Abstract
Antibody responses of pigs to defined Erns fragments, after classical swine fever virus (CSFV) infection, were studied by using an enzyme-linked immunosorbent assay (ELISA). Selection of various Erns fragments was based on an immunodominant Erns region encompassing three overlapping antigenic regions, amino acids 65 to 145 (Ernsaa 65-145) (AR1), 84 to 160 (Ernsaa 84-160) (AR2), and 109 to 220 (Ernsaa 109-220) (AR3), identified earlier by our group (M. Lin, E. Trottier, J. Pasick, and M. Sabara, J. Biochem., in press). Defined Erns fragments, including AR1, AR2, AR3, Ernsaa 65-160 (AR12), Ernsaa 84-220 (AR23), Ernsaa 65-220 (AR123), Ernsaa 109-145 (the consensus region defined by the three overlapping regions), and Ernsaa 109-160 (a fragment 15 amino acids larger than the consensus region), were expressed in Escherichia coli, purified by nickel chelate affinity chromatography, and used to measure antibody responses in 20 sera serially collected from pigs experimentally infected with CSFV. Based on the optimum cutoffs determined by receiver operating characteristic analysis after testing 238 negative field sera from Canadian sources, all the Erns fragments were capable of distinguishing positive from negative antibody responses with sensitivities ranging between 75 and 90% and specificities ranging between 83.2 and 100%. Detection of antibody responses to refolded Ernsaa 109-145 and Ernsaa 109-160 by ELISA (this study) but not by Western blots (Lin et al., in press) indicated that the epitopes within the consensus region are conformational. When cutoff values were raised to give a specificity of 100%, four Erns fragments (AR2, AR23, Ernsaa 109-145, and Ernsaa 109-160) offered much higher sensitivities (75 to 90%) than those obtained with other fragments (20 to 65%). Ernsaa 109-145 and Ernsaa 109-160 were capable of detecting antibody responses in infected pigs as early as 7 days postinfection. Demonstration of antibody responses to either one of the four fragments can thus be an alternative to use of the full-length protein in ELISA for serological diagnosis of CSFV infection. An advantage of such a test would be its utilization for serological survey in a classical swine fever-free country (e.g., Canada) in biocontainment level 2 laboratories.
Classical swine fever virus (CSFV) is an enveloped positive-stranded RNA virus (20) of the genus Pestivirus of the Flaviviridae family (34). Other viruses classified in this genus are Bovine viral diarrhea virus and Border disease virus of sheep. CSFV is highly contagious and can cause a fatal disease in pigs. The disease, characterized by fever and hemorrhage, can be acute, chronic, or subclinical with substantial economic loss. The classical swine fever outbreak in The Netherlands during 1997 and 1998 resulted in destruction of more than 10 million pigs at a cost of more than 2 billion U.S. dollars (25). Similar to other members of the genus, the 12.5-kb CSFV genome contains a single large open reading frame encoding a polyprotein precursor of approximately 4,000 amino acids (aa). The precursor is cleaved co- and posttranslationally by cellular and viral proteases into structural proteins C, Erns, E1, and E2 and nonstructural proteins NS2, NS3, NS4A, NS4B, NS5A, and NS5B (26).
The biochemical, biological, and functional properties of the envelope glycoprotein Erns have been studied in some detail. This protein forms a disulfide-bridged homodimer in the virion (31, 15) and is highly modified by N-linked glycosylation, which contributes about 50% to the molecular mass of Erns (28, 36). The glycoprotein is both virion associated and secreted, since it is found both on the surface of pestivirus-infected cells and in the culture medium (28). Erns does not contain a potential membrane-spanning domain, yet its C-terminal domain can translocate the full-length protein across eukaryotic cell membranes (13). The protein can bind to several cell types and inhibits CSFV and bovine viral diarrhea virus infection in cell culture, suggesting that Erns is involved in attachment to or entry of the viruses into susceptible cells (7). This interaction is believed to be mediated with cell surface glycoaminoglycans, such as heparan sulfate (9, 10). The Erns protein contains RNase activity (5, 6, 30, 36), which is a unique feature for a viral surface protein. Homology in two stretches of the Erns sequence to members of the Rh/T2/S RNase superfamily provides a structural basis for the RNase activity (15, 30). The biological function of Erns RNase activity is not fully understood; however, destruction of the RNase activity by mutations gives rise to viruses that are more cytopathic in culture and attenuated in vivo (8, 19). Antibodies that inhibit RNase activity also tend to neutralize CSFV infectivity (36). Although cytotoxicity is a feature of other soluble ribonucleases (29), the link has yet to be established for Erns.
Antibodies directed against Erns, E2, and NS3 have been demonstrated in infected animals (14, 16, 21, 22, 24), with Erns and E2 capable of inducing neutralizing antibodies (11, 12, 32). It has been shown that antibodies to full-length Erns or even a 37-mer peptide derived from its C-terminal end (aa 191 to 227) could be used as an indicator of CSFV infection in pigs (14, 21). Recent studies have indicated that an Erns-based enzyme-linked immunosorbent assay (ELISA) can be used as a companion diagnostic test to identify CSFV-infected pigs in herds vaccinated with the E2 subunit marker vaccine (4, 21). However, very little is known about the structural immunogenic organization of Erns. This information would be invaluable for the development of a serological diagnostic test with high sensitivity and specificity. Recently, we have mapped an immunodominant region encompassing three overlapping antigenic regions (ARs) that induce antibody responses during CSFV infection: aa 65 to 145 (Ernsaa 65-145) (AR1), aa 84 to 160 (Ernsaa 84-160) (AR2), and 109 to 220 (Ernsaa 109-220) (AR3) (17). Assignment of these antigenic regions correlates well with the three-dimensional structural model of Erns derived from disulfide bond connectivity and homology modeling (15). Interestingly, the consensus region of the three Erns antigenic regions contained one complete and one partial T-cell epitope sequence as described previously (1). In this study, we measured and compared the antibody responses of pigs to AR1, AR2, AR3, combinations of two or three individual overlapping regions, and the consensus region after experimental infection. The data reported in this study support the selection of an Erns fragment for early detection of antibody in CSFV-infected pigs.
