The impact of postexercise leucine ingestion on processes of skeletal muscle breakdown in older adults is not well understood. Additional postexercise leucine ingestion appears to further reduce autophagy, but it does not interfere with the increase in ubiquitin proteasome system markers or the breakdown of intact proteins in skeletal muscle of older men. Postexercise leucine ingestion may promote a healthier protein pool and favorable muscle adaptations in older adults through greater accretion of myofibrillar proteins.
Keywords: leucine, skeletal muscle, protein breakdown, autophagy, fractional breakdown rate
Abstract
Essential amino acid (EAA) ingestion enhances postexercise muscle protein synthesis, and, in particular, the anabolic response of older adults appears sensitive to the quantity of ingested leucine. The effect of leucine ingestion on muscle breakdown following resistance exercise (RE) is less understood. The purpose of this study was to identify the impact of postexercise leucine ingestion on the ubiquitin proteasome and autophagosomal-lysosomal systems following acute RE in older men. Subjects (72 ± 2 yr) performed RE and 1 h postexercise ingested 10 g of EAA containing a leucine quantity similar to quality protein (control, 1.8 g leucine, n = 7) or enriched in leucine (leucine, 3.5 g leucine, n = 8). Stable isotope infusion and muscle biopsies (vastus lateralis) obtained at rest and 2, 5, and 24 h postexercise were used to examine protein content (Western blot), mRNA expression (RT-quantitative PCR), and muscle protein fractional breakdown rate (FBR). Muscle-specific RING finger 1 mRNA increased in both groups at 2 and 5 h (P < 0.05). LC3 mRNA increased, and the LC3BII-to-LC3BI ratio decreased at all postexercise time points in control (P < 0.05). Conversely, LC3 mRNA only increased at 2 h, and the LC3BII-to-LC3BI ratio only decreased at 2 and 5 h in leucine (P < 0.05). Tumor necrosis factor receptor-associated factor-6 mRNA increased (P < 0.05) in control at 5 h. FBR was not statistically different between groups or from basal 24 h postexercise (P > 0.05). These data indicate that ingesting a larger quantity of leucine following RE may further reduce postexercise skeletal muscle autophagy in older men; however, it does not appear to influence the acute postexercise elevation in markers of the ubiquitin proteasome system or the breakdown of intact proteins.
NEW & NOTEWORTHY The impact of postexercise leucine ingestion on processes of skeletal muscle breakdown in older adults is not well understood. Additional postexercise leucine ingestion appears to further reduce autophagy, but it does not interfere with the increase in ubiquitin proteasome system markers or the breakdown of intact proteins in skeletal muscle of older men. Postexercise leucine ingestion may promote a healthier protein pool and favorable muscle adaptations in older adults through greater accretion of myofibrillar proteins.
aging is characterized by a gradual loss of skeletal muscle mass (24), which contributes to reductions in whole muscle strength and function and has debilitating consequences for older adults. Specifically, the collective loss of muscle mass and function with aging, commonly referred to as sarcopenia (3), is associated with impaired physical function, mobility impairments, and a reduced ability to perform activities of daily living. This muscle dysfunction places older adults at an increased risk for falls, frailty, and dependence (3).
Skeletal muscle adaptation is dependent on a dynamic interplay between changes in muscle protein synthesis and muscle protein breakdown. Not only does this relationship dictate changes in skeletal muscle mass, but the turnover of muscle proteins also represents an important process to promote/preserve a healthier intramuscular protein pool (37, 51). In young individuals, ingesting essential amino acids (EAA) and/or protein shortly following a bout of resistance exercise (RE) facilitates a positive net protein balance largely through the enhancement of protein synthesis rather than a reduction in protein breakdown, although this has been largely studied only during the immediate hours after exercise (1, 36). On the other hand, while postexercise EAA ingestion appears to be a necessary strategy to elicit a protein synthesis response to exercise in older adults (9), the extent to which postexercise EAA ingestion impacts the processes that regulate muscle protein breakdown following exercise remains much less defined in older adults, in particular over an extended postexercise time course.
The breakdown of skeletal muscle protein following exercise is facilitated through both the ubiquitin proteasome (39) and the autophagosomal-lysosomal systems (13). While the ubiquitin proteasome system is largely responsible for the degradation of the myofibrillar proteins (44), the autophagosomal-lysosomal system involves the degradation of intracellular proteins, macromolecules, and organelles through the formation and subsequent degradation of autophagosomes (11). Both of these degradation pathways appear to be centrally controlled by the transcription factor forkhead box O3a (FoxO3a) (27, 42, 56). Specifically, FoxO3a is responsible for increasing the expression of two well-known E3 ubiquitin ligases, muscle-specific RING finger-1 (MuRF1) and muscle atrophy F-box, (MAFbx; atrogin-1) (42), as well as several genes involved in the induction of autophagy (27, 56). The activity of FoxO3a is inhibited through phosphorylation by Akt, which leads to the relocation of FoxO3a out of the nucleus and into the cytosol to inhibit its transcriptional activity. In addition to regulation through Akt/FoxO3a, recent data indicate that tumor necrosis factor (TNF) receptor-associated factor-6 (TRAF6) may coordinate the activity of the ubiquitin proteasome and autophagosomal-lysosomal systems in skeletal muscle (34). TRAF6 can be activated by fibroblast growth factor-inducible-14 (Fn14) (33), which is a TNF-like weak inducer of apoptosis receptor that has recently been shown to increase in expression in human skeletal muscle following RE (40).
The amino acid leucine has received considerable attention as a stimulator of growth-related cell signaling and translation initiation (15). In particular, aging muscle appears to have an anabolic sensitivity to different quantities of ingested leucine (19), and studies indicate that older adults may require greater than 2 g of ingested leucine to elicit a substantial protein synthesis response (7). Alternatively, leucine ingestion has also been shown in animal models to suppress protein breakdown (28, 29), and we have demonstrated in younger adults that higher doses of ingested leucine, in the absence of previous exercise, may reduce muscle protein breakdown, perhaps through a reduction in autophagy (15). While the sensitivity of aging muscle to the anabolic effect of leucine is well documented, the extent to which different quantities of ingested leucine impact the processes that regulate breakdown in aging skeletal muscle remains relatively unexplored. Therefore, the purpose of this study was to identify the impact of postexercise EAA ingestion on the molecular regulation of the ubiquitin proteasome and autophagosomal-lysosomal systems in skeletal muscle following acute RE in older adults, and in particular examine the role of postexercise leucine ingestion. We hypothesized that greater quantities of postexercise leucine ingestion would be associated with an attenuated overall induction of skeletal muscle protein breakdown, as assessed through various molecular markers.
MATERIALS AND METHODS
Fifteen healthy older men volunteered for this study. All participants were considered recreationally active but not engaged in a regularly scheduled exercise-training program. Screening was performed as previously described for these participants (6), with clinical history, physical examination, and laboratory tests, including complete blood count with differential, liver, and kidney function tests, coagulation profile, fasting blood glucose, oral glucose tolerance test, hepatitis B and C screening, HIV testing, thyroid-stimulating hormone, urinalysis, and drug screening. Maximal knee extensor muscle strength was determined for each subject on two separate occasions using a 1-repetition maximum (1 RM) performed on a leg extension device (Cybex-VR2, Medway, MA), as previously described (6). All participants gave informed, written consent before participation in the study, which was approved by the Institutional Review Board of the University of Texas Medical Branch (in compliance with the Declaration of Helsinki, as revised in 1983).
Study design.
Subjects were randomized to one of two treatment groups, control (n = 7) or leucine (n = 8) (Table 1). Both groups completed an identical 2-day experimental trial, with the exception of the composition of the EAA beverage (6, 15) that was ingested following RE (Table 2).
Table 1.
Subject characteristics
| Control | Leucine | |
|---|---|---|
| n | 7 | 8 |
| Age, yr | 74 ± 2 | 71 ± 3 |
| Height, cm | 174 ± 2 | 172 ± 3 |
| Weight, kg | 80 ± 4 | 80 ± 3 |
| Body mass index, kg/m2 | 26 ± 1 | 27 ± 1 |
| 1 RM, kg | 69 ± 2 | 76 ± 3 |
| 1 RM/leg lean mass | 3.9 ± 0.1 | 4.0 ± 0.2 |
Values are means ± SE. 1 RM, 1-repetition maximum (leg extension); 1 RM/leg lean mass, 1 RM (kg) divided by leg lean mass (kg; derived from duel-energy X-ray absorptiometry).
