Abstract
Background:
Salmonella enterica serovar Typhi, as causative agent of typhoid fever, is one of the most important endemic pathogens. Non-typhoidal Salmonella serovars, including Typhimurium, Infantis, and Enteritidis are amongst the most prevalent serotypes worldwide and in developing areas such as Iran. The aim of this study was to apply a uniplex PCR for rapid detection of Salmonella spp., and a multiplex PCR for the simultaneous detection of the four most common Salmonella serovars in Iran.
Methods:
Current research was done in 2010 at Molecular Biology Research Center, Baqiyatallah University of Medical Sciences, Tehran, Iran. For detection of Salmonella spp a pair of primers was used to replicate a chromosomal sequence. Four other sets of primers were also designed to amplify the target genes of four Salmonella species including S. typhi, and three non-typhoidal Salmonella spp (S. enteritidis, S. infantis, and S. typhimurium). The assay specificity was investigated by testing 15 different Salmonella serovars and 8 other additional non-Salmonella species.
Results:
The Salmonella genus-specific PCR yielded the expected DNA band of 404 bp in all Salmonella spp., strains tested. The uniplex and multiplex PCR assays produced also the expected fragments of 489 bp, 304 bp, 224 bp, and 104 bp for serovars Typhi, Enteritidis, Typhimurium, and Infantis, respectively. Each species-specific primer pair set did not show any cross-reactivity when tested on other Salmonella serovars or other non- but related- Salmonella strains.
Conclusion:
Both uniplex and multiplex PCR protocols had a good specificity. They can provide an important tool for the rapid and simultaneous detection and differentiation of the four most prevalent Salmonella serovars in Iran.
Keywords: Salmonella spp, Rapid detection, Multiplex PCR
Introduction
Salmonella is considered as one of the most important causes of acute gastroenteritis and food-borne infections worldwide (1). Gastroenteritis and diarrheal diseases remain one of the most important health problems worldwide, especially in developing countries (2–3). Salmonella species are prevalent throughout of the world (4) including Iran (5). This organism can cause a range of clinical outcomes, from an auto-limited gastroenteritis to a systemic infection that compromises the patient’s life (6). The genus Salmonella belongs to the family Enterobacteriaceae and consists of two species, S. enterica and S. bongori (7). S. enterica is divided into the following six subspecies: S. enterica subsp. enterica (I), S. enterica subsp. salamae (II), S. enterica subsp. arizonae (IIIa), S. enterica subsp. diarizonae (IIIb), S. enterica subsp. houtenae (IV) and S. enterica subsp. ndica (VI) (8).
At present, over 2600 serovars have been identified, but 99% of the Salmonella strains isolated from humans and warm-blooded animals usually belong to group I (S. enterica subsp. enterica) (9).
Systemic infection caused by Salmonella is usually host-dependent (10). Young peoples who have a weakened immune system and animals under the stress are more vulnerable to systemic Salmonella infections (11). Salmonella enterica servar Typhi (S. typhi) causes only systemic infection –typhoid fever–in humans (10, 12).
Typhoid fever as an endemic infectious disease in many developing countries is most important health problems (13) and sometimes is imported in developed countries from travelers or migrants (14, 15). Infections caused by Salmonella spp. are increasing in Iran (16).
The US Centers for Disease Control and Prevention (CDC) reported that Nontyphoidal Salmonellae were responsible for around 1.4 million cases of gastrointestinal disease with the mortality rates of 500 deaths each year in United States (17). Among the non-typhoidal Salmonella serovars, Typhimurium, Infantis, and Enteritidis are amongst the most prevalent serotypes worldwide and in developing areas such as Iran, Southeast Asia and Africa (18–24).
Culture-based microbiological procedures and biochemical/ serological tests are most commonly used methods for detection and identification of Salmonella spp. however, these methods are time-consuming resulting in delay in diagnosis, treatment and control of infections. Hence, there is a need to develop “quick and cheap” molecular methods for a rapid and simultaneous detection of the most common Salmonella serovars (16, 25, 26). In recent years, some new molecular methods are introduced for rapid detection of Salmonella particularly those techniques with good reproducibility and rapidity. PCR and similar nucleotide-based methods have become potentially powerful alternative approaches in microbiological diagnostics because of their higher user-friendliness, rapidity, reproducibility, accuracy and affordability (12, 16, 26–28).