MATERIALS AND METHODS
Protein expression clones.
Protein expression constructs pET217-143, pET133-142, pET167-218, pET217-142, pET133-218, pET217-218, pET167-143, pET167-142, and pET134-131 were generated (17) to code for Ernsaa 65-145 (AR1), Ernsaa 84-160 (AR2), Ernsaa 109-220 (AR3), Ernsaa 65-160 (AR12), Ernsaa 84-220 (AR23), Ernsaa 65-220 (AR123), Ernsaa 109-145, Ernsaa 109-160, and Ernsaa 121-227E1aa 1-7, respectively. Recombinant proteins expressed from the constructs have an N-terminal fusion of 49 to 58 residues, including a six-histidine tag, and an additional fusion of 8 residues at the C-terminal end.
Sera.
Twenty sera from pigs experimentally infected with CSFV (3) were provided by the National Centre for Foreign Animal Diseases (Winnipeg, Manitoba, Canada) and are listed in Table 1. A total of 238 pig serum samples from Canadian herds (classical swine fever [CSF] free) were used for this study.
TABLE 1.
Sera from pigs experimentally infected with various CSFV strains
| Serum | Time of testinga | CSFV strain used |
|---|---|---|
| S#1 HC HyperImmune Serum P15-93 (302-08) 94-09-15 | 15 dpc 4 | NA |
| S#2 HC HyperImmune Serum P16-93 94-09-21 | 21 dpc 4 | NA |
| S#3 HC Antiserum P43-83 83-08-16 | 17 dpc 5 | NA |
| S#4 HC Standard Serum P78-82 83-04-18 | 166 dpi | NA |
| S#5 HC P12-93 94-01-12 | 8 dpc | Alfort |
| S#6 HC P17-93 93-12-29 | 56 dpi | Glentorf |
| S#7 HC P18-93 93-12-29 | 56 dpi | Glentorf |
| S#8 HC P19-93 93-12-30 | 57 dpi | Glentorf |
| S#9 HC P4-92 92-06-02 | 7 dpi | Glentorf |
| S#10 HC P4-92 92-06-30 | 35 dpi | Glentorf |
| S#11 HC P5-92 92-06-02 | 7 dpi | Glentorf |
| S#12 HC P5-92 92-06-09 | 14 dpi | Glentorf |
| S#13 HC P5-92 92-06-16 | 21 dpi | Glentorf |
| S#14 HC P5-92 92-06-30 | 35 dpi | Glentorf |
| S#15 HCV HyperImmune Serum P222 64-06-02 | NA | NA |
| S#16 HCV Antiserum L-1 P154 66-06-02 | 210 dpi | Lapinized |
| S#17 HCV Antisera NT P155 66-06-22 | 210 dpc | Lapinized |
| S#18 HCV Immune Sera 51PIC/41 P2427 P3C 69-01-16 | 71 dpi | NA |
| S#19 HCV Antiserum P54-83 83-10-24 | NAb | NA |
| S#20 HC PA03 98-12-14 | 7 dpi | Alfort |
Animals tested n dpc were infected with one CSFV strain followed by challenge with other CSFV strains during the course of infection.
NA, not available.
Expression of recombinant Erns fragments.
Escherichia coli BL21(DE3)pLysS cells (Novagen, Madison, Wis.) harboring one of the above expression constructs were cultured at 37°C with shaking in 2 to 4 liters of Luria-Bertani broth supplemented with 30 μg of kanamycin/ml until the optical density at 590 nm (OD590) reached between 0.5 and 1.0. Isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 1 mM, and the cells were allowed to express the recombinant proteins for 3 h. The cells were harvested by centrifugation at 10,000 × g for 20 min.
Purification of recombinant proteins.
Cell pellets were resuspended in phosphate-buffered saline (PBS) (pH 7.2) with 1 mM phenylmethylsulfonyl fluoride and lysed with a French press at 1,500 lb/in2. The homogenates were spun at 27,000 × g for 20 min at 4°C, and the pellet fraction was resuspended in protein extraction buffer (6 M guanidine hydrochloride, 0.1 M NaH2PO4, 0.01 M Tris [pH 8.0]). The solubilized sample was spun again at 27,000 × g for 20 min at 4°C, and the supernatant was loaded onto a column (1 by 3.5 cm) of nickel-nitrilotriacetic acid agarose (QIAGEN, Santa Clarita, Calif.). The column was washed with buffer A (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris [pH 8.0]), followed by buffer B (buffer A plus 0.5 M NaCl [pH 6.3]) and buffer C (buffer B plus 5 mM imidazole [pH 5.9]). The denatured protein was then refolded on the column by washes with Tris-buffered saline (TBS) (pH 7.4) containing 1 M urea, followed by TBS (pH 7.4). The recombinant protein was eluted and collected with fractions of 1 ml from the column using TBS (pH 7.4) containing 200 mM imidazole. The protein fragment Ernsaa 109-160 was further purified by anion exchange chromatography on a column (1.5 by 4 cm) of Q-Sepharose (Amersham Biosciences, Baie d'Urfe, Quebec, Canada) equilibrated with 50 mM phosphate buffer pH (7.8) and eluted over a 0 to 1 M NaCl linear gradient (60 ml) in phosphate buffer. Proteins were quantified by using the Bradford method (2) with bovine serum albumin as a standard.
Optimization of ELISA conditions.
Nunc Maxisorp ELISA plates were coated with 1 or 2 μg of purified antigen/ml to determine the optimal concentration of antigen. The purified protein in 0.06 M carbonate buffer (sodium bicarbonate and sodium carbonate [pH 9.6]) was aliquoted into each well (100 μl) and allowed to incubate at room temperature overnight (18 to 20 h). The plates were then frozen at −20°C and were thawed before use. Plates were washed with PBS (pH 7.2) containing 0.05% Tween 20 (PBST). A positive CSFV-infected pig serum (17 days postinfection [dpi]) and a CSFV-negative pig serum were diluted 1/50 and 1/100 in PBST; 100 μl was applied to each well and incubated for 1 h. The plates were washed and incubated for 1 h with horseradish peroxidase-rabbit anti-swine immunoglobulin G (whole molecule) antibody (1/2,000 dilution in PBST), 100 μl/well. The plates were washed and developed for 10 min with shaking in a solution (100 μl/well) of 1 mM 2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid) and 0.015% H2O2 in 50 mM citrate buffer (26 mM trisodium citrate, 24 mM citric acid [pH 4.5]). The absorbances at 414 nm were measured on a Labsystems Multiskan bichromatic plate photometer. In addition, bovine serum albumin (3%) as a blocking reagent and the presence of the divalent cation chelating agents EDTA and EGTA (7.5 mM each; pH 6.3) during the serum incubation step were examined to determine whether these reagents could improve the assay.
Detection of pig antibodies to Erns fragments by ELISA.
Based on optimization results, all ELISAs were performed with the purified antigen coated at 2 μg/ml and the pig sera diluted 1/100 in PBST containing 7.5 mM of each of EDTA and EGTA (pH 6.3) to reduce the background signal. Twenty CSFV antisera from experimentally infected pigs representing a variety of animals, virus strains, and days postinfection (Table 1) were tested with all nine defined Erns fragments. Also, 238 negative pig sera were tested for each protein.
Data analysis.
Receiver operating characteristic (ROC) analysis of the ELISA results was performed using the statistical program MedCalc, version 7.3 (MedCalc Software, Mariakerke, Belgium). The values for the area under the ROC curve, the 95% confidence interval (CI) for the area, and cutoff points were derived from the ROC curve analysis. An area of X means that a randomly selected individual from the positive group has a test value larger than that for a randomly chosen individual from the negative group 100X % of the time. A test that gives a perfect separation of the values of the two groups has an area of 1, whereas a noninformative test that cannot distinguish between the two groups has an area of 0.5. A 95% CI for the area, which does not include the 0.5 value, indicates that the test has an ability to distinguish between the two groups.
RESULTS
Selection of assay parameters.
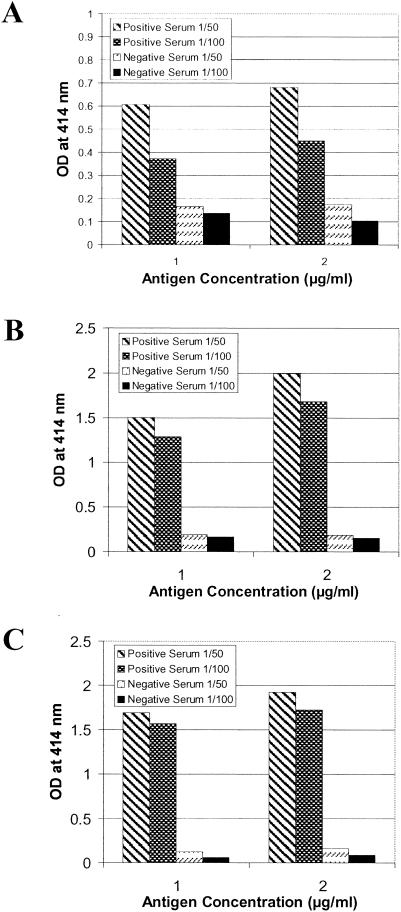
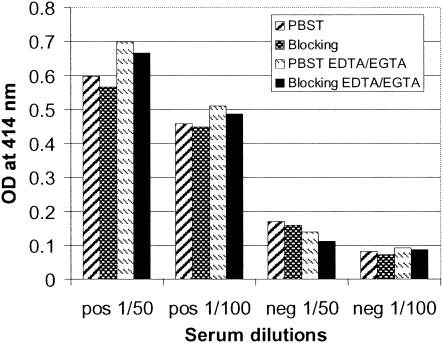
Pig sera at 1:50 and 1:100 dilutions were tested for the presence of CSFV-specific antibodies with ELISA at Erns fragment antigen concentrations of 1 or 2 μg/ml (Fig. 1). Several recombinant proteins, Ernsaa 112-227E1aa 1-7, Ernsaa 109-160, and Ernsaa 109-145, were used in the initial evaluation. For the positive serum, the antigens at 2 μg/ml consistently showed higher OD414 values than those obtained at 1 μg/ml. The negative serum, in contrast, did not demonstrate any measurable difference in reading when the antigen coating concentration was altered. Thus, the coating concentration of a selected antigen was set at 2 μg/ml to maximize the detection of positive antibody response in sera from infected pigs. For the majority of the proteins used in this evaluation step (at 2 μg/ml), the positive serum at either a 1:50 dilution or a 1:100 dilution gave consistent OD414 values greater than 1.00. This indicates that positive antibody responses to selected Erns fragments are well detected at a serum dilution of 1:100. One exception was Ernsaa 112-227E1aa 1-7, which at a 1:50 serum dilution did not exceed a reading of 0.70 and which at 1:100 dropped to below 0.50. The negative sera gave only slightly lower readings at 1:100 than at 1:50. To further optimize the assay conditions, bovine serum albumin (BSA) (3%) as a blocking reagent and divalent cation chelating agents EDTA and EGTA (7.5 mM each; pH 6.3), which have been shown to reduce nonspecific serum protein interaction in enzyme immunoassay (23), were explored in the assay (Fig. 2). BSA did not significantly alter the OD414 readings of either the positive or negative serum. Thus, this blocking reagent was excluded from the ELISA assays. The presence of EDTA and EGTA during the serum incubation step increased the OD414 values for the positive sera and slightly decreased the readings for the negative sera (Fig. 2). Based on these results, subsequent ELISA procedures were standardized to use the coating antigen at 2 μg/ml and serum samples at a 1:100 dilution in PBS containing the chelating agents EDTA and EGTA at 7.5 mM each (pH 6.3).
FIG. 1.
Optimization of the concentration of coating antigens and dilutions of serum for ELISA. Several recombinant proteins, Ernsaa 112-227E1aa 1-7 (A), Ernsaa 109-160 (B), and Ernsaa 109-145 (C), at 1 and 2 μg/ml, were tested for reactivity with one positive serum (S#3) and one negative serum at 1/50 and 1/100 dilutions, respectively. Each bar represents the mean value of two determinations.
FIG. 2.
Influence of BSA (blocking reagent) and divalent cation chelating agents on ELISA. Ernsaa 112-227E1aa 1-7 (2 μg/ml) was immobilized on an ELISA plate and reacted with one positive serum (S#3) and one negative serum at 1/50 and 1/100 dilutions. Four assay conditions were examined for each serum dilution: (i) dilution of pig sera in PBST; (ii) blocking of the plate with 3% BSA in PBST prior to adding pig serum (diluted in PBST); (iii) dilution of pig serum in PBST containing EDTA/EGTA (7.5 mM each; pH 6.3); and (iv) blocking of the plate with 3% BSA in PBST prior to adding pig serum (diluted in PBST containing EDTA and EGTA).
Antibody responses to individual antigenic regions (AR1, AR2, and AR3) of Erns.
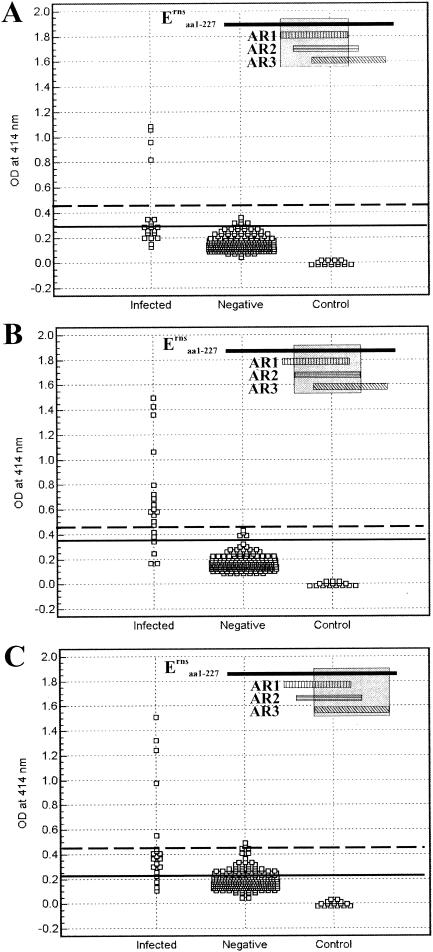
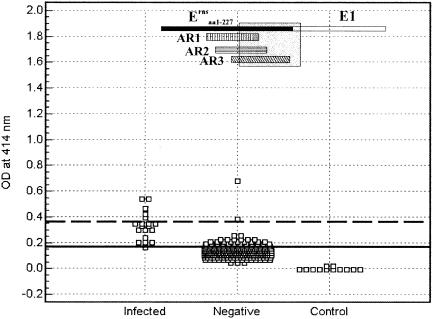
The reactivity of antibodies from CSFV-infected pigs with Erns fragments AR1, AR2, and AR3 was examined by using ELISA (Fig. 3). Two hundred thirty-eight pig serum samples from Canadian herds (CSF free) were also analyzed in parallel to establish a cutoff that distinguishes positive antibody responses from negative responses. An optimum cutoff at OD414, corresponding with the highest accuracy (i.e., minimal false-negative and false-positive results), of 0.199, 0.32, and 0.294 was determined by ROC analysis for AR1, AR2, and AR3, respectively. Based on these cutoffs, the serological specificities were 83.2, 98.3 and 92.4% for AR1, AR2, and AR3, respectively; 18 (90%), 17 (85%), and 15 (75%) out of 20 serum samples from 16 infected pigs (Table 1) were identified as having positive antibody responses to these fragments. The ROC analysis yielded an area under the curve (AUC) of 0.898, with a 95% CI from 0.854 to 0.932 for AR1, 0.952 (95% CI = 0.919 to 0.975) for AR2, and 0.841 (95% CI = 0.790 to 0.883) for AR3. These values can be interpreted as described in Materials and Methods. Higher cutoff values that resulted in 100% specificity (no false positives) resulted in detection of 4 (20%), 14 (70%), and 5 (25%) out of 20 infected pig serum samples for AR1, AR2, and AR3, respectively.
FIG. 3.
Detection of antibody responses of pigs to AR1, AR2, and AR3 of Erns by ELISA. Twenty serum samples from CSFV-infected pigs and 238 negative pig sera were analyzed for reactivity with immobilized AR1 (A), AR2 (B), and AR3 (C), using the optimized conditions as described in the text. Dilution buffer without testing serum served as a control. The optimum cutoff determined by ROC analysis is indicated by a solid line. A higher cutoff eliminating all false positives (100% specificity) is indicated by a dashed line. The location of each Erns fragment relative to the full-length protein is marked (shaded) in the insets.
Antibody responses to Erns fragments encompassing two to three overlapping individual antigenic regions.
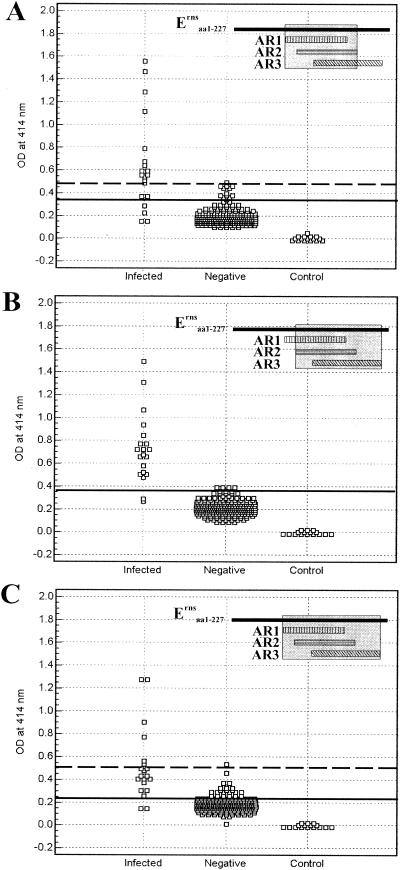
Antibody responses in CSFV-infected pigs to the Erns fragments corresponding to overlapping antigenic regions AR12, AR23, and AR123 were evaluated by ELISA using the same serum samples as described above (Fig. 4). The ROC analysis derived an optimum cutoff of 0.361, 0.4, and 0.247 for AR12, AR23, and AR123, respectively. Based on these cutoffs, the serological specificity was 95.8, 100 and 90.8% for AR12, AR23, and AR123, respectively; 16 (80%), 18 (90%), and 18 (90.0%) out of 20 serum samples from infected pigs were identified as having positive antibody responses to these fragments. The ROC analysis yielded an AUC of 0.911 (95% CI = 0.870 to 0.943) for AR12, 0.984 (95% CI = 0.960 to 0.995) for AR23, and 0.910 (95% CI = 0.868 to 0.942) for AR123. Higher cutoff values that resulted in 100% specificity (no false-positive results) resulted in detection of 13 (65%), 18 (90%), and 6 (30.0%) out of 20 serum samples from infected pigs for AR12, AR23, and AR123, respectively.
FIG. 4.
Detection of antibody responses of pigs to AR12, AR23, and AR123 of Erns by ELISA. Erns fragments corresponding to overlapping antigenic regions AR12 (A), AR23 (B), and AR123 (C) were used as coating antigens. The assay conditions and data presentation are as described in the legend to Fig. 3.
Antibody responses to the consensus region of overlapping AR1, AR2, and AR3.
Reactivities of antibodies in sera from CSFV-infected pigs to the consensus region (Ernsaa 109-145) defined by AR1, AR2, and AR3 and Ernsaa 109-160, a fragment that is 15 amino acids larger than the consensus region, are shown in Fig. 5. Optimum cutoff values were determined by ROC analysis to be 0.366 and 0.287 for Ernsaa 109-145 and Ernsaa 109-160, respectively. Based on these cutoffs, the serological specificity was 99.6 and 99.6% for Ernsaa 109-145 and Ernsaa 109-160, respectively; 16 (80%) and 18 (90%) out of 20 serum samples from infected pigs exhibited positive antibody responses to these Erns fragments. The ROC analysis yielded an AUC of 0.937 (95% CI = 0.900 to 0.963) for Ernsaa 109-145 and 0.979 (95% CI = 0.953 to 0.993) for Ernsaa 109-160. Higher cutoff values that resulted in 100% specificity (no false positive) resulted in detection of 15 (75%) and 16 (80%) out of 20 infected pig serum samples for Ernsaa 109-145 and Ernsaa 109-160, respectively.
FIG. 5.
Detection of antibody responses to the consensus region of overlapping AR1, AR2, and AR3. Ernsaa 109-145 (A), corresponding to the consensus region defined by AR1, AR2, and AR3, and Ernsaa 109-160 (B), a fragment that is 15 amino acids larger than the consensus region, were used as coating antigens. The assay conditions and data presentation are as described in the legend to Fig. 3.
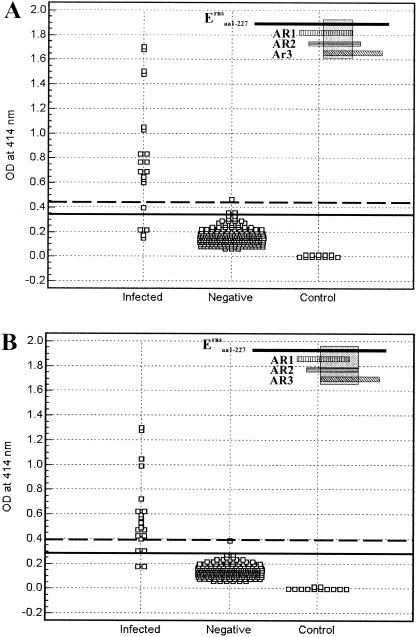
Antibody responses of CSFV-infected pigs to the C-terminal end (37 aa) of Erns were demonstrated earlier by Langedijk et al. (14). Reactivity of an Erns fragment (Ernsaa 121-227E1aa 1-7) containing this C-terminal end with sera from CSFV-infected pigs and negative pig sera were also evaluated in the present study. As shown in Fig. 6, the Ernsaa 121-227E1aa 1-7 fragment exhibits a weaker reaction with pig antisera than other defined Erns fragments.
FIG. 6.
Detection of antibody responses to the Ernsaa 121-227E1aa 1-7 fragment containing the C-terminal end (37 residues). The assay conditions and data presentation are as described in the legend to Fig. 3.
DISCUSSION
Several proteins were previously reported to elicit a humoral immune response in pigs after infection with CSFV (14, 16, 18, 21, 22, 27, 24, 33, 35). Of these immunogenic proteins, E2 and Erns are particularly of importance for the development of diagnostic tests and/or subunit marker vaccine (4). Erns-based ELISA can be a companion diagnostic test for CSFV-infected pigs in an E2-vaccinated herd (4, 21). In this study, we evaluated antibody responses of CSFV-infected pigs to nine defined Erns fragments corresponding to each of the three previously identified overlapping ARs (17), combinations of two to three overlapping ARs, and the consensus region of the three ARs. The study has generated information useful for designing a diagnostic test to improve the serological detection of CSFV infection.
ROC analysis revealed that there is a significant difference in the antibody responses to individual ARs. When the cutoffs were raised above the optimum value calculated by ROC analysis to completely eliminate the false-positive reactions in negative samples, AR1 and AR3 reacted with only a few sera of CSFV-infected pigs, such as S#3, S#7, S#18, S#19, and S#14 (reactive with AR3 only). All of these reactive sera were collected at 35 days postinfection (dpi) or later. This tends to suggest that antibody responses to AR1 or AR3 appear during the late stages of infection. In contrast, AR2 could be used to detect antibodies in a much greater number of sera from infected animals, ranging from 14 to 210 dpi (S#12 and S#16). This indicates that antibody responses directed against AR2 occurred during both early and late infection. Antibody responses to the C-terminal end (37 aa) of Erns were previously demonstrated to be an indicator of CSFV infection (14). For comparison, antibody responses to the C-terminal end (37 residues)-containing fragment Ernsa a121-227E1aa 1-7 were also determined (Fig. 6). This fragment, though not recognized by antisera from CSFV-infected pigs on Western blots (17), was able to react with anti-CSFV antisera on ELISA after refolding and eluting from a Ni-agarose column using the method previously described (16). Reaction of Ernsaa 121-227E1aa 1-7 with pig antisera was much weaker than that of other defined Erns fragments, indicating that this region is not immunodominant. This further supports the immunodominant nature of an Erns region encompassing three overlapping antigenic regions, AR1, AR2, and AR3 (17).
As expected, the combination of overlapping AR1 and AR2 or AR2 and AR3 (i.e., AR12 or AR23) allowed detection of more positive antibody responses in the 20 serum samples from CSFV-infected pigs than AR1 or AR3 alone at the cutoff of 100% specificity (Fig. 4). Compared to AR2, AR12 resulted in a similar number of positive antibody responses, while use of AR23 detected more positive responses. This indicates that AR2 makes a major contribution to the sensitivity of detection from the overlapping combination; the contribution of AR1 is insignificant, while AR3 appears to enhance the detection sensitivity. Surprisingly, the number of antibody responses to AR123 is similar to that with AR1 or AR3 but is much smaller than that with AR2. The reason for this unexpected result is not clear. Perhaps some antibody binding sites could have been masked by the additional sequences surrounding AR2.
Our previous data showed that Ernsaa 109-145, the consensus region defined by AR1, AR2, and AR3, and Ernsaa 109-160, a fragment 15 amino acids larger than the consensus region, were not recognized by CSFV antisera on Western blots (17). The present data show that both Ernsaa 109-145 and Ernsaa 109-160, after refolding and eluting from a Ni-agarose column, were recognized by antibodies in sera from CSFV-infected animals in ELISA. This result provides evidence that the epitopes located within the consensus region are conformational. This is consistent with the proposed three-dimensional model of Erns (15), which predicts the cysteine-rich consensus region to be on a large surface-exposed loop between helix 6 and helix 7 and likely to form two disulfide bonds in this region. A comparison of the ELISA results from Ernsaa 109-145 and Ernsaa 109-160 with a larger Erns fragment (AR123) that contains the consensus sequences reveals that the consensus region detected more positive serum samples than AR123 at a 100% specificity cutoff. This, together with other results, suggests that extra residues added to the N-terminal end of Ernsaa 109-145 reduce the binding of antibodies to Ernsaa 109-145, possibly by masking or altering the epitopes within the consensus region. Use of relatively smaller fragments, AR23 and AR2, both of which contain Ernsaa 109-145, detected a similar number of antibody responses in pigs after infection compared to use of Ernsaa 109-145. This provides further support that additional N-terminal amino acid sequences interfere with binding of antibodies to Ernsaa 109-145.
The 20 serum samples from infected pigs used in the study cover various days postinfection (Table 1). Inspection of the ELISA data (OD414) associated with each of the 20 serum samples (Fig. 3, 4, and 5) has allowed us to make the interesting observation that AR2, AR23, Ernsaa 109-145, and Ernsaa 109-160 tend to detect a nearly full spectrum of antibody responses, including those during early infection. In particular, antibody responses to AR23 or Ernsaa 109-160 can be detected as early as 7 dpi. These small Erns fragments offer an advantage over the entire protein, which can only be used to detect CSFV-specific antibodies in vaccinated or unvaccinated pigs as early as 14 dpi (21). None of the nine defined Erns fragments was capable of detecting anti-Erns antibodies in sera S#5 and S#20 of infected pigs. These two sera were collected in early infection (8 days postchallenge [dpc] and 7 dpi), and most likely anti- Erns antibodies had not yet been induced. Other Erns fragments tend to detect only antibodies arising in the later stages of infection. Exploration of this striking difference will allow development of an Erns fragment-based ELISA for the serological detection of CSFV infection at various stages of infection. Selection of a small Erns fragment, such as Ernsaa 109-145 and Ernsaa 109-160 or a synthetic peptide corresponding to Ernsaa 109-145, would offer a diagnostic antigen superior to the full-length protein. Production of recombinant Erns fragments in large quantity from this study will facilitate the development and validation of a test that can be used for serological survey in a CSF-free country (e.g., Canada) in biocontainment level 2 laboratories.
Acknowledgments
We acknowledge technical assistance from Jose Riva and Maria Mallory. The automatic DNA sequencing services were provided by University of Ottawa Biotechnology Research Institute (Ottawa, Ontario, Canada) and Canadian Molecular Research Services, Inc. (Ottawa, Ontario, Canada).
This work was supported in part by Diachemix Corporation (Grayslake, Ill.).
REFERENCES
- 1.Armengol, E., K. H. Wiesmuller, D. Wienhold, M. Buttner, E. Pfaff, G. Jung, and A. Saalmuller. 2002. Identification of T-cell epitopes in the structural and non-structural proteins of classical swine fever virus. J. Gen. Virol. 83:551-560. [DOI] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Clavijo, A., E. M. Zhou, S. Vydelingum, and R. Heckert. 1998. Development and evaluation of a novel antigen capture assay for the detection of classical swine fever virus antigens. Vet. Microbiol. 60:155-168. [DOI] [PubMed] [Google Scholar]
- 4.de Smit, A. J. 2000. Laboratory diagnosis, epizootiology, and efficacy of marker vaccines in classical swine fever: a review. Vet. Q. 22:182-188. [DOI] [PubMed] [Google Scholar]
- 5.Hausmann, Y., G. Roman-Sosa, H. J. Thiel, and T. Rumenapf. 2004. Classical swine fever virus glycoprotein Erns is an endoribonuclease with an unusual base specificity. J. Virol. 78:5507-5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hulst, M. M., G. Himes, E. Newbigin, and R. J. Moormann. 1994. Glycoprotein E2 of classical swine fever virus: expression in insect cells and identification as a ribonuclease. Virology 200:558-565. [DOI] [PubMed] [Google Scholar]
- 7.Hulst, M. M., and R. J. Moormann. 1997. Inhibition of pestivirus infection in cell culture by envelope proteins Erns and E2 of classical swine fever virus: Erns and E2 interact with different receptors. J. Gen. Virol. 78:2779-2787. [DOI] [PubMed] [Google Scholar]
- 8.Hulst, M. M., F. E. Panoto, A. Hoekman, H. G. van Gennip, and R. J. Moormann. 1998. Inactivation of the RNase activity of glycoprotein Erns of classical swine fever virus results in a cytopathogenic virus. J. Virol. 72:151-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hulst, M. M., H. G. van Gennip, and R. J. Moormann. 2000. Passage of classical swine fever virus in cultured swine kidney cells selects virus variants that bind to heparan sulfate due to a single amino acid change in envelope protein Erns. J. Virol. 74:9553-9561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hulst, M. M., H. G. van Gennip, A. C. Vlot, E. Schooten, A. J. de Smit, and R. J. Moormann. 2001. Interaction of classical swine fever virus with membrane-associated heparan sulfate: role for virus replication in vivo and virulence. J. Virol. 75:9585-9595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hulst, M. M., D. F. Westra, G. Wensvoort, and R. J. Moormann. 1993. Glycoprotein E1 of hog cholera virus expressed in insect cells protects swine from hog cholera. J. Virol. 67:5435-5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konig, M., T. Lengsfeld, T. Pauly, R. Stark, and H. J. Thiel. 1995. Classical swine fever virus: independent induction of protective immunity by two structural glycoproteins. J. Virol. 69:6479-6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langedijk, J. P. 2002. Translocation activity of C-terminal domain of pestivirus Erns and ribotoxin L3 loop. J. Biol. Chem. 277:5308-5314. [DOI] [PubMed] [Google Scholar]
- 14.Langedijk, J. P., W. G. Middel, R. H. Meloen, J. A. Kramps, and J. A. de Smit. 2001. Enzyme-linked immunosorbent assay using a virus type-specific peptide based on a subdomain of envelope protein Erns for serologic diagnosis of pestivirus infections in swine. J. Clin. Microbiol. 39:906-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langedijk, J. P., P. A. van Veelen, W. M. Schaaper, A. H. de Ru, R. H. Meloen, and M. M. Hulst. 2002. A structural model of pestivirus Erns based on disulfide bond connectivity and homology modeling reveals an extremely rare vicinal disulfide. J. Virol. 76:10383-10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin, M., F. Lin, M. Mallory, and A. Clavijo. 2000. Deletions of structural glycoprotein E2 of classical swine fever virus strain alfort/187 resolve a linear epitope of monoclonal antibody WH303 and the minimal N-terminal domain essential for binding immunoglobulin G antibodies of a pig hyperimmune serum. J. Virol. 74:11619-11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin, M., E. Trottier, J. Pasick, and M. Sabara. J. Biochem., in press. [DOI] [PubMed]
- 18.Liu, J. J., M. L. Wong, P. F. Chen, and T. J. Chang. 1998. Cloning, expression and sequence analysis of the classical swine fever virus nucleocapsid protein. Virus Genes 16:225-234. [DOI] [PubMed] [Google Scholar]
- 19.Meyers, G., A. Saalmuller, and M. Buttner. 1999. Mutations abrogating the RNase activity in glycoprotein Erns of the pestivirus classical swine fever virus lead to virus attenuation. J. Virol. 73:10224-10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moennig, V. 1988. Characteristics of the virus, p. 55-58. In B. Liess (ed.), Classical swine fever and related viral infections. Martinus Nijhoff Publishing, Boston, Mass.
- 21.Moormann, R. J., A. Bouma, J. A. Kramps, C. Terpstra, and H. J. De Smit. 2000. Development of a classical swine fever subunit marker vaccine and companion diagnostic test. Vet. Microbiol. 73:209-219. [DOI] [PubMed] [Google Scholar]
- 22.Moser, C., N. Ruggli, J. D. Tratschin, and M. A. Hofmann. 1996. Detection of antibodies against classical swine fever virus in swine sera by indirect ELISA using recombinant envelope glycoprotein E2. Vet. Microbiol. 51:41-53. [DOI] [PubMed] [Google Scholar]
- 23.Nielsen, K., L. Kelly, D. Gall, P. Smith, J. Bosse, P. Nicoletti, and W. Kelly. 1994. The use of divalent cation chelating agents (EDTA/EGTA) to reduce non-specific serum protein interaction in enzyme immunoassay. Vet. Res. Commun. 18:433-437. [DOI] [PubMed] [Google Scholar]
- 24.Paton, D. J., G. Ibata, S. Edwards, and G. Wensvoort. 1991. An ELISA detecting antibody to conserved pestivirus epitopes. J. Virol. Methods 31:315-324. [DOI] [PubMed] [Google Scholar]
- 25.Pluimers, F. H., P. W. de Leeuw, J. A. Smak, A. R. Elbers, and J. A. Stegeman. 1999. Classical swine fever in The Netherlands 1997-1998: a description of organisation and measures to eradicate the disease. Prev. Vet. Med. 42:139-155. [DOI] [PubMed] [Google Scholar]
- 26.Rice, M. C., and B. D. Lindenbach. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 27.Rumenapf, T., R. Stark, G. Meyers, and H. J. Thiel. 1991. Structural proteins of hog cholera virus expressed by vaccinia virus: further characterization and induction of protective immunity. J. Virol. 65:589-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rumenapf, T., G. Unger, J. H. Strauss, and H. J. Thiel. 1993. Processing of the envelope glycoproteins of pestiviruses. J. Virol. 67:3288-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schein, C. H. 1997. From housekeeper to microsurgeon: the diagnostic and therapeutic potential of ribonucleases. Nat. Biotechnol. 15:529-536. [DOI] [PubMed] [Google Scholar]
- 30.Schneider, R., G. Unger, R. Stark, E. Schneider-Scherzer, and H. J. Thiel. 1993. Identification of a structural glycoprotein of an RNA virus as a ribonuclease. Science 261:1169-1171. [DOI] [PubMed] [Google Scholar]
- 31.Thiel, H. J., R. Stark, E. Weiland, T. Rumenapf, and G. Meyers. 1991. Hog cholera virus: molecular composition of virions from a pestivirus. J. Virol. 65:4705-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Rijn, P. A., A. Bossers, G. Wensvoort, and R. J. Moormann. 1996. Classical swine fever virus (CSFV) envelope glycoprotein E2 containing one structural antigenic unit protects pigs from lethal CSFV challenge. J. Gen. Virol. 77:2737-2745. [DOI] [PubMed] [Google Scholar]
- 33.Weiland, E., R. Ahl, R. Stark, F. Weiland, and H. J. Thiel. 1992. A second envelope glycoprotein mediates neutralization of a pestivirus, hog cholera virus. J. Virol. 66:3677-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wengler, G. 1991. Classification and nomenclature of viruses. Fifth report of the international committee on taxonomy of viruses. Arch. Virol. Suppl. 2:223-233. [Google Scholar]
- 35.Wensvoort, G., M. Bloemraad, and C. Terpstra. 1988. An enzyme immunoassay employing monoclonal antibodies and detecting specifically antibodies to classical swine fever virus. Vet. Microbiol. 17:129-140. [DOI] [PubMed] [Google Scholar]
- 36.Windisch, J. M., R. Schneider, R. Stark, E. Weiland, G. Meyers, and H. J. Thiel. 1996. RNase of classical swine fever virus: biochemical characterization and inhibition by virus-neutralizing monoclonal antibodies. J. Virol. 70:352-358. [DOI] [PMC free article] [PubMed] [Google Scholar]