levels reach a peak between the 8-cell and morula stages [2, 3]. In bovine embryos, TEAD4 mRNA and protein expression drastically changed before and after the morula stage and resembled the corresponding expression dynamics observed in murine embryos.
There are two conflicting reports regarding the distribution of Tead4 protein at the morula and blastocyst stages in mice. One report demonstrated that although Tead4 is expressed in all cells, Tead4 cannot induce Cdx2 expression in the inner cells. This is because the role of Tead4 as a transcription factor depends on the existence of its coactivator, Yap, in the nucleus [6]. In the inner cells of a morula and the ICM, the transition of Yap into the nucleus is blocked by position-dependent Hippo/Lats signaling, and Tead4 does not regulate the expression of TE-related genes at this location. The other repor proposed that Tead4 is not localized in a nucleus of the inner cells of a morula and the ICM and that the TE-specific transcriptional program is not activated in the inner cells [12]. Similarly, Home and coworkers [12] performed immunofluorescent staining for TEAD4 in bovine blastocysts and showed that nuclei of ICM lacked the protein expression of TEAD4. However, in the present study, we observed a TEAD4 nuclear signal in the ICM and thus do not agree with the results of the previous study. Although the exact reasons for the contrasting results are unclear, one plausible reason might be due to the process during immunofluorescent staining. In fact, other groups have demonstrated that nuclear localization of Tead4 in the murine ICM could be detected using the anti-Tead4 antibody that was used in the study by Home and coworkers [12] and in the present study [15, 16].
Tead4 is known as one of the most crucial factors for TE differentiation. Besides Tead4, genetic evidence suggests that Cdx2 and Gata3 also have regulatory roles in TE differentiation. However, loss of Tead4 leads to more severe phenotypes than loss of Cdx2 and Gata3 [17, 18] and results in arrested embryonic development at the morula stage [2, 3]. Therefore, Tead4 is thought to be an upstream regulator of TE differentiation. In bovine embryos, CDX2 downregulation does not prevent blastocyst formation but causes aberrations in expansion, proliferation, and elongation of the TE lineage [9, 19, 20]. If TEAD4 plays a role in regulating the expression of TE-specific genes in bovine embryos, TEAD4 downregulation is expected to result in more serious defects compared to CDX2 downregulation. However, in the present study, TEAD4-downregulated embryos correctly formed blastocoels and showed normal TE expansion. Furthermore, TEAD4 downregulation had no influence not only on the ICM-related genes, OCT-4 and NANOG, but also on the TE-related genes, CDX2, GATA3, and IFNT. CDX2 expression level in control siRNA-injected embryos was significantly lower than that in uninjected embryos. However, in our previous study [9], injection of control siRNA did not result in CDX2 downregulation. Although the reason for the above result remains unclear, we suspect that injection of control siRNA will not downregulate CDX2 expression. These results suggest that in contrast to murine embryos, TEAD4 might be not an indispensable factor for the formation and differentiation of TE in bovine embryos.
Kaneko and DePamphilis [8] reported that Tead4 deficient murine embryos are capable of forming blastocyst when cultured under low oxidative stress conditions. They also proposed that Tead4 is required to prevent oxidative stress during early embryonic development. Glucose and O2 consumption are increased in murine embryos between the morula and blastocyst stages [21,22,23], implying that generation of mitochondrial reactive oxygen species (ROS) is also sharply increased [24]. Tead4 is involved in preventing excess accumulation of ROS in murine embryos [8]. In bovine embryos, the similar patterns of glucose and O2 consumption have been reported [25,26,27]. In general, 5% O2 is adopted as a standard culture condition for bovine embryos to protect them from oxidative stress [28]. Although TEAD4-downregulated embryos were able to develop to the blastocyst stage under a low O2 condition, it is possible that problems that occur in TEAD4-downregulated embryos were rescued by low O2 tension. We therefore observed the developmental capacity of TEAD4-downregulated embryos under the atmospheric oxygen condition. However, there was no significant effect of TEAD4 downregulation on embryonic development, which implies that TEAD4 might not exert protective effect from oxidative stress in bovine embryos. However, knowledge regarding the relationship between TEAD4 and energy homeostasis in mammalian embryos is limited, and further studies are required to investigate this relationship.