Multiplex PCR is considered as a rapid molecular approach for simultaneous detection of several targets a single amplification reaction. This technique is frequently evaluated in order to assess the possible presence of microbial pathogens causing foodborne diseases (12, 25, 29– 34).
The aim of this study was to apply a uniplex PCR for rapid detection of Salmonella spp., and a multiplex PCR for the simultaneous detection of the four most common Salmonella serovars in Iran.
Materials and Methods
Bacterial strains
Twenty-three strains including 15 Salmonella spp and 8 non-Salmonella spp were used for the test. Salmonella isolates included 12 clinical and 3 standard strains belonging to different serovars. Clinical Salmonella isolates were recovered from patients with Salmonella infections admitted to including Children’s Medical Center and Baqiyatallah Hospital in Tehran, Iran, during 2008–2010.
As shown in Table 1, the bacterial positive controls used in this study were S. typhi (ATCC 19430), S. typhimurium (ATCC 14028), S. infantis (clinical strains) and S. enteritidis (ATCC 4931). Additional bacterial pathogens including Campylobacter jejuni (ATCC 33560), Escherichia coli (ATCC 25922), Enterococcus faecalis (PTCC 1393), Klebsiella oxytoca (ATCC 68831), Shigella sonnei (ATCC 9290), Vibrio cholerae (PTCC 1611), Proteus mirabilis (PTCC 1076), and Serratia marcescens (ATCC 14223) were used to check the specificity of the assay.
Table 1:
Salmonella species and serovars and non-Salmonella micro organisms included in this study
| Strains | Reference | Salmonella spp-specific PCR results | S. Typhimurium-specific PCR results | S. Typhi - specific PCR results | S. Enteritidis - specific PCR results | S. Infantis-specific PCR results |
|---|---|---|---|---|---|---|
| Salmonella spp. | ||||||
| S. Albany | Clinical strain | + | − | − | − | − |
| S. Enteritidis | ATCC 4931 | + | − | − | + | − |
| S. Hadar | Clinical strain | + | − | − | − | − |
| S. Haifa | Clinical strain | + | − | − | − | − |
| S. Havana | Clinical strain | + | − | − | − | − |
| S. Infantis | Clinical strain | + | − | − | − | + |
| S. Kentucky | Clinical strain | + | − | − | − | − |
| S. Mnchen | Clinical strain | + | − | − | − | − |
| S. Newport | Clinical strain | + | − | − | − | − |
| S. Orion | Clinical strain | + | − | − | − | − |
| S. Paratyphi B | Clinical strain | + | − | − | − | − |
| S. Reading | Clinical strain | + | − | − | − | − |
| S. Richmond | Clinical strain | + | − | − | − | − |
| S. Typhi | ATCC 19430 | + | − | + | − | − |
| S. Typhimurium | ATCC 14028 | + | + | − | − | − |
| Non-Salmonella organisms | ||||||
| Campylobacter jejuni | ATCC 33560 | − | − | − | − | − |
| Enterococcus faecalis | PTCC 1393 | − | − | − | − | − |
| Escherichia coli | ATCC 25922 | − | − | − | − | − |
| Klebsiella oxytoca | ATCC 68831 | − | − | − | − | − |
| Shigella sonnei | ATCC 9290 | − | − | − | − | − |
| Vibrio cholerae | PTCC 1611 | − | − | − | − | − |
| Proteus mirabilis | PTCC 1076 | − | − | − | − | − |
| Serratia marcescens | ATCC 14223 | − | − | − | − | − |
Identification of the references and clinical strains was confirmed by culture, biochemical testing by the API test system (BioMérieux, Marcy-l’Étoile, France) and slide agglutination with serovar specific antisera (Staten Serum Institute, Copenhagen, Denmark) (5, 28). All strains were grown on Trypticase Soy Broth (TSB) and incubated at 37 °C for 18 to 24 h to obtain a fresh culture prior to DNA extraction.
Bacterial DNA extraction
Three ml of an overnight culture of each Salmonella isolate in LB broth were centrifuged at 9000 rpm for 10 min.
Genomic DNA of the Salmonella strains was extracted using a DNA extraction Kit (Roche, Germany) according to the manufacturer’s instruction. The supernatant containing the DNA was transferred to a clean tube and stored at −20 °C until used for PCR.
