Abstract
We isolated an arbovirus from bovine blood in Indonesia. The arbovirus was obtained from the plasma of a cow showing no clinical symptoms in West Java in February 2014, and was identified as Akabane virus (AKAV) by AKAV-specific RT-PCR and subsequent sequence analysis. Phylogenetic analysis based on partial S segment indicated the AKAV isolate, WJ-1SA/P/2014, was most closely related with two isolates from Israel and Turkey reported in 2001 and 2015, respectively, and that WJ-1SA/P/2014 isolate belongs to AKAV genogroup Ib. This is the first isolation of AKAV from Indonesia.
Keywords: Akabane virus, arbovirus, cattle, genogroup
Akabane virus (AKAV) is an arthropod-borne virus (arbovirus) which was first isolated from Aedes vexans and Culex tritaeniorhynchus in Japan in 1959 [21, 22, 26]. The AKAV belongs to the genus Orthobunyavirus in the family Bunyaviridae, and its genome consists of three-segmented (L, M and S RNA segments), negative single-stranded RNA [8]. The L RNA segment encodes the L protein, which has RNA polymerase activity for replication and transcription of the viral genome. The M RNA segment encodes two viral envelope glycoproteins, Gn and Gc, responsible for viral neutralization and a non-structural protein NSm in the form of a precursor polypeptide which is processed by post-translational cleavage. The S RNA segment encodes the nucleocapsid (N) protein and a smaller nonstructural (NSs) protein in overlapping reading frames. The AKAV is transmitted principally by Culicoides biting midges [14, 26, 30], and it is widely distributed from the tropical to temperate zones of the world, except for the Americas [21]. The AKAV causes Akabane disease when susceptible pregnant cattle, sheep and goats are infected [21]. Clinical signs of the Akabane disease are mainly abortion, stillbirth and congenital abnormalities characterized by arthrogryposis-hydranencephaly syndrome [21]. Bovine cases of Akabane viral encephalomyelitis have also been reported in Japan and Korea [13, 17, 19].
In the Asia-Pacific region, AKAV has been isolated in Japan, South Korea, Taiwan, China, Vietnam and Australia [5, 9, 12, 13, 15, 19, 29]. Therefore, it is possible that AKAV is actively circulating in the tropical zones of Indonesia, although there has been no report about isolation and/or characterization of AKAV in that country. In the present study, we conducted virus isolation from bovine blood to understand the activity of AKAV and other related arboviruses in Indonesia.
Anticoagulated blood samples were collected from 34 heads of cattle in seven herds in West Java, Indonesia, from January 2014 to February 2014. The ages of the cattle were categorized as follows: <6 months (2 heads), 6‒12 months (20 heads), 13‒24 months (10 heads) and >24 months (6 heads). The six cows that were >24 months old suffered spontaneous abortion on the day of sampling, but all the other cattle showed no clinical symptoms at the sampling. Blood samples were centrifuged at 1,940 ×g for 15 min at 4°C on the day of sampling, and then, plasma was stored at ‒80°C until use. The blood cells were washed three times with sterilized phosphate-buffered saline (PBS), then resuspended in PBS and stored at ‒80°C until use.
Baby hamster kidney (BHK-21) and hamster lung (HmLu-1) cells were cultured in Dulbecco’s minimum essential medium (MEM) (GIBCO, Great Island, NY, U.S.A.) supplemented with 0.015% sodium bicarbonate and 5% fetal bovine serum. The two cell lines cultured on 24-well plates were washed three times with Earl’s solution, and then, 200 µl of each sample (plasma or blood cells) was inoculated in four wells, and the plates were incubated at 37°C for 1 hr. After the incubation, the inoculum was replaced with 1 ml of Dulbecco’s MEM supplemented with 0.015% sodium bicarbonate and 10 µg/ml gentamicin, and then, the plates were incubated at 37°C again and observed for cytopathic effects (CPEs) over seven days. Passages were conducted twice for the supernatant of the first inoculated culture. As a result, one of the 20 plasma samples collected from the 6‒12 month-old cattle showed CPE after the first passage to HmLu-1 cells. The supernatant of the cell culture showing the CPE was collected, and then, RNA was extracted. All the other samples collected in the present study showed no CPEs.
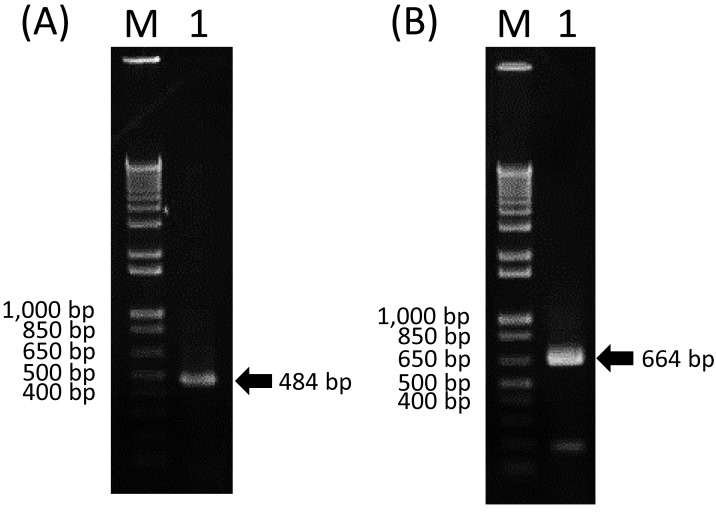
The RNA sample was used as a template for an RT-PCR assay to identify the virus. The RT-PCR assay was conducted by using six primer sets targeting arboviruses (Table 1) and the OneStep RT-PCR Kit (Qiagen, Valencia, CA, U.S.A.) according to the following program [11, 18, 20, 28]: 50°C for 30 min; 95°C for 15 min; 35 cycles of 94°C for 30 sec, 55°C for 30 sec and 72°C for 1 min; and then 72°C for 10 min. Products were then subjected to agarose gel electrophoresis (1.5%) in TAE buffer, and the gel was stained with 0.5 µg/ml ethidium bromide in TAE buffer and observed under UV light. As a result, the RNA sample tested positive with a primer set for broad-range detection of Simbu serogroup viruses targeting a 484-nt fragment of the S segment (Fig. 1A) [11], and the sample also tested positive with another primer set for AKAV-specific detection targeting a 664-nt fragment of the M segment (Fig. 1B) [28]. On the other hand, the RNA sample tested negative with the other primer sets for the detection of Aino, Peaton, bluetongue and bovine ephemeral fever viruses.
Table 1. Oligonucleotide primers used in this study.
| Target virusa) | Primer | Sequence (5′-3′) | Position | Purpose | Reference |
|---|---|---|---|---|---|
| Simbu serogroup | AKAI206F | CACAACCAAGTGTCGATCTTA | 206–226 | RT-PCR and sequencing | [11, 20] |
| SimbuS637-656 | GAGAATCCAGATTTAGCCCA | 689–670 | |||
| AKAV | AKAVM-F | AAGCAAGAGGAATGCAGCTCTACA | 1799–1822 | RT-PCR and sequencing | [28] |
| AKAVM-R | CTGTTTTGAGGAGTCGAATAGACC | 2462–2439 | |||
| AINOV | AINOVM-F | TGCTATAGCCCCTTCATACATTGG | 2769–2792 | RT-PCR | [28] |
| AINOVM-R | TGGCATGTTTGCAGTGGTTACAGT | 3336–3313 | |||
| PEAV | PEAVM-F | CCTTCCATACGCCATTTAGGTGA | 2104–2126 | RT-PCR | [28] |
| PEAVM-R | TGCTCATCACATTCAGATGA | 2591–2572 | |||
| BTV | BTVL3-1 | CCTGATGTTTCCAGGACAAATTATACTC | 1055–1082 | RT-PCR | [20] |
| BTVL3-2 | CCGATAAAGGCAAACCAAAGCGAAATCC | 1763–1736 | |||
| BEFV | BEF-AO-F | GAATCATTATGGGATCGGATC | 1140–1160 | RT-PCR | [18] |
| BEF-AO-R | CCAACCTACAACAGCAGATAAAAC | 1587–1564 |
a) AKAV: Akabane virus; AINOV: Aino virus; PEAV: Peaton virus; BTV: Bluetongue virus; BEFV: Bovine ephemeral fever virus.
Fig. 1.
Agarose gel electrophoresis of RT-PCR products amplified by using the primers for broad-range detection of Simbu serogroup viruses (panel A) and the primers for specific detection of AKAV (panel B). Lane 1, supernatant of HmLu-1 cell culture that showed CPE. M, molecular mass ladder.