In the present study, we demonstrated that TEAD4 might be dispensable for embryonic development and TE differentiation in bovine embryos. Results suggest that mechanisms regulating embryonic development until the blastocyst stage and differentiation mechanisms of ICM and TE vary among species. TE differentiation in bovine embryos remains incomplete at the blastocyst stage, but is completed at the elongation stage [19]. Although we did not determine the exact function of TEAD4 until the blastocyst stage, investigating the role of TEAD4 from the blastocyst to elongation stage is necessary. In the present study, we downregulated TEAD4 expression using RNA interference. However, this method does not completely eliminate the expression of the target gene. Therefore, the possibility that the remaining TEAD4 expression was sufficient to maintain the normal development and differentiation cannot be ruled out. Further studies are required to elucidate functions of TEAD4 in bovine embryos.
Acknowledgments
This work was supported by JSPS KAKENHI Grant Number 26292162.
References
- 1.Pedersen RA, Wu K, Bałakier H. Origin of the inner cell mass in mouse embryos: cell lineage analysis by microinjection. Dev Biol 1986; 117: 581–595. [DOI] [PubMed] [Google Scholar]
- 2.Nishioka N, Yamamoto S, Kiyonari H, Sato H, Sawada A, Ota M, Nakao K, Sasaki H. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech Dev 2008; 125: 270–283. [DOI] [PubMed] [Google Scholar]
- 3.Yagi R, Kohn MJ, Karavanova I, Kaneko KJ, Vullhorst D, DePamphilis ML, Buonanno A. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development 2007; 134: 3827–3836. [DOI] [PubMed] [Google Scholar]
- 4.Ralston A, Cox BJ, Nishioka N, Sasaki H, Chea E, Rugg-Gunn P, Guo G, Robson P, Draper JS, Rossant J. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development 2010; 137: 395–403. [DOI] [PubMed] [Google Scholar]
- 5.Wu G, Gentile L, Fuchikami T, Sutter J, Psathaki K, Esteves TC, Araúzo-Bravo MJ, Ortmeier C, Verberk G, Abe K, Schöler HR. Initiation of trophectoderm lineage specification in mouse embryos is independent of Cdx2. Development 2010; 137: 4159–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishioka N, Inoue K, Adachi K, Kiyonari H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki N, Makita R, Kurihara H, Morin-Kensicki EM, Nojima H, Rossant J, Nakao K, Niwa H, Sasaki H. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell 2009; 16: 398–410. [DOI] [PubMed] [Google Scholar]
- 7.Sasaki H. Mechanisms of trophectoderm fate specification in preimplantation mouse development. Dev Growth Differ 2010; 52: 263–273. [DOI] [PubMed] [Google Scholar]
- 8.Kaneko KJ, DePamphilis ML. TEAD4 establishes the energy homeostasis essential for blastocoel formation. Development 2013; 140: 3680–3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakurai N, Takahashi K, Emura N, Fujii T, Hirayama H, Kageyama S, Hashizume T, Sawai K. The necessity of OCT-4 and CDX2 for early development and gene expression involved in differentiation of inner cell mass and trophectoderm lineages in bovine embryos. Cell Reprogram 2016; 18: 309–318. [DOI] [PubMed] [Google Scholar]
- 10.Sakurai T, Sakamoto A, Muroi Y, Bai H, Nagaoka K, Tamura K, Takahashi T, Hashizume K, Sakatani M, Takahashi M, Godkin JD, Imakawa K. Induction of endogenous interferon tau gene transcription by CDX2 and high acetylation in bovine nontrophoblast cells. Biol Reprod 2009; 80: 1223–1231. [DOI] [PubMed] [Google Scholar]
- 11.Fujii T, Moriyasu S, Hirayama H, Hashizume T, Sawai K. Aberrant expression patterns of genes involved in segregation of inner cell mass and trophectoderm lineages in bovine embryos derived from somatic cell nuclear transfer. Cell Reprogram 2010; 12: 617–625. [DOI] [PubMed] [Google Scholar]
- 12.Home P, Saha B, Ray S, Dutta D, Gunewardena S, Yoo B, Pal A, Vivian JL, Larson M, Petroff M, Gallagher PG, Schulz VP, White KL, Golos TG, Behr B, Paul S. Altered subcellular localization of transcription factor TEAD4 regulates first mammalian cell lineage commitment. Proc Natl Acad Sci USA 2012; 109: 7362–7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamashita S, Abe H, Itoh T, Satoh T, Hoshi H. A serum-free culture system for efficient in vitro production of bovine blastocysts with improved viability after freezing and thawing. Cytotechnology 1999; 31: 123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bavister BD, Leibfried ML, Lieberman G. Development of preimplantation embryos of the golden hamster in a defined culture medium. Biol Reprod 1983; 28: 235–247. [DOI] [PubMed] [Google Scholar]
- 15.Hirate Y, Cockburn K, Rossant J, Sasaki H. Tead4 is constitutively nuclear, while nuclear vs. cytoplasmic Yap distribution is regulated in preimplantation mouse embryos. Proc Natl Acad Sci USA 2012; 109: E3389–E3390, author reply :E3391–E3392; author reply E3391−3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rayon T, Menchero S, Nieto A, Xenopoulos P, Crespo M, Cockburn K, Cañon S, Sasaki H, Hadjantonakis AK, de la Pompa JL, Rossant J, Manzanares M. Notch and hippo converge on Cdx2 to specify the trophectoderm lineage in the mouse blastocyst. Dev Cell 2014; 30: 410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma GT, Roth ME, Groskopf JC, Tsai FY, Orkin SH, Grosveld F, Engel JD, Linzer DI. GATA-2 and GATA-3 regulate trophoblast-specific gene expression in vivo. Development 1997; 124: 907–914. [DOI] [PubMed] [Google Scholar]
- 18.Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 2005; 132: 2093–2102. [DOI] [PubMed] [Google Scholar]
- 19.Berg DK, Smith CS, Pearton DJ, Wells DN, Broadhurst R, Donnison M, Pfeffer PL. Trophectoderm lineage determination in cattle. Dev Cell 2011; 20: 244–255. [DOI] [PubMed] [Google Scholar]
- 20.Goissis MD, Cibelli JB. Functional characterization of CDX2 during bovine preimplantation development in vitro. Mol Reprod Dev 2014; 81: 962–970. [DOI] [PubMed] [Google Scholar]
- 21.Houghton FD, Thompson JG, Kennedy CJ, Leese HJ. Oxygen consumption and energy metabolism of the early mouse embryo. Mol Reprod Dev 1996; 44: 476–485. [DOI] [PubMed] [Google Scholar]
- 22.Johnson MT, Mahmood S, Patel MS. Intermediary metabolism and energetics during murine early embryogenesis. J Biol Chem 2003; 278: 31457–31460. [DOI] [PubMed] [Google Scholar]
- 23.Trimarchi JR, Liu L, Porterfield DM, Smith PJ, Keefe DL. Oxidative phosphorylation-dependent and -independent oxygen consumption by individual preimplantation mouse embryos. Biol Reprod 2000; 62: 1866–1874. [DOI] [PubMed] [Google Scholar]
- 24.Dumollard R, Carroll J, Duchen MR, Campbell K, Swann K. Mitochondrial function and redox state in mammalian embryos. Semin Cell Dev Biol 2009; 20: 346–353. [DOI] [PubMed] [Google Scholar]
- 25.de Souza DK, Salles LP, Rosa e Silva AA. Aspects of energetic substrate metabolism of in vitro and in vivo bovine embryos. Braz J Med Biol Res 2015; 48: 191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khurana NK, Niemann H. Energy metabolism in preimplantation bovine embryos derived in vitro or in vivo. Biol Reprod 2000; 62: 847–856. [DOI] [PubMed] [Google Scholar]
- 27.Thompson JG, Partridge RJ, Houghton FD, Cox CI, Leese HJ. Oxygen uptake and carbohydrate metabolism by in vitro derived bovine embryos. J Reprod Fertil 1996; 106: 299–306. [DOI] [PubMed] [Google Scholar]
- 28.Harvey AJ. The role of oxygen in ruminant preimplantation embryo development and metabolism. Anim Reprod Sci 2007; 98: 113–128. [DOI] [PubMed] [Google Scholar]