Genomic PCR targets and primers
Five sets of primers were designed to amplify the target genes of Salmonella spp and of four Salmonella serovars, and the three most prevalent nontyphoidal Salmonella serovars in Iran. The list of the primers and their sequences are presented in Table 2. To avoid cross-reactivity with Salmonella related bacteria and within each other Salmonella serovar, genus and species specific regions of the Salmonella genome were considered to design the primers, respectively. The target genes included invA for Salmonella spp., STY4669 (a hypothetical protein) for Typhi; STMO159 (a putative restriction endonuclease) for Typhimurium; SEN1383 (a hypothetical protein) for Enteritidis, and M. sini (a modification methylase) for Infantis.
Table 2:
The primers and their sequences used in the study
| Bacterial strains | Target gene | Sequence of the primers | PCR Size (bp) |
|---|---|---|---|
| Salmonella spp | invA-secretory protein | F: 5′-GTATTGTTGATTAATGAGATCCG-3′ R: 5′-ATATTACGCACGGAAACACGTT-3′ |
404 |
| S. Typhi | STY4669 - hypothetical protein | F: 5′-TGTCCGCTGTCTGAAGTCATC-3′ R: 5′-ATCTCAGGCAAACTCACAAGGG-3′ |
489 |
| S. Typhimurium | STM0159 - restriction endonuclease | F: 5′-ATGATGCCTTTTGCTGCTTT-3′ R: 5′-TCCCAGCTCATCCAAAAA-3′ |
224 |
| S. Enteritidis | SEN1383 - hypothetical protein | F: 5′-TGTGTTTTATCTGATGCAAGAGG-3′ R: 5′-TGAACTACGTTCGTTCTTCTGG-3′ |
304 |
| S. Infantis | M.SinI -modification methylase | F: 5′-CACAATGAACGTGGTGAAGG-3′. R: 5′-CGTCCCGCGAACATATTATT-3′ |
184 |
Uniplex PCR and Multiplex PCR
A uniplex PCR assay was first evaluated using bacterial colonies isolated from pure cultures of clinical strains of each Salmonella serovar. The concentration of each primer pair, magnesium chloride concentration, and primer annealing temperature were optimized before checking for the specificity of the primer pairs. The optimized PCR was carried out by using a total volume of 25 μL containing 1× PCR buffer, 1 mM MgCl2, 1 U Taq DNA polymerase (Fermentas, Lithuania), 200 μM dNTPs (Fermentas, Lithuania), 0.5 μM of each primers and 2.5 μL of DNA template. The PCR program used for amplification consisted of 5 min at 95 °C, followed by 30 cycles of 60 sec at 95 °C of denaturing temperature, 60 sec at 57 °C of annealing temperature, and 5 min at 72 °C of extension temperature. At the end of the 30 cycles, a 10 min extension at 72 °C was used. Each multiplex PCR mixture in one reaction was prepared by using a total volume of 25 μL containing 0.5 μM of each primer (five pairs), 2,5 μL PCR buffer 10X, 1.5 U Taq DNA polymerase (Fermentas, Lithuania), 1.5 mM MgCl2, 200 μM dNTPs (Fermentas, Lithuania) and 1 μL DNA template. The multiplex PCR was carried out through 30 cycles following a pre-heat step at 95 °C for 5 min. Each cycle consisted of denaturation at 95 °C for 60 sec, annealing at 57 °C for 1min, and extension at 72 °C for 1min. After the 30 cycles, samples were maintained at 72 °C for 10 min. Sterile distilled water was included in each PCR assay as a negative control. The amplified DNA was separated by 1.5% agarose gel electrophoresis, stained with ethidium bromide, and visualized by UV transillumination. To prevent any contamination, reaction mixture preparation, DNA amplification and gel migration were done in separate rooms.
Results
Both uni and multiplex PCR assays produced the expected fragments bands when applied on standard and clinical strains of Salmonella.
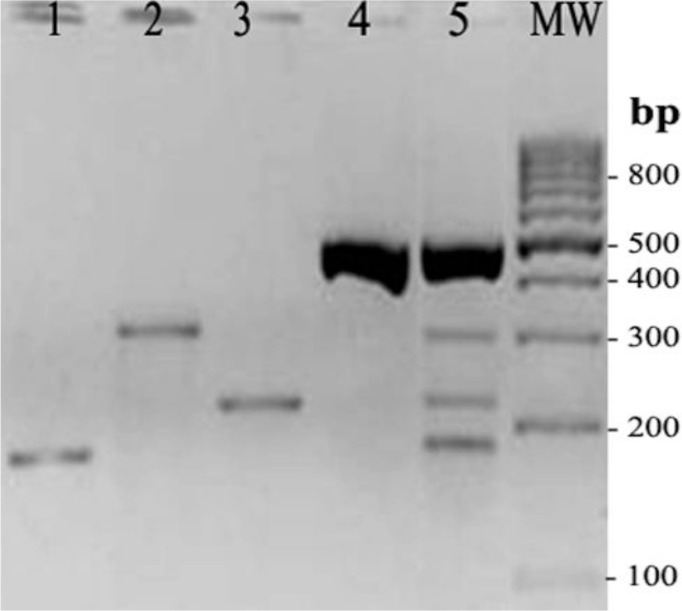
Uniplex PCR targeting for Salmonella genus specific gene showed the expected amplified DNA band in all Salmonella spp. strains tested and any non-specific reaction with other non Salmonella spp., strains was seen. Fig. 1 shows the specific band of 404 bp obtained from some Salmonella serovars tested. Multiplex PCR assays produced the expected fragments of 489 bp, 304 bp, 224 bp, and 104 bp for serovars Typhi, Enteritidis, Typhimurium and Infantis, respectively. No amplification products were observed with any of the other Salmonella serovars as well as with non Salmonella strains indicating the good specificity of the test. Fig. 2 shows the specifically amplified bands obtained by multiplex PCR on the four Salmonella serovars.
Fig. 1:
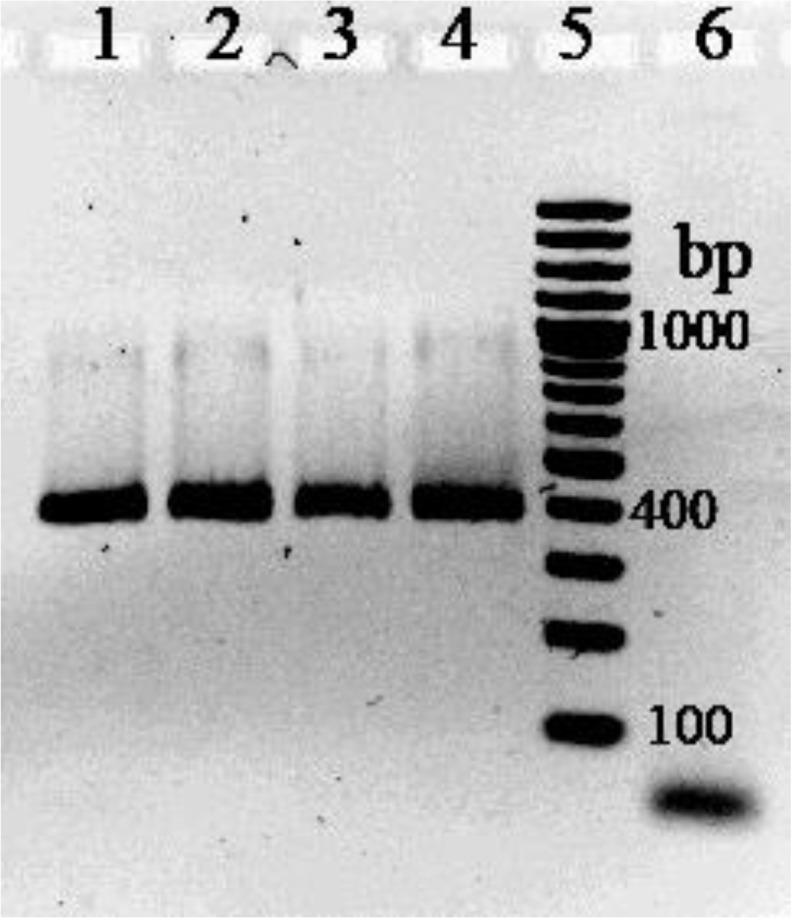
Salmonella spp specific PCR. Lanes 1–4: PCR products from some representative strains of S. Infantis, S. Enteritidis, S. Typhimurium and S. Typhi, respectively. Lane 5: molecular weight (100bp DNA ladder). Lane 6: E. coli
Fig. 2:
Multiplex PCR. Lanes 1–4: uniplex PCR of some representative strains of S. Infantis, S. Enteritidis, S. Typhimurium and S. Typhi, respectively. Lane 5: multiplex PCR for the same four Salmonella serovars in a single PCR tube. MW: molecular weight (100 bp DNA ladder)
Discussion
The invasion protein A gene is a good target for rapid and specific detection of Salmonella spp. This protein plays a part in cytoskeletal rearrangements for internalization of Salmonella spp. (35).