Table 2.
Essential amino acid beverage composition for the control and leucine groups
| Amino Acid | Control, g | Leucine, g |
|---|---|---|
| Histidine | 1.10 | 0.80 |
| Lysine | 1.55 | 1.20 |
| Methionine | 0.30 | 0.30 |
| Threonine | 1.45 | 1.00 |
| Phenylalanine | 1.55 | 1.40 |
| Valine | 1.20 | 1.00 |
| Isoleucine | 1.00 | 0.80 |
| Leucine | 1.85 | 3.50 |
| Total | 10.0 | 10.0 |
All experimental trials were conducted at the Institute for Translational Sciences Clinical Research Center (ITS-CRC) of the University of Texas Medical Branch. Subjects were admitted to the ITS-CRC the evening before the experimental trial and fed a standard dinner and a snack at 2200. All subjects were studied following an overnight fast under basal conditions and were asked to refrain from exercise for 24 h before the experimental trial. All experimental trials were conducted during the same time of day to avoid potential circadian changes. Whole and segmental body composition of all subjects was measured with a dual-energy X-ray absorptiometry scan (Hologic QDR 4500W, Bedford, MA).
At ~0600 on the morning of day 1 of the experimental trial, an 18-gauge polyethylene catheter was inserted into the antecubital vein for tracer infusion, and a separate catheter was inserted into a hand vein for heated arterialized blood sampling. After background blood sampling, primed continuous infusions of l-[15N]phenylalanine and l-[1-13C]leucine (Cambridge Isotope laboratory, Tewksbury, MA) were initiated and maintained at a constant rate. l-[15N]phenylalanine was infused for 3.5 h for the measurement of fractional breakdown rate (FBR, see below) as described (13), while l-[1-13C]leucine was infused for the duration of the experimental trial day for determination of blood and intracellular leucine concentrations. Each tracer was dissolved in sterile 0.9% saline and passed through a 2-μm filter. The priming doses for l-[15N]phenylalanine and l-[1-13C]leucine were 2 and 4.8 μmol/kg, respectively. The constant infusion rates for l-[15N]phenylalanine and l-[1-13C]leucine were 0.05 and 0.08 μmol·kg−1·min−1, respectively. Two hours into the tracer infusion, a basal muscle biopsy was obtained under sterile procedures and local anesthesia (1% lidocaine) from the lateral portion of the vastus lateralis using a 5-mm Bergström biopsy needle with suction. The muscle tissue was immediately blotted and frozen in liquid nitrogen and stored at −80°C until analysis. A second muscle biopsy was obtained from the same incision 2.5 h following the first biopsy (~1 h following termination of the l-[15N]phenylalanine tracer infusion).
Following the second biopsy, subjects were escorted to a Cybex leg extension machine on which they performed eight sets of 10 repetitions of bilateral leg extension RE at a mean intensity of ~65% (average of all 8 sets) of their predetermined 1 RM with 3 min of rest between sets, as our laboratory has previously described (4, 6, 9). On completion of the exercise bout, the subjects returned to their hospital bed and rested supine for the remainder of the study. At 1 h postexercise, subjects in the control group ingested 10 g of EAAs containing 1.85 g of leucine, whereas subjects in the leucine group ingested 10 g of EAAs containing 3.5 g of leucine (see below). Muscle biopsies were performed at 2 and 5 h postexercise (1 and 4 h postbeverage, respectively). Following collection of the 5-h postexercise muscle biopsy, day 1 was concluded, and subjects were given a standard lunch. Subjects were also fed a similar dinner and snack on the night before day 1.
The next morning (day 2), after an overnight fast, catheters were inserted at 0600 as described above for the primed, continuous infusions of l-[15N]phenylalanine and l-[1-13C]leucine and for arterialized blood sampling. l-[15N]phenylalanine was again continuously infused for 3.5 h, whereas l-[1-13C]leucine was infused for the duration of the experimental trial day. A fifth and sixth muscle biopsy were obtained at 2 and 4.5 h after initiation of the tracer infusion. The last biopsy time corresponded to 24 h following completion of exercise (~1 h following termination of the l-[15N]phenylalanine infusion).
Nutrient solution composition.
Ingested EAAs for both groups were provided in a double-blind fashion. The composition of the EAA mixture ingested by the control group was representative of high-quality protein and contained 18% leucine, whereas the mixture ingested by the leucine group contained 35% leucine (Table 2), as our laboratory has employed in previous studies (4, 5, 8, 9, 15). EAAs were individually weighed and dissolved in 350 ml of a noncaloric, noncaffeinated solution.
Cytosolic and nuclear extraction.
Frozen muscle tissue was weighed (34.4 ± 0.2 mg), placed in buffer, homogenized (1:9 wt/vol), and centrifuged at 3,400 g for 10 min at 4°C, followed by removal of the supernatant, which was used for Western blotting of cytosolic proteins (p/t Akt, FoxO3a, Beclin-1, LC3B). The resulting pellet was then suspended in isolation buffer (1 M sucrose, 1 M Tris·HCl, 1 M KCl, 0.5 M EDTA, pH 7.4) containing protease and phosphatase inhibitors and centrifuged for 10 min at 4°C and 700 g. After three series of PBS buffer suspensions and centrifugations at 15,000 g for 5 min at 4°C, the pellet was resuspended and agitated on ice for 2 × 20 min and in a 4°C sonication bath in high-salt buffer (1:4 wt/vol). The slurry was centrifuged at 15,000 g for 10 min at 4°C, and the supernatant was taken as the nuclear fraction and used for Western blotting of nuclear proteins (FoxO3a). Our laboratory has previously validated nuclear and cytosolic fractioning using these procedures (41).
Immunoblot analysis.
Total protein concentrations were determined in cytosolic and nuclear fractions using the Bradford assay (Smartspec Plus, Bio-Rad, Hercules, CA). Samples were diluted (1:1) in a 2 × sample buffer mixture containing 125 mM Tris, pH 6.8, 25% glycerol, 2.5% SDS, 2.5% β-mercaptoethanol, and 0.002% bromophenol blue and then boiled for 3 min at 100°C. Equal amounts of total protein (cytosol, 50 μg; nuclear, 20 μg) were loaded into each lane, and the samples were separated by electrophoresis (150 V for 60 min) on a polyacrylamide gel (Criterion, Bio-Rad) based on size of the target protein. Each sample was loaded in duplicate, and each gel contained an internal loading control and molecular weight ladder (Precision Plus, Bio-Rad). Following electrophoresis, protein was transferred to a polyvinylidene difluoride membrane (Bio-Rad) at 50 V for 60 min. Blots were then blocked for 1 h in 5% nonfat dry milk and incubated with primary antibody overnight at 4°C (total Akt, 1:1,000; AktThr308, 1:500; FoxO3a, 1:500; LC3B, 1:1,000; Beclin-1, 1:1,000). The next morning, blots were incubated with anti-rabbit IgG horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature. Blots were then incubated in a chemiluminescent solution (ECL plus, Amersham BioSciences, Piscataway, NJ) for 5 min, and optical density measurements were obtained with a phosphoimager (ChemiDoc, Bio-Rad), and densitometric analysis was performed using Quantity One 4.5.2 software (Bio-Rad). Immunoblot data were normalized to an internal loading control, which was loaded on all gels for comparison across blots, and data are adjusted to represent fold change from basal. The proportion of FoxO3a protein in the nucleus relative to the cytosol was determined as characterized previously (50) by the ratios of the normalized optical density measurements for the nucleus and cytosol at each biopsy time point. These relative optical density indexes were used to calculate fold change in the relative location of FoxO3a protein (nuclear-cytosolic) from basal. All antibodies were purchased from Cell Signaling Technologies (Beverly, MA).
RNA extraction and semiquantitative real-time PCR.
RNA isolation, cDNA synthesis, and real-time quantitative PCR were performed as our laboratory has previously described (10). Briefly, tissue was weighed (19.1 ± 0.2 mg) and homogenized with a handheld homogenizing dispenser (T10 Basic Ultra Turrax, IKA, Wilmington, NC) in 1 ml of TRI-Reagent. RNA concentration was determined using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE), and RNA was DNase treated using a commercially available kit (DNA-free, Ambion, Austin, TX). A total of 1 μg of RNA was reverse transcribed into cDNA according to the directions provided by the manufacturer (iScript, Bio-Rad, Hercules, CA). Real-time quantitative PCR was carried out with a CFX Connect Real-Time PCR Detection System (Bio-Rad). cDNA was analyzed with SYBR green fluorescence (iQ SYBR green supermix; Bio-Rad). Primer sequences not previously published (13) were designed using the National Center for Biotechnology Information database and carefully optimized (Table 3). β2-Microglobulin was utilized as a normalization/housekeeping gene. Relative fold changes were determined from the Ct values using the 2–ΔΔCt method (25).
Table 3.
Primer sequences used for real-time PCR (previously unpublished)
| Primer Sequence (5′-3′) |
||||
|---|---|---|---|---|
| mRNA Target | Accession No. | Forward | Reverse | Product Size, bp |
| ATG7 | NM_006395 | GCTCTTCCTTACTTCTTA | ATTGTTATCTTCGTCCTT | 104 |
| BECN1 | NM_003766 | GATACCGACTTGTTCCTTACG | TATAACGGCAGCTCCTTAGAT | 76 |
| BNIP3 | NM_004052 | CATCGGATTGGGGATCTATATT | ATCCACTAACGAACCAAGTC | 94 |
| GABARAP | NM_007278 | TGCCGGTGATAGTAGAAAAG | GTAGAACTGACCAACTGTGA | 98 |
| LAMP2B | NM_013995 | GTATTCTACAGCCCAAGAGTGTT | AATCAAGCCTGAAAGACCAGC | 85 |
| LC3 | NM_022818 | CCGCACCTTCGAACAAAGAG | AAGCTGCTTCTCACCCTTGT | 103 |
| CISD2 | NM_001008388 | TTCGCTAGGCTCACAGTTTCA | TCTTCGGGAGGAATGGACGA | 100 |
| RUNX1 | NM_001001890 | CTTTCAAGGTGGTGGCCCTA | CATGGCTGCGGTAGCATTTC | 110 |
| Fn14 | NM_016639 | GAAGTTCACCACCCCCATA | GATGAATGAATGATGAGTGGG | 116 |
| TRAF6 | NM_145803 | GGTCCGGAATTTCCAGGAAA | CATTTTAGCAGTCAGCTCCCG | 88 |
ATG7, autophagy related 7; BECN1, Beclin-1; BNIP3, BCL2/adenovirus E1B 19-kDa interacting protein 3; GABARAP, GABA(A) receptor-associated protein; LAMP2B, lysosomal-associated membrane protein 2 transcript variant B; LC3, microtubule associated protein 1 light chain 3; CISD2, CDGSH iron-sulfur domain-containing protein 2; RUNX1, runt-related transcription factor 1; Fn14, fibroblast growth factor-inducible 14; TRAF6, tumor necrosis factor receptor-associated factor 6.
Determination of leucine concentrations.
Concentrations of leucine were determined in blood and muscle intracellular fluid using [1-13C]leucine tracer enrichments and l-[5,5,5-2H3]leucine as the internal standard, as previously described (52). All tracer measurements were determined via gas chromatography-mass spectrometry (6890 Plus GC, 5973N MSD, 7683 autosampler, Agilent Technologies, Palo Alto, CA). Basal blood and muscle intracellular leucine concentrations, taken as the average of two preexercise blood and muscle samples, respectively, were subtracted from all time points, such that data are presented as absolute change from basal leucine concentration for both blood and muscle.
Calculation of FBR.
The FBR of muscle proteins was measured only at basal and 24 h postexercise as the ingestion of EAAs and associated changes in intracellular amino acid concentration (6) could have violated assumptions associated with this methodology (52). FBR was determined at these time points with l-[15N]phenylalanine using the precursor-product method (55), where the blood and muscle intracellular dilutions following cessation of tracer infusion are used to model FBR, and formulas, as our laboratory has previously described (13, 17). Briefly, this method requires measurement of intracellular free phenylalanine enrichment at steady state and after 1 h of tracer decay, along with frequent arterialized blood sampling, in addition to the free and bound phenylalanine content of the muscle. The first biopsy of day 1 and day 2 (biopsies 1 and 5, respectively) were used to establish intracellular free phenylalanine enrichment at steady state for day 1 (basal) and day 2 (24 h postexercise), while the second biopsy of each day (biopsies 2 and 6, respectively) were used to assess intracellular free phenylalanine enrichment after 1 h of tracer decay for day 1 and day 2, respectively. Arterialized blood was frequently sampled (5- to 10-min intervals) during the 1 h of tracer decay in an identical fashion for the measurement on day 1 and day 2. It was assumed that arterial blood is the only source of tracer entering the muscle intracellular free pool, such that there is no tracer recycling (55). Due to failure in two catheters during the breakdown period of day 1, FBR was determined for seven control and six leucine subjects.
Statistical analysis.
All data were tested for normality through skewness and kurtosis analyses and visual inspection of the normality plots using SPSS version 22 (IBM). For nonnormally distributed data, a natural log (ln) transformation was performed before statistical analyses. A two-way ANOVA with repeated measures on the time factor was used to test group by time differences. A Tukey’s post hoc analysis was used to determine specific differences within an ANOVA. An independent t-test was used to test for differences in subject characteristics between groups. Data analyses were conducted using SigmaStat version 12.0 (Systat Software). Significance for all analyses was set a priori at P < 0.05. For presentation of transformed data, appropriate back transformations were conducted, and data are presented as geometric mean ± 95% confidence interval. Transformed data are noted in respective figure and table captions. Nontransformed data are presented as means ± SE.
RESULTS
There were no group differences for 1 RM (Table 1) or for %1 RM performed during the exercise trial (average of all 8 sets: control, 64 ± 1%; leucine, 65 ± 1%) (P > 0.05). Similarly, there were no group differences for total weight lifted (sum of all exercise sets: control, 3,532 ± 152 kg; leucine, 3,969 ± 172 kg) or total weight lifted relative to leg lean mass (control, 198 ± 3 kg/kg leg lean mass; leucine, 208 ± 10 kg/kg leg lean mass) (P > 0.05) during the exercise trial.
Representative blots for all proteins are presented in Fig. 1. Cytosolic Akt phosphorylation (Thr308) was increased in both groups at 2 h postexercise (main effect of time, P = 0.06), whereas no changes in Akt phosphorylation from basal were observed at 5 h or 24 h postexercise in either group (Fig. 2A) (P > 0.05). No group differences in cytosolic Akt phosphorylation (Thr308) were observed at any time point (P > 0.05).
Fig. 1.
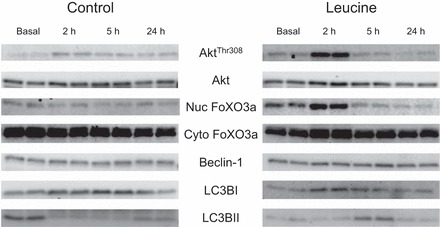
Representative immunoblot protein images for basal, and 2, 5, and 24 h following the combination of resistance exercise and postexercise ingestion of 10 g of essential amino acids containing 1.8 g of leucine (control) or 3.5 g of leucine (leucine). All samples were loaded in duplicate. Nuc, nuclear fraction; Cyto, cytosolic fraction; FoxO3a, forkhead box O3a; LC3, microtubule associated protein 1 light chain 3.
Fig. 2.
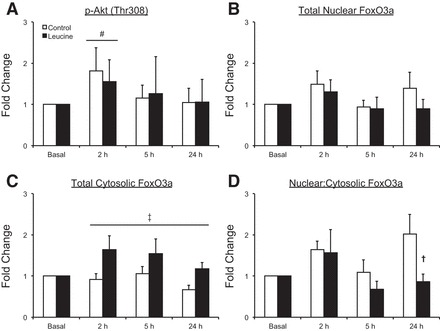
Time course of AktThr308 phosphorylation (A), forkhead box O3a (FoxO3a) protein in the nucleus (B), FoxO3a protein in the cytosol (C), and the abundance of FoxO3a protein in the nucleus relative to the cytosol (D) following the combination of resistance exercise and postexercise ingestion of 10 g of essential amino acids containing 1.8 g of leucine (control) or 3.5 g of leucine (leucine). Values represent fold change from basal. Values for AktThr308 were transformed and are presented as geometric means ± 95% confidence interval. All other values are presented as means ± SE; control, N = 7; leucine, N = 8. #Main effect of time, P = 0.06 vs. basal. ‡Main effect of group, P < 0.05. †P < 0.05 between groups. Representative blots are presented in Fig. 1.
Total nuclear FoxO3a protein was unchanged from basal (P > 0.05) in both groups (Fig. 2B), and no group differences in total nuclear FoxO3a protein were observed at any time point (P > 0.05). Total cytosolic FoxO3a protein was not statistically changed from basal in either group (P > 0.05); however, the leucine group had higher levels of total cytosolic FoxO3a protein during the postexercise period (main effect of group, P < 0.05) (Fig. 2C). No changes from basal in the ratio of nuclear to cytosolic FoxO3a protein were observed for either group (P > 0.05); however, this ratio was higher in the control group than in the leucine group at 24 h postexercise (P < 0.05) (Fig. 2D).
mRNA expression of MuRF1 was elevated above basal values in both groups at 2 h and 5 h postexercise (time effects, P < 0.05) and returned to basal values at 24 h postexercise in both groups (Fig. 3A). MAFbx mRNA expression was unchanged from basal values in both groups and at all time points (P > 0.05) (Fig. 3B). No group differences were observed for MuRF1 or MAFbx mRNA expression at any time point (P > 0.05).
Fig. 3.
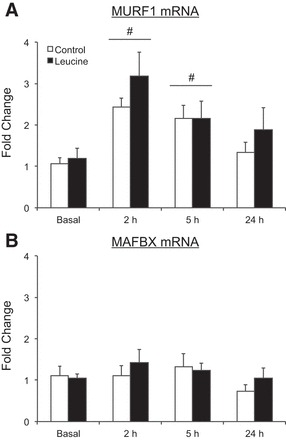
Time course of MuRF1 (A) and MAFbx (B) mRNA expression following the combination of resistance exercise and postexercise ingestion of 10 g of essential amino acids containing 1.8 g of leucine (control) or 3.5 g of leucine (leucine). Values represent fold change from basal and are presented as means ± SE; control, N = 7; leucine, N = 8. #Main effect of time, P < 0.05 vs. basal.
mRNA expression for several autophagy-related markers are displayed in Table 4. LC3 mRNA expression [necessary for autophagosome formation and degradation (47)] was elevated above basal values only at 2 h postexercise in the leucine group (P < 0.05), whereas LC3 mRNA expression was elevated above basal values at 2, 5, and 24 h in the control group (P < 0.05). Furthermore, the mRNA expression of CDGSH iron-sulfur domain-containing protein 2 (CISD2), a negative regulator of autophagy (2), was increased at 24 h postexercise only in the leucine group (P < 0.05). Alternatively, runt-related transcription factor 1 (RUNX1) mRNA expression, which has also been shown to be associated with reduced autophagy (48), was increased at 5 h and 24 h postexercise in both groups (P < 0.05), with a tendency for RUNX1 mRNA expression to be increased at 2 h postexercise in the leucine group (P = 0.08). No group or time differences (P > 0.05) were observed for the mRNA expression of additional positive regulators of autophagy: ATG7 [supports induction of autophagy (31)], BECN1 [supports autophagosome formation (18)], BNIP3 [supports induction of autophagy (54)], GABARAP [supports autophagosome formation (30)], or LAMP2B [supports fusion of autophagosome with the lysosome (46)].
Table 4.
mRNA analyses of positive and negative regulators of autophagy in the skeletal muscle of older men under basal conditions and after resistance exercise and the ingestion of 10 g of essential amino acids containing 1.8 g (control) or 3.5 g of leucine (leucine)
| Basal |
2 h |
5 h |
24 h |
|||||
|---|---|---|---|---|---|---|---|---|
| mRNA | Control | Leucine | Control | Leucine | Control | Leucine | Control | Leucine |
| Positive regulators of autophagy | ||||||||
| ATG7 | 1.08 ± 0.15 | 1.07 ± 0.14 | 1.06 ± 0.15 | 1.21 ± 0.15 | 1.04 ± 0.15 | 1.06 ± 0.17 | 1.08 ± 0.05 | 1.29 ± 0.23 |
| BECN1 | 1.01 ± 0.07 | 0.92 ± 0.05 | 1.07 ± 0.06 | 0.98 ± 0.05 | 1.25 ± 0.13 | 1.01 ± 0.08 | 1.22 ± 0.10 | 1.11 ± 0.11 |
| BNIP3 | 1.02 ± 0.09 | 1.09 ± 0.18 | 1.09 ± 0.10 | 1.13 ± 0.20 | 1.12 ± 0.11 | 1.13 ± 0.11 | 1.05 ± 0.10 | 0.96 ± 0.17 |
| GABARAP | 1.07 ± 0.16 | 1.02 ± 0.08 | 1.09 ± 0.15 | 1.29 ± 0.23 | 1.20 ± 0.16 | 1.18 ± 0.14 | 1.09 ± 0.16 | 1.22 ± 0.22 |
| LAMP2B | 1.08 ± 0.15 | 1.05 ± 0.12 | 1.19 ± 0.16 | 1.31 ± 0.23 | 1.25 ± 0.17 | 1.11 ± 0.10 | 0.96 ± 0.13 | 0.99 ± 0.21 |
| LC3 | 1.04 ± 0.12 | 1.02 ± 0.09 | 1.51 ± 0.20* | 1.39 ± 0.18* | 1.60 ± 0.18* | 1.29 ± 0.11 | 1.52 ± 0.19* | 1.32 ± 0.18 |
| Negative regulators of autophagy | ||||||||
| CISD2 | 1.03 ± 0.10 | 0.99 ± 0.12 | 0.95 ± 0.13 | 1.05 ± 0.14 | 1.06 ± 0.13 | 1.00 ± 0.09 | 1.35 ± 0.16 | 1.42 ± 0.25* |
| RUNX1 | 1.07 ± 0.95 | 1.16 ± 0.46 | 2.13 ± 1.05 | 4.51 ± 2.89† | 5.10 ± 2.84* | 6.91 ± 4.66* | 7.03 ± 3.69* | 12.02 ± 7.63* |
Values represent relative fold changes determined from the Ct values using the 2–ΔΔCt method (25). Data for runt-related transcription factor 1 (RUNX1) were transformed and are presented as geometric means ± 95% confidence interval. All other data are presented as means ± SE. ATG7, autophagy related 7; BECN1, Beclin-1; BNIP3, BCL2/adenovirus E1B 19-kDa interacting protein 3; GABARAP, GABA(A) receptor-associated protein; LAMP2B, lysosomal-associated membrane protein 2 transcript variant B; LC3, microtubule associated protein 1 light chain 3; CISD2, CDGSH iron-sulfur domain-containing protein 2.
P < 0.05 vs. basal.
P = 0.08 vs. basal.
Beclin-1 protein, which supports autophagosome formation (18), was unchanged from basal values in both groups at all time points (P > 0.05) (Fig. 4A). LC3BI protein (cytosolic isoform involved in autophagosome formation) was also unchanged from basal values in both groups (P > 0.05) (Fig. 4B). On the other hand, LC3BII, which is the conjugated, membrane-bound form of LC3BI that is recruited to autophagosomes during later stages of autophagy (47), was reduced from basal values at 2 h postexercise in both groups (main effect, P < 0.05), and it was lower in the control group compared with the leucine group at 5 h postexercise (P < 0.05) (Fig. 4C). The ratio of LC3BII to LC3BI (LC3BII/LC3BI) was reduced below basal values at 2 h and 5 h in the leucine group (P < 0.05), whereas this ratio was reduced at 2, 5, and 24 h in the control group (P < 0.05) (Fig. 4D). Furthermore, LC3BII/LC3BI was lower in the control group compared with the leucine group at 5 h and 24 h postexercise (P < 0.05).
Fig. 4.
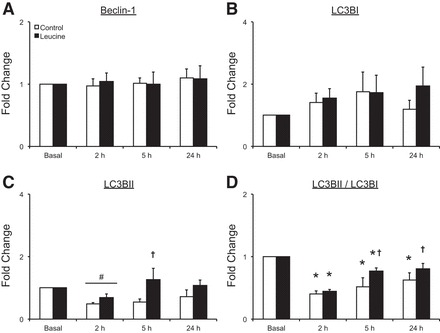
Time course of Beclin-1 protein (A), LC3 (microtubule associated protein 1 light chain 3) BI protein (B), LC3BII protein (C), and the ratio of LC3BII to LC3BI protein (D) following the combination of resistance exercise and postexercise ingestion of 10 g of essential amino acids containing 1.8 g of leucine (control) or 3.5 g of leucine (leucine). Values represent fold change from basal and are presented as means ± SE; control, N = 7; leucine, N = 8. #Main effect of time, P < 0.05 vs. basal. †P < 0.05 between groups. *P < 0.05 vs. basal. Representative blots are presented in Fig. 1.
The mRNA expression of Fn14 was increased at all postexercise time points in both groups (P < 0.05) (Fig. 5A). No group differences were observed for Fn14 mRNA expression at any time point (P > 0.05). TRAF6 mRNA expression was increased at 5 h in the control group (P < 0.05), whereas no changes in TRAF6 mRNA expression were observed at any time point for the leucine group (P > 0.05) (Fig. 5B). No group differences were observed for TRAF6 mRNA expression at any time point (P > 0.05).
Fig. 5.
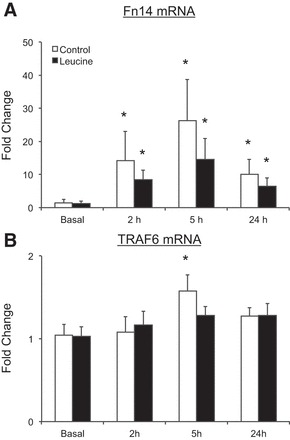
Time course of fibroblast growth factor-inducible 14 (Fn14; A) and tumor necrosis factor receptor-associated factor 6 (TRAF6; B) mRNA expression following the combination of resistance exercise and postexercise ingestion of 10 g of essential amino acids containing 1.8 g of leucine (control) or 3.5 g of leucine (leucine). Values represent fold change from basal. Values for Fn14 were transformed and are presented as geometric means ± 95% confidence interval. Values for TRAF6 are presented as means ± SE; control, N = 7; leucine, N = 8. *P < 0.05 vs. basal.
Blood leucine concentrations were elevated from basal only at 2 h postexercise in the control group (P < 0.05), whereas blood leucine concentrations were elevated at 2 h and 3 h postexercise in the leucine group (P < 0.05) (Table 5). Furthermore, the elevation from basal values in blood leucine concentrations at 2 h postexercise was greater in the leucine group (P < 0.05). Muscle intracellular leucine concentrations were not changed from basal at any time point in the control group (P > 0.05) (Table 5). Conversely, muscle intracellular leucine concentrations were elevated in the leucine group at 2 h postexercise (P < 0.05). Furthermore, the leucine group demonstrated a greater change in intracellular leucine concentrations at 2 h postexercise compared with control (P < 0.05).
Table 5.
Absolute change from basal for blood and muscle intracellular leucine concentrations after resistance exercise and the ingestion of 10 g of essential amino acids containing 1.8 g (control) or 3.5 g of leucine (leucine)
| Time Post-Resistance Exercise |
||||||
|---|---|---|---|---|---|---|
| Group | 1 h | 2 h | 3 h | 4 h | 5 h | 24 h |
| Blood leucine, absolute change from basal | ||||||
| Control | −12 ± 4 | 303 ± 42* | 135 ± 27 | 74 ± 13 | 53 ± 11 | 12 ± 6 |
| Leucine | −16 ± 3 | 663 ± 117*† | 225 ± 26* | 123 ± 13 | 97 ± 14 | 5 ± 3 |
| Muscle intracellular leucine, absolute change from basal | ||||||
| Control | 41 ± 15 | 9 ± 8 | −17 ± 12 | |||
| Leucine | 291 ± 50*† | 46 ± 11 | −10 ± 9 | |||
Values are means ± SE in μmol/l.
Different from basal, P < 0.05.
Group difference, P < 0.05.
Skeletal muscle FBR did not change from basal levels in either group, and no group differences were observed at basal or 24 h postexercise (P > 0.05) (Fig. 6).
Fig. 6.
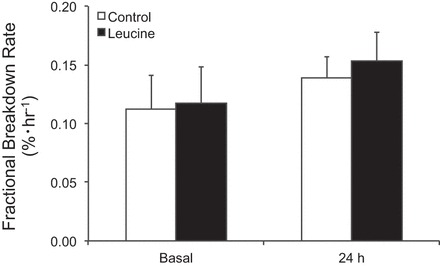
Skeletal muscle fractional breakdown rate following the combination of resistance exercise and postexercise ingestion of 10 g of essential amino acids containing 1.8 g of leucine (control) or 3.5 g of leucine (leucine).
DISCUSSION
Leucine has been characterized as a potent stimulator of muscle protein synthesis; however, its influence on postexercise protein breakdown in human skeletal muscle has received less attention. The results from the present study indicate that markers of the ubiquitin proteasome system are acutely elevated following the combination of RE and EAA ingestion in older men, and that the quantity of ingested leucine following RE appears to have minimal effects on this acute response or on the breakdown of intact proteins (as measured via FBR). On the other hand, while both groups showed a similar reduction in the LC3BII/LC3BI and increase in LC3 mRNA expression at 2 h postexercise, indicating reduced autophagosome formation in both groups, differences in these markers between groups at later time points suggest that larger quantities of ingested leucine may also facilitate a greater suppression of autophagosome degradation over a 24-h postexercise time course.
Our laboratory has previously demonstrated in young and older adults that RE, in the absence of postexercise nutrition, stimulates an acute upregulation of the ubiquitin proteasome system (13), the primary pathway responsible for myofibrillar protein breakdown (44). In the present study, coupling RE with postexercise EAA ingestion produced similar responses in the time course of Akt phosphorylation and the expression of MuRF1 and MAFbx mRNA as our laboratory’s previous findings (13) and those of others (45). While the present study was focused on different postexercise leucine quantities and did not include an RE-only group (e.g., no postexercise nutrition), comparison to these previous findings indicate that postexercise EAA ingestion has minimal influence on markers of the ubiquitin proteasome system that are responsive to RE in older adults. Furthermore, although the effects of ingested amino acids on muscle protein breakdown appear limited (1, 16), some data indicate that leucine may reduce protein breakdown (28, 29). While we did observe subtle differences in the cellular location of FoxO3a, expression of MuRF1 and MAFbx mRNA was similar in both groups. Thus it appears that greater amounts of ingested leucine do not interfere with the response of the ubiquitin proteasome system to RE, at least at the time points examined. Given that our laboratory has previously demonstrated in these subjects that leucine ingestion enhances myofibrillar protein synthesis over this postexercise time course (6), these data demonstrate that the accretion of myofibrillar proteins may be enhanced with higher postexercise leucine ingestion (i.e., similar response of the ubiquitin proteasome system and enhanced myofibrillar protein synthesis).
In agreement with previous human studies (12, 13, 43), we observed a reduced LC3BII/LC3BI in both groups 2 h post RE, largely due to a reduction in LC3BII. A change in the LC3BII/LC3BI alone can be difficult to interpret (21), as it can be indicative of either a reduction in autophagy flux (less conversation of LC3BI to LC3BII) or an increase in autophagy flux (increased degradation of LC3BII in the lysosome or reconversion to LC3BI). In the present study, the reduced LC3BII/LC3BI in both groups 2 h postexercise was coupled with limited changes in many autophagy-related genes (Table 4). Collectively, we interpret these findings to indicate that autophagy, specifically the formation of autophagosomes, was reduced from basal in both groups 2 h post-RE. Furthermore, this response was similar between groups, suggesting that the quantity of ingested leucine does not impact this reduction in autophagosome formation in skeletal muscle during the immediate few hours postexercise.
Interestingly, whereas the control group demonstrated a reduced LC3BII/LC3BI up to 24 h postexercise, the leucine group experienced a shorter time course that was coupled with a higher LC3BII/LC3BI at 5 and 24 h postexercise and higher LC3BII protein at 5 h compared with the control group. It is interesting to speculate that the higher LC3BII protein and LC3BII/LC3BI in the leucine group relative to the control group at later postexercise time points could signify less relative degradation of LC3BII, and thus a greater reduction in the degradation of formed autophagosomes compared with control. Indeed, the higher LC3BII/LC3BI in the leucine group relative to control 24 h postexercise coincided with a group-specific upregulation of CISD2, which negatively regulates autophagy (2). Furthermore, a greater reduction in autophagosome degradation in the leucine group would be supported by greater intracellular leucine availability, which reduces autophagic activity (15, 53), and sustained activation of mammalian target of rapamycin signaling (14), which our laboratory has previously published in these subjects (6). In addition, the control group demonstrated a sustained increase in the expression of LC3 mRNA up to 24 h postexercise relative to the leucine group (increased only at 2 h). Recent speculation suggests that a feedback mechanism, perhaps at the lysosome, may coordinate the later stages of autophagy (degradation) with the initial stages (autophagosome formation) (14). Thus, while it appears autophagy flux was generally reduced after exercise in both groups (relative to basal), the sustained upregulation of LC3 mRNA specific to the control group may indicate a greater suppression of autophagosome degradation in the leucine group.
Recent findings indicate that both the ubiquitin proteasome and autophagosomal-lysosomal systems may be regulated by the E3 ubiquitin ligase, TRAF6 (23, 34). Animal studies have demonstrated that knockout of TRAF6 prevents starvation-induced muscle atrophy, in conjunction with reduced activity of the ubiquitin proteasome and autophagosomal-lysosomal systems (33). TRAF6 can be activated by Fn14 (23), and recent data highlight that RE independently increases skeletal muscle Fn14 mRNA expression in both young and older adults (38, 40). In agreement with these studies, Fn14 mRNA expression was substantially increased in the present study, and this response was unaffected by the quantity of leucine ingested. On the other hand, TRAF6 mRNA was only increased in the control group, which occurred 5 h postexercise. Indeed, TRAF6 appears to be sensitive to nutrient status (32), and our data indicate that greater postexercise leucine ingestion may suppress this increase in TRAF6 mRNA. Thus it is interesting to speculate that suppression of TRAF6 in the leucine group could be responsible for what appears to be greater suppression of autophagosome degradation relative to control. Further research is warranted to more precisely uncover the role of TRAF6 in mediating postexercise autophagy in human skeletal muscle.
We did not observe a significant increase in FBR 24 h postexercise in either group. Interestingly, the ~27% overall increase observed in the present study 24 h post-RE is somewhat greater than the increase previously presented by our laboratory (13) and others (35). While the breakdown of intact proteins may have been influenced at earlier time points (35), the study design did not allow for the inclusion of earlier time points, as the influx of ingested EAAs could have violated assumptions of the methodology (see materials and methods). Furthermore, despite what appears to be a greater reduction in autophagosome degradation in the leucine group 24 h postexercise, no difference in FBR was observed between groups. FBR measures the breakdown of intact muscle proteins (52), whereas autophagy is associated with the breakdown of not only proteins, but also organelles and other macromolecules. Thus changes in autophagy flux following RE may be diluted by other processes (i.e., ubiquitin proteasome system) and not captured in the measurement of FBR post-RE. Alternatively, reductions in skeletal muscle autophagy following RE and nutrient ingestion could be specific to nonprotein sources.
The transcription factor RUNX1 has also been implicated in the regulation of muscle atrophy. Although the precise mechanisms have yet to be uncovered, knockout of RUNX1 increases autophagy (48), highlighting that RUNX1 may act as a negative regulator of autophagy. Interestingly, and despite potential differences in autophagosome degradation, RUNX1 mRNA was increased in both groups at 5 h and 24 h postexercise, which may be a response to reduce autophagosome formation, as opposed to degradation (48). In addition, RUNX1 has been identified in young individuals as a transcription factor involved in the remodeling of skeletal muscle to exercise (20, 26). Our data indicate that RE with postexercise EAA ingestion provides a stimulus to increase skeletal muscle RUNX1 mRNA expression in older men. Furthermore, RUNX1 has also been identified as a regulator of myosin heavy chain (MHC) IIa expression in skeletal muscle (48). RE can facilitate a transition toward MHC IIa fibers (49), and it has been shown by some to preferentially increase MHC IIa fiber cross-sectional area in older adults (22). Thus increased RUNX1 expression could alternatively be related to the transcription of MHC IIa. Given that older adults experience a preferential atrophy of MHC IIa fibers (24), continued investigation into the role of RUNX1 in the regulation of skeletal muscle adaption of older adults to exercise and nutrition is warranted.
In conclusion, we demonstrate that the combination of RE and postexercise EAA ingestion appears to upregulate the activity of the ubiquitin proteasome system while reducing autophagy in the skeletal muscle of older men. Furthermore, a larger quantity of leucine ingested following exercise appeared to result in a greater reduction in the degradation of autophagosomes over the time course studied. However, the quantity of ingested leucine had minimal effects on markers of the ubiquitin proteasome system or on the breakdown of intact protein 24 h postexercise. Given the robust ability for postexercise leucine ingestion to stimulate muscle protein synthesis, these data indicate that accretion of myofibrillar proteins is likely also enhanced by strategic postexercise leucine ingestion. This response could promote a healthier protein pool and more favorable muscle quality-based adaptations. More research is needed to better understand the interaction among exercise, nutrition, and aging on skeletal muscle breakdown and autophagy, and, in particular, some focus should be given to more precisely uncover the functions of TRAF6 and RUNX1 in the context of muscle adaptation to exercise and nutrition in older adults.
GRANTS
This work was supported by National Institutes of Health (NIH)/National Institute of Arthritis and Musculoskeletal and Skin Diseases Grant R01AR049877, NIH/National Institute on Aging Grants P30AG024832 and R01AG030070, NIH Clinical and Translational Science Award (5UL1TR001439) from the National Center for Advancing Translational Sciences, and Intramural Funds from Arizona State University. This trial was registered at clinicaltrials.gov as NCT00891696.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
J.M.D., E.V., and B.B.R. conception and design of research; J.M.D., P.T.R., D.M.G., M.S.B., D.K.W., A.C.D., E.V., and B.B.R. performed experiments; J.M.D., A.C.D., and B.B.R. analyzed data; J.M.D., P.T.R., and B.B.R. interpreted results of experiments; J.M.D. prepared figures; J.M.D. drafted manuscript; J.M.D., P.T.R., D.M.G., M.S.B., D.K.W., A.C.D., E.V., and B.B.R. edited and revised manuscript; J.M.D., P.T.R., D.M.G., M.S.B., D.K.W., A.C.D., E.V., and B.B.R. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank the staff of the University of Texas Medical Branch Institute for Translational Sciences Clinical Research Center, and Junfung Hao and Ming-Qian Zheng for technical assistance.
Present addresses: D. K. Walker, Center for Translational Research in Aging & Longevity, Texas A&M University, College Station, TX; P. T. Reidy, Department of Physical Therapy, University of Utah, Salt Lake City, UT.
REFERENCES
- 1.Biolo G, Tipton KD, Klein S, Wolfe RR. An abundant supply of amino acids enhances the metabolic effect of exercise on muscle protein. Am J Physiol Endocrinol Metab 273: E122–E129, 1997. [DOI] [PubMed] [Google Scholar]
- 2.Chang NC, Nguyen M, Shore GC. BCL2-CISD2: an ER complex at the nexus of autophagy and calcium homeostasis? Autophagy 8: 856–857, 2012. doi: 10.4161/auto.20054. [DOI] [PubMed] [Google Scholar]
- 3.Cruz-Jentoft AJ, Baeyens JP, Bauer JM, Boirie Y, Cederholm T, Landi F, Martin FC, Michel JP, Rolland Y, Schneider SM, Topinková E, Vandewoude M, Zamboni M; European Working Group on Sarcopenia in Older People . Sarcopenia: European consensus on definition and diagnosis: Report of the European Working Group on Sarcopenia in Older People. Age Ageing 39: 412–423, 2010. doi: 10.1093/ageing/afq034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dickinson JM, Drummond MJ, Coben JR, Volpi E, Rasmussen BB. Aging differentially affects human skeletal muscle amino acid transporter expression when essential amino acids are ingested after exercise. Clin Nutr 32: 273–280, 2013. doi: 10.1016/j.clnu.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dickinson JM, Gundermann DM, Walker DK, Reidy PT, Borack MS, Drummond MJ, Arora M, Volpi E, Rasmussen BB. Leucine-enriched amino acid ingestion after resistance exercise prolongs myofibrillar protein synthesis and amino acid transporter expression in older men. J Nutr 144: 1694–1702, 2014. doi: 10.3945/jn.114.198671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dickinson JM, Gundermann DM, Walker DK, Reidy PT, Borack MS, Drummond MJ, Arora M, Volpi E, Rasmussen BB. Leucine-enriched amino acid ingestion after resistance exercise prolongs myofibrillar protein synthesis and amino acid transporter expression in older men. J Nutr 144: 1694–1702, 2014. doi: 10.3945/jn.114.198671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dickinson JM, Volpi E, Rasmussen BB. Exercise and nutrition to target protein synthesis impairments in aging skeletal muscle. Exerc Sport Sci Rev 41: 216–223, 2013. doi: 10.1097/JES.0b013e3182a4e699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dreyer HC, Drummond MJ, Pennings B, Fujita S, Glynn EL, Chinkes DL, Dhanani S, Volpi E, Rasmussen BB. Leucine-enriched essential amino acid and carbohydrate ingestion following resistance exercise enhances mTOR signaling and protein synthesis in human muscle. Am J Physiol Endocrinol Metab 294: E392–E400, 2008. doi: 10.1152/ajpendo.00582.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drummond MJ, Dreyer HC, Pennings B, Fry CS, Dhanani S, Dillon EL, Sheffield-Moore M, Volpi E, Rasmussen BB. Skeletal muscle protein anabolic response to resistance exercise and essential amino acids is delayed with aging. J Appl Physiol (1985) 104: 1452–1461, 2008. doi: 10.1152/japplphysiol.00021.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drummond MJ, Fry CS, Glynn EL, Timmerman KL, Dickinson JM, Walker DK, Gundermann DM, Volpi E, Rasmussen BB. Skeletal muscle amino acid transporter expression is increased in young and older adults following resistance exercise. J Appl Physiol (1985) 111: 135–142, 2011. doi: 10.1152/japplphysiol.01408.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunn WA., Jr Autophagy and related mechanisms of lysosome-mediated protein degradation. Trends Cell Biol 4: 139–143, 1994. doi: 10.1016/0962-8924(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 12.Fritzen AM, Madsen AB, Kleinert M, Treebak JT, Lundsgaard AM, Jensen TE, Richter EA, Wojtaszewski J, Kiens B, Frøsig C. Regulation of autophagy in human skeletal muscle: effects of exercise, exercise training and insulin stimulation. J Physiol 594: 745–761, 2016. doi: 10.1113/JP271405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fry CS, Drummond MJ, Glynn EL, Dickinson JM, Gundermann DM, Timmerman KL, Walker DK, Volpi E, Rasmussen BB. Skeletal muscle autophagy and protein breakdown following resistance exercise are similar in younger and older adults. J Gerontol A Biol Sci Med Sci 68: 599–607, 2013. doi: 10.1093/gerona/gls209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallagher LE, Williamson LE, Chan EY. Advances in autophagy regulatory mechanisms. Cells 5: 24, 2016. doi: 10.3390/cells5020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glynn EL, Fry CS, Drummond MJ, Timmerman KL, Dhanani S, Volpi E, Rasmussen BB. Excess leucine intake enhances muscle anabolic signaling but not net protein anabolism in young men and women. J Nutr 140: 1970–1976, 2010. doi: 10.3945/jn.110.127647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenhaff PL, Karagounis LG, Peirce N, Simpson EJ, Hazell M, Layfield R, Wackerhage H, Smith K, Atherton P, Selby A, Rennie MJ. Disassociation between the effects of amino acids and insulin on signaling, ubiquitin ligases, and protein turnover in human muscle. Am J Physiol Endocrinol Metab 295: E595–E604, 2008. doi: 10.1152/ajpendo.90411.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gundermann DM, Walker DK, Reidy PT, Borack MS, Dickinson JM, Volpi E, Rasmussen BB. Activation of mTORC1 signaling and protein synthesis in human muscle following blood flow restriction exercise is inhibited by rapamycin. Am J Physiol Endocrinol Metab 306: E1198–E1204, 2014. doi: 10.1152/ajpendo.00600.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ 18: 571–580, 2011. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katsanos CS, Kobayashi H, Sheffield-Moore M, Aarsland A, Wolfe RR. A high proportion of leucine is required for optimal stimulation of the rate of muscle protein synthesis by essential amino acids in the elderly. Am J Physiol Endocrinol Metab 291: E381–E387, 2006. doi: 10.1152/ajpendo.00488.2005. [DOI] [PubMed] [Google Scholar]
- 20.Keller P, Vollaard NB, Gustafsson T, Gallagher IJ, Sundberg CJ, Rankinen T, Britton SL, Bouchard C, Koch LG, Timmons JA. A transcriptional map of the impact of endurance exercise training on skeletal muscle phenotype. J Appl Physiol (1985) 110: 46–59, 2011. doi: 10.1152/japplphysiol.00634.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, Bamber BA, Bassham DC, Bergamini E, Bi X, Biard-Piechaczyk M, Blum JS, Bredesen DE, Brodsky JL, Brumell JH, Brunk UT, Bursch W, Camougrand N, Cebollero E, Cecconi F, Chen Y, Chin LS, Choi A, Chu CT, Chung J, Clarke PG, Clark RS, Clarke SG, Clavé C, Cleveland JL, Codogno P, Colombo MI, Coto-Montes A, Cregg JM, Cuervo AM, Debnath J, Demarchi F, Dennis PB, Dennis PA, Deretic V, Devenish RJ, Di Sano F, Dice JF, Difiglia M, Dinesh-Kumar S, Distelhorst CW, Djavaheri-Mergny M, Dorsey FC, Dröge W, Dron M, Dunn WA Jr, Duszenko M, Eissa NT, Elazar Z, Esclatine A, Eskelinen EL, Fésüs L, Finley KD, Fuentes JM, Fueyo J, Fujisaki K, Galliot B, Gao FB, Gewirtz DA, Gibson SB, Gohla A, Goldberg AL, Gonzalez R, González-Estévez C, Gorski S, Gottlieb RA, Häussinger D, He YW, Heidenreich K, Hill JA, Høyer-Hansen M, Hu X, Huang WP, Iwasaki A, Jäättelä M, Jackson WT, Jiang X, Jin S, Johansen T, Jung JU, Kadowaki M, Kang C, Kelekar A, Kessel DH, Kiel JA, Kim HP, Kimchi A, Kinsella TJ, Kiselyov K, Kitamoto K, Knecht E, Komatsu M, Kominami E, Kondo S, Kovács AL, Kroemer G, Kuan CY, Kumar R, Kundu M, Landry J, Laporte M, Le W, Lei HY, Lenardo MJ, Levine B, Lieberman A, Lim KL, Lin FC, Liou W, Liu LF, Lopez-Berestein G, López-Otín C, Lu B, Macleod KF, Malorni W, Martinet W, Matsuoka K, Mautner J, Meijer AJ, Meléndez A, Michels P, Miotto G, Mistiaen WP, Mizushima N, Mograbi B, Monastyrska I, Moore MN, Moreira PI, Moriyasu Y, Motyl T, Münz C, Murphy LO, Naqvi NI, Neufeld TP, Nishino I, Nixon RA, Noda T, Nürnberg B, Ogawa M, Oleinick NL, Olsen LJ, Ozpolat B, Paglin S, Palmer GE, Papassideri I, Parkes M, Perlmutter DH, Perry G, Piacentini M, Pinkas-Kramarski R, Prescott M, Proikas-Cezanne T, Raben N, Rami A, Reggiori F, Rohrer B, Rubinsztein DC, Ryan KM, Sadoshima J, Sakagami H, Sakai Y, Sandri M, Sasakawa C, Sass M, Schneider C, Seglen PO, Seleverstov O, Settleman J, Shacka JJ, Shapiro IM, Sibirny A, Silva-Zacarin EC, Simon HU, Simone C, Simonsen A, Smith MA, Spanel-Borowski K, Srinivas V, Steeves M, Stenmark H, Stromhaug PE, Subauste CS, Sugimoto S, Sulzer D, Suzuki T, Swanson MS, Tabas I, Takeshita F, Talbot NJ, Tallóczy Z, Tanaka K, Tanaka K, Tanida I, Taylor GS, Taylor JP, Terman A, Tettamanti G, Thompson CB, Thumm M, Tolkovsky AM, Tooze SA, Truant R, Tumanovska LV, Uchiyama Y, Ueno T, Uzcátegui NL, van der Klei I, Vaquero EC, Vellai T, Vogel MW, Wang HG, Webster P, Wiley JW, Xi Z, Xiao G, Yahalom J, Yang JM, Yap G, Yin XM, Yoshimori T, Yu L, Yue Z, Yuzaki M, Zabirnyk O, Zheng X, Zhu X, Deter RL. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4: 151–175, 2008. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kosek DJ, Kim JS, Petrella JK, Cross JM, Bamman MM. Efficacy of 3 days/wk resistance training on myofiber hypertrophy and myogenic mechanisms in young vs. older adults. J Appl Physiol (1985) 101: 531–544, 2006. doi: 10.1152/japplphysiol.01474.2005. [DOI] [PubMed] [Google Scholar]
- 23.Kumar A, Bhatnagar S, Paul PK. TWEAK and TRAF6 regulate skeletal muscle atrophy. Curr Opin Clin Nutr Metab Care 15: 233–239, 2012. doi: 10.1097/MCO.0b013e328351c3fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lexell J, Taylor CC, Sjöström M. What is the cause of the ageing atrophy? Total number, size and proportion of different fiber types studied in whole vastus lateralis muscle from 15- to 83-year-old men. J Neurol Sci 84: 275–294, 1988. doi: 10.1016/0022-510X(88)90132-3. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25: 402–408, 2001. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.MacNeil LG, Glover E, Bergstra TG, Safdar A, Tarnopolsky MA. The order of exercise during concurrent training for rehabilitation does not alter acute genetic expression, mitochondrial enzyme activity or improvements in muscle function. PLoS One 9: e109189, 2014. doi: 10.1371/journal.pone.0109189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, Goldberg AL, Schiaffino S, Sandri M. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab 6: 458–471, 2007. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 28.Nagasawa T, Kido T, Yoshizawa F, Ito Y, Nishizawa N. Rapid suppression of protein degradation in skeletal muscle after oral feeding of leucine in rats. J Nutr Biochem 13: 121–127, 2002. doi: 10.1016/S0955-2863(01)00209-1. [DOI] [PubMed] [Google Scholar]
- 29.Nakashima K, Ishida A, Yamazaki M, Abe H. Leucine suppresses myofibrillar proteolysis by down-regulating ubiquitin-proteasome pathway in chick skeletal muscles. Biochem Biophys Res Commun 336: 660–666, 2005. doi: 10.1016/j.bbrc.2005.08.138. [DOI] [PubMed] [Google Scholar]
- 30.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130: 165–178, 2007. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 31.Neel BA, Lin Y, Pessin JE. Skeletal muscle autophagy: a new metabolic regulator. Trends Endocrinol Metab 24: 635–643, 2013. doi: 10.1016/j.tem.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paul PK, Bhatnagar S, Mishra V, Srivastava S, Darnay BG, Choi Y, Kumar A. The E3 ubiquitin ligase TRAF6 intercedes in starvation-induced skeletal muscle atrophy through multiple mechanisms. Mol Cell Biol 32: 1248–1259, 2012. doi: 10.1128/MCB.06351-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paul PK, Gupta SK, Bhatnagar S, Panguluri SK, Darnay BG, Choi Y, Kumar A. Targeted ablation of TRAF6 inhibits skeletal muscle wasting in mice. J Cell Biol 191: 1395–1411, 2010. doi: 10.1083/jcb.201006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paul PK, Kumar A. TRAF6 coordinates the activation of autophagy and ubiquitin-proteasome systems in atrophying skeletal muscle. Autophagy 7: 555–556, 2011. doi: 10.4161/auto.7.5.15102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phillips SM, Tipton KD, Aarsland A, Wolf SE, Wolfe RR. Mixed muscle protein synthesis and breakdown after resistance exercise in humans. Am J Physiol Endocrinol Metab 273: E99–E107, 1997. [DOI] [PubMed] [Google Scholar]
- 36.Rasmussen BB, Tipton KD, Miller SL, Wolf SE, Wolfe RR. An oral essential amino acid-carbohydrate supplement enhances muscle protein anabolism after resistance exercise. J Appl Physiol (1985) 88: 386–392, 2000. [DOI] [PubMed] [Google Scholar]
- 37.Rattan SI, Clark BF. Intracellular protein synthesis, modifications and aging. Biochem Soc Trans 24: 1043–1049, 1996. doi: 10.1042/bst0241043. [DOI] [PubMed] [Google Scholar]
- 38.Raue U, Jemiolo B, Yang Y, Trappe S. TWEAK-Fn14 pathway activation after exercise in human skeletal muscle: insights from two exercise modes and a time course investigation. J Appl Physiol (1985) 118: 569–578, 2015. doi: 10.1152/japplphysiol.00759.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raue U, Slivka D, Jemiolo B, Hollon C, Trappe S. Proteolytic gene expression differs at rest and after resistance exercise between young and old women. J Gerontol A Biol Sci Med Sci 62: 1407–1412, 2007. doi: 10.1093/gerona/62.12.1407. [DOI] [PubMed] [Google Scholar]
- 40.Raue U, Trappe TA, Estrem ST, Qian HR, Helvering LM, Smith RC, Trappe S. Transcriptome signature of resistance exercise adaptations: mixed muscle and fiber type specific profiles in young and old adults. J Appl Physiol (1985) 112: 1625–1636, 2012. doi: 10.1152/japplphysiol.00435.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reidy PT, Walker DK, Dickinson JM, Gundermann DM, Drummond MJ, Timmerman KL, Cope MB, Mukherjea R, Jennings K, Volpi E, Rasmussen BB. Soy-dairy protein blend and whey protein ingestion after resistance exercise increases amino acid transport and transporter expression in human skeletal muscle. J Appl Physiol (1985) 116: 1353–1364, 2014. doi: 10.1152/japplphysiol.01093.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117: 399–412, 2004. doi: 10.1016/S0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwalm C, Jamart C, Benoit N, Naslain D, Prémont C, Prévet J, Van Thienen R, Deldicque L, Francaux M. Activation of autophagy in human skeletal muscle is dependent on exercise intensity and AMPK activation. FASEB J 29: 3515–3526, 2015. doi: 10.1096/fj.14-267187. [DOI] [PubMed] [Google Scholar]
- 44.Solomon V, Goldberg AL. Importance of the ATP-ubiquitin-proteasome pathway in the degradation of soluble and myofibrillar proteins in rabbit muscle extracts. J Biol Chem 271: 26690–26697, 1996. doi: 10.1074/jbc.271.43.26690. [DOI] [PubMed] [Google Scholar]
- 45.Stefanetti RJ, Zacharewicz E, Della Gatta P, Garnham A, Russell AP, Lamon S. Ageing has no effect on the regulation of the ubiquitin proteasome-related genes and proteins following resistance exercise. Front Physiol 5: 30, 2014. doi: 10.3389/fphys.2014.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanaka Y, Guhde G, Suter A, Eskelinen EL, Hartmann D, Lüllmann-Rauch R, Janssen PM, Blanz J, von Figura K, Saftig P. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature 406: 902–906, 2000. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- 47.Tanida I. Autophagosome formation and molecular mechanism of autophagy. Antioxid Redox Signal 14: 2201–2214, 2011. doi: 10.1089/ars.2010.3482. [DOI] [PubMed] [Google Scholar]
- 48.Wang X, Blagden C, Fan J, Nowak SJ, Taniuchi I, Littman DR, Burden SJ. Runx1 prevents wasting, myofibrillar disorganization, and autophagy of skeletal muscle. Genes Dev 19: 1715–1722, 2005. doi: 10.1101/gad.1318305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williamson DL, Gallagher PM, Carroll CC, Raue U, Trappe SW. Reduction in hybrid single muscle fiber proportions with resistance training in humans. J Appl Physiol (1985) 91: 1955–1961, 2001. [DOI] [PubMed] [Google Scholar]
- 50.Williamson DL, Raue U, Slivka DR, Trappe S. Resistance exercise, skeletal muscle FOXO3A, and 85-year-old women. J Gerontol A Biol Sci Med Sci 65: 335–343, 2010. doi: 10.1093/gerona/glq005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolfe RR. The underappreciated role of muscle in health and disease. Am J Clin Nutr 84: 475–482, 2006. [DOI] [PubMed] [Google Scholar]
- 52.Wolfe RR, Chinkes DL. Isotopic Tracers in Metabolic Research: Principles and Practice of Kinetic Analysis. Hoboken, NJ: Wiley, 2005, p. 1–474. [Google Scholar]
- 53.Yan X, Sun Q, Ji J, Zhu Y, Liu Z, Zhong Q. Reconstitution of leucine-mediated autophagy via the mTORC1-Barkor pathway in vitro. Autophagy 8: 213–221, 2012. doi: 10.4161/auto.8.2.18563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang J, Ney PA. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ 16: 939–946, 2009. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang XJ, Chinkes DL, Sakurai Y, Wolfe RR. An isotopic method for measurement of muscle protein fractional breakdown rate in vivo. Am J Physiol Endocrinol Metab 270: E759–E767, 1996. [DOI] [PubMed] [Google Scholar]
- 56.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab 6: 472–483, 2007. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]