In a previous study, PCR using the invA primers has shown to be specific for detection of Salmonella spp (36). Our result confirmed the finding of previous reports, which indicated, this gene is present in nearly all Salmonella spp. (34, 35).
We also designed a new multiplex PCR with four sets of primers to identify Typhi and the most common non-typhoidal Salmonella serovars in our country. Accordingly, the designed method was able to detect all the Salmonella strains under study. Any non-specific reaction with other non S. enterica strains was not observed confirming that, this assay is specific for detection of Salmonella spp.
Several molecular techniques such as Multiplex PCR were evaluated for detection of foodborne pathogen organisms including Salmonella spp. However, among them; Multiplex PCR is considered as a rapid approach for simultaneous detection and identification of several targets in a mix-contaminated sample (12, 15, 25, 29–32). Optimization of reaction parameters and, in particular, of annealing temperature is very critical in this technique. The annealing temperature of 57 °C proved to be optimal for detection and differentiation of the four Salmonella serovars under study. The assessment of multiplex PCR analysis using standard and clinical samples showed that this method PCR was reliable for simultaneous detection of Salmonella strains belonging to different species. PCR results obtained from the clinical samples were consistent with results from the standard strains.
Some researchers have introduced some multiplex PCR for rapid detection and differentiation of endemic and prevalent Salmonella serovars in their countries (6,12,14,15,18).
Salmonella typhi and paratyphi A could be detected and differentiated using five sets of primer pairs targeting against the viaB, prt, tyv, fliC-d, and fliC-a genes (33). The conventional multiplex PCR evaluated to detect the causative agents of typhoid and paratyphoid fever based on tyv (rfbE), prt (rfbS) and invA genes. The multiplex PCR was indicated a potentially valuable tool for rapid diagnosis of typhoid Salmonella (15).
Multiplex PCR was a simple, inexpensive and sensitive molecular method for the rapid detection of the most prevalent Salmonella serovars in human clinical samples. They used six different primers for amplification of some target genes in Salmonella spp., DT104 and U302, S. C2 serogroup and Salmonella serovar 4,5,12:i:- and found their multiplex PCR assay can consider as an appropriate alternative approach for subtyping of Salmonella serovars (27).
Salmonella isolates identified in chicken abattoirs from Sao Paulo State, Brazil by a multiplex-PCR using three sets of primers targeting the invA, pefA, and sefA gene sequences from Salmonella spp., Typhimurium and Enteritidis, respectively (31).
The multiplex PCR designed to detect and identify Salmonella spp,. Enteritidis and Typhimurium strain isolated from fecal samples of diseased cattle, sheep and chicken in Egypt. They selected three PCR target sequences, which were invA specific for the genus Salmonella, spvA specific for Enteritidis and fliC gene specific for Typhimurium. Their results showed that Typhimurium and Enteritidis could be distinguished in less than 4 h (34).
A multiplex PCR introduced to detect Salmonella genus and serovars in refrigerated carcasses and chicken viscera and showed that the multiplex PCR was able to detect the presence of these serovars in a short period of time (25).
A multiplex PCR assay established to identify the seven major serovars of Salmonella, i.e., Typhimurium, Choleraesuis, Infantis, Hadar, Enteritidis, Dublin and Gallinarum circulating in Japan. Multiplex PCR had sufficient simplicity and specificity to identify the seven Salmonella serovars, and therefore, had the potential for use as rapid screening methods (12).
Ngan et al developed an mPCR assay for specific detection of serovars Typhi and Paratyphi A. They evaluated their assays with 124 clinical and reference strains of Salmonella serovars and found that S. enterica serovars Typhi and Paratyphi A can be detected with high specificity and sensitivity. They showed that mPCR could prove to be a useful diagnostic tool for the detection and differentiation of serovars Typhi and Paratyphi A (37).
Recently, an mPCR was described for identification of Salmonella spp. They could differentiate simultaneously different serovars of S. enterica (38).
We tested our method on DNA extracted from pure bacterial colonies. It is the major limitation of the study. To achieve a powerful assay for rapid detection and differentiation of Salmonella serovars, it is better to evaluate our method directly on clinical samples in the future.
The second limitation was to apply the test for rapid detection of other Salmonella serovars, however, they are not prevalent in our country. Moreover, optimization of multiplex PCR would be more difficult for simultaneous detection of more than 4 different bacterial species.
The choice of a multiplex PCR method, will depend on the type of microorganisms particularly those are most prevalent and the aim and scope of the study.
With the new more sophisticated molecular technologies, a new era seems to be opening in the field of diagnostic and molecular epidemiology of infectious diseases in which a single method will likely apply for detection of different targets (39).
Conclusion
The technique presented here showed to be specific for detection of Salmonella spp. and differentiation of the four Salmonella serovars tested. No false positive results occurred during the assay indicating that target genes used in the study were specific for these Salmonella serovars. The multiplex PCR using four primers sets was able to detect four serovars of Salmonella simultaneously in a single reaction by the combinations of the different size amplicons without any cross-reactivity. The method presented here showed a good specificity and proved to be able to provide an important diagnostic tool for the rapid and simultaneous detection of the four most prevalent serovars of Salmonella in Iran.
Ethical considerations
Ethical issues (Including plagiarism, informed consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc.) have been completely observed by the authors.
Acknowledgements
The authors declare that there is no conflict of interests.
References
- 1. Ranjbar R, Ahmadi M, Memariani M. (2016). Multiple-locus variable-number tandem repeat analysis (MLVA) for genotyping of Salmonella enterica subspecies enterica serotype Infantis isolated from human sources. Microb Pathog, 100: 299–304. [DOI] [PubMed] [Google Scholar]
- 2. Kumar SG, Subita L. (2012). Diarrhoeal diseases in developing countries: A situational analysis. Kathmandu Univ Med J (KUMJ), 10(38): 83–88. [DOI] [PubMed] [Google Scholar]
- 3. Ranjbar R, Salimkhani E, Sadeghifard N, Yazdi JZ, Morovvati S, Jonaidi N, Izadi M. (2007). An outbreak of gastroenteritis of unknown origin in Tehran, July 2003. Pak J Biol Sci, 10( 7): 1138–40. [DOI] [PubMed] [Google Scholar]
- 4. Ahmed AM, Younis EE, Ishida Y, Shimamoto T. (2009). Genetic basis of multidrug resistance in Salmonella enterica serovars Enteritidis and Typhimurium isolated from diarrheic calves in Egypt. Acta Trop, 111( 2): 144–9. [DOI] [PubMed] [Google Scholar]
- 5. Ranjbar R, Giammanco GM, Aleo A, Plano MR, Naghoni A, Owlia P, Mammina C. (2010). Characterization of the first extended-spectrum b-lactamase producing nontyphoidal Salmonella strains isolated in Tehran, Iran. Foodborne Pathog Dis, 7( 1): 91–5. [DOI] [PubMed] [Google Scholar]
- 6. Cardona-Castro N, Sánchez-Jiménez M, Lavalett L, Múñoz N, Moreno J. (2009). Development and evaluation of a multiplex polymerase chain reaction assay to identify Salmonella serogroups and serotypes. Diagn Microbiol Infect Dis, 65( 3): 327–30. [DOI] [PubMed] [Google Scholar]
- 7. Lopez FE, Mercedes Pescaretti ML, Morero R, Delgado MA. (2012). Salmoenlla Typhimurium general virulence factors: A battle of David against Goliath?. Food Res Int, 45( 2): 842–851. [Google Scholar]
- 8. Hendriksen SW, Orsel K, Wagenaar JA, Miko A, van Duijkeren E. (2004). Animal–to-human transmission of Salmonella typhimurium DT104A variant. Emerg Infect Dis, 10( 12): 2225–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stevens MP, Humphrey TJ, Maskell DJ. (2009). Molecular insights into farm animal and zoonotic Salmonella infections. Philos Trans R Soc Lond B Biol Sci, 364( 1530): 2709–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ruby T, McLaughlin L, Gopinath S, Monack D. (2012). Salmonella’s long-term relationship with its host. FEMS Microbiol Rev, 36( 3): 600–15. [DOI] [PubMed] [Google Scholar]
- 11. Yue M, Rankin SC, Blanchet RT, Nulton JD, Edwards RA, Schifferli DM. (2012). Diversification of the Salmonella Fimbriae: A Model of Macro- and Microevolution. PLoS One, 7 (6): e38596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Akiba M, Kusumoto M, Iwata T. (2011). Rapid identification of Salmonella enterica serovars, Thyphimurim, Choleraesuis, Infantis, Hadar, Enteritidis, Dublin and Gallinarum, by multiplex PCR. J Microbiol Methods, 85( 1): 9–15. [DOI] [PubMed] [Google Scholar]
- 13. Ranjbar R, Naghoni A, Farshad S, Lashini H, Najafi A, Sadeghifard N, Mammina C. (2014). Use of TaqMan® real-time PCR for rapid detection of Salmonella enterica serovar Typhi. Acta Microbiol Immunol Hung, 61( 2): 121–30. [DOI] [PubMed] [Google Scholar]
- 14. Karami A, Ranjbar R, Ahmadi Z, Safiri Z. (2007). Rapid Detection of Different Serovares of Salmonella entrica by Multiplex PCR. Iran J Publ Health, 36: 38–42. [Google Scholar]
- 15. Aarts HJ, Vos P, Larsson JT, van Hoek AH, Huehn S, Weijers T, Grønlund HA, Malorny B. (2011). A multiplex ligation detection assay for the characterization of Salmonella enterica strains. Int J Food Microbiol. 145 Suppl 1: S68– 78. [DOI] [PubMed] [Google Scholar]
- 16. Ranjbar R, Naghoni A, Yousefi S, Ahmadi A, Jonaidi N, Panahi Y. (2013). The Study of genetic relationship among third generation cephalosporin-resistant. Salmonella enterica strains by ERIC-PCR. Open Microbiol J, 7: 142–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV. (1999). Food-related illness and death in the United States. Emerg Infect Dis, 5( 5): 607–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Soumet C, Ermel G, Rose V, Rose N, Drouin P, Salvat G, Colin P. (1999). Identification by multiplex PCR based assay of Salmonella typhimurium and Salmonella enteritidis strains from environmental swabs of poultry houses. Lett Appl Microbiol, 29( 1): 1–6. [DOI] [PubMed] [Google Scholar]
- 19. Ranjbar R, Giammanco GM, Farshad S, Owlia P, Aleo A, Mammina C. (2011). Serotypes, antibiotic resistance and class 1 integrons in Salmonella isolates from pediatric cases of enteritis in Tehran, Iran. Foodborne Pathog Dis, 8( 4): 547–53. [DOI] [PubMed] [Google Scholar]
- 20. Herikstad H, Motarjemi Y, Tauxe RV. (2002). Salmonella surveillance: a global survey of public health serotyping. Epidemiol Infect, 129: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Salehi TZ, Tadjbakhsh H, Atashparvar N, Nadalian MG, Mahzounieh MR. (2007). Detection and identification of Salmonella typhimurium in bovine diarrhoeic fecal samples by immunomagnetic separation and multiplex PCR assay. Zoonoses Public Health, 54( 6–7): 231–6. [DOI] [PubMed] [Google Scholar]
- 22. Morpeth SC, Ramadhani HO, Crump JA. (2009). Invasive non-Typhi Salmonella disease in Africa. Clin Infect Dis, 49: 606–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Feasey NA, Dougan G, Kingsley RA, Heyderman RS, Gordon MA. (2012). Invasive non-typhoidal salmonella disease: an emerging and neglected tropical disease in Africa. Lancet, 379( 9835): 2489–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Van TT, Nguyen HN, Smooker PM, Coloe PJ. (2012). The antibiotic resistance characteristics of non-typhoidal Salmonella enterica isolated from food-producing animals, retail meat and humans in South East Asia. Int J Food Microbiol, 154( 3): 98–106. [DOI] [PubMed] [Google Scholar]
- 25. de Freitas CG, Santana AP, da Silva PH, Gonçalves VS, Barros Mde A, Torres FA, Murata LS, Perecmanis S. (2010). PCR multiplex for detection of Salmonella Enteritidis, Typhi and Typhimurium and occurrence in poultry meat. Int J Food Microbiol, 139( 1–2): 15–22. [DOI] [PubMed] [Google Scholar]
- 26. Chen J, Zhang L, Paoli GC, Shi C, Tu SI, Shi X. (2010). A real-time PCR method for the detection of Salmonella enterica from food using a target sequence identified by comparative genomic analysis. Int J Food Microbiol, 137( 2–3): 168–74. [DOI] [PubMed] [Google Scholar]
- 27. Alvarez J, Sota M, Vivanco AB, Perales I, Cisterna R, Rementeria A, Garaizar J. (2004). Development of a multiplex PCR technique for detection and epidemiological typing of salmonella in human clinical samples. J Clin Microbiol, 42( 4): 1734–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Naghoni A, Ranjbar R, Tabaraie B, Farshad S, Owlia P, Safiri Z, Mammina C. (2010). High prevalence of integron-mediated resistance in clinical isolates of Salmonella enterica. Jpn J Infect Dis, 63( 6): 417–21. [PubMed] [Google Scholar]
- 29. Kong RY, Lee SK, Law TW, Law SH, Wu RS. (2002). Rapid detection of six types of bacterial pathogens in marine waters by multiplex PCR. Water Res, 36( 11): 2802–12. [DOI] [PubMed] [Google Scholar]
- 30. Kim S, Frye JG, Hu J, Fedorka-Cray PJ, Gautom R, Boyle DS. (2006). Multiplex PCR-based method for identification of common clinical serotypes of Salmonella enterica subsp. enterica. J Clin Microbiol, 44( 10): 3608–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cortez AL, Carvalho AC, Ikuno AA, Bürger KP, Vidal-Martins AM. (2006). Identification of Salmonella spp. isolates from chicken abattoirs by multiplex-PCR. Res Vet Sci, 81( 3): 340–4. [DOI] [PubMed] [Google Scholar]
- 32. Amini K, Zahraei-Salehi T, Nikbakht Gh, Ranjbar R, Amini J, Ashrafganjooei SB. (2010). Molecular detection of invA and spv virulence genes in Salmonella enteritidis isolated from human and animals in Iran. Afr J Microbiol Res, 4( 21): 2202–10. [Google Scholar]
- 33. Hirose K, Itoh K, Nakajima H, Kurazono T, Yamaguchi M, Moriya K, Ezaki T, Kawamura Y, Tamura K, Watanabe H. (2002). Selective amplification of tyv (rfbE), prt (rfbS), viaB, and fliC genes by multiplex PCR for identification of Salmonella enterica serovars Typhi and Paratyphi A. J Clin Microbiol, 40( 2): 633–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nashwa MH, Mahmoud AH, Sami SA. (2009). Application of Multiplex Polymerase Chain Reaction (MPCR) for Identification and Characterization of Salmonella enteritidis and Salmonella Typhimurium. J Appl Sci Res, 5( 12): 2343–2348. [Google Scholar]
- 35. Swamy SC, Barnhart HM, Lee MD, Dreesen DW. (1996). Virulence determinants invA and spvC in Salmonella isolated from poultry products, wastewater, and human sources. Appl Environ Microbiol, 62( 10): 3768–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oliveira SD, Rodenbusch CR, Cé MC, Rocha SL, Canal CW. (2003). Evaluation of selective and non-selective enrichment PCR procedures for Salmonella detection. Lett Appl Microbiol, 36( 4): 217–21. [DOI] [PubMed] [Google Scholar]
- 37. Ngan GJ, Ng LM, Lin RT, Teo JW. (2010). Development of a novel multiplex PCR for the detection and differentiation of Salmonella enterica serovars Typhi and Paratyphi A. Res Microbiol, 161( 4): 243–8. [DOI] [PubMed] [Google Scholar]
- 38. Zhu C, Yue M, Rankin S, Weill FX, Frey J, Schifferli DM. (2015). One-step identification of five prominent chicken Salmonella serovars and biotypes. J Clin Microbiol, 53( 12): 3881–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ranjbar R, Karami A, Farshad S, Giammanco GM, Mammina C. (2014). Typing methods used in the molecular epidemiology of microbial pathogens: a how-to guide. New Microbiol, 37( 1): 1–15. [PubMed] [Google Scholar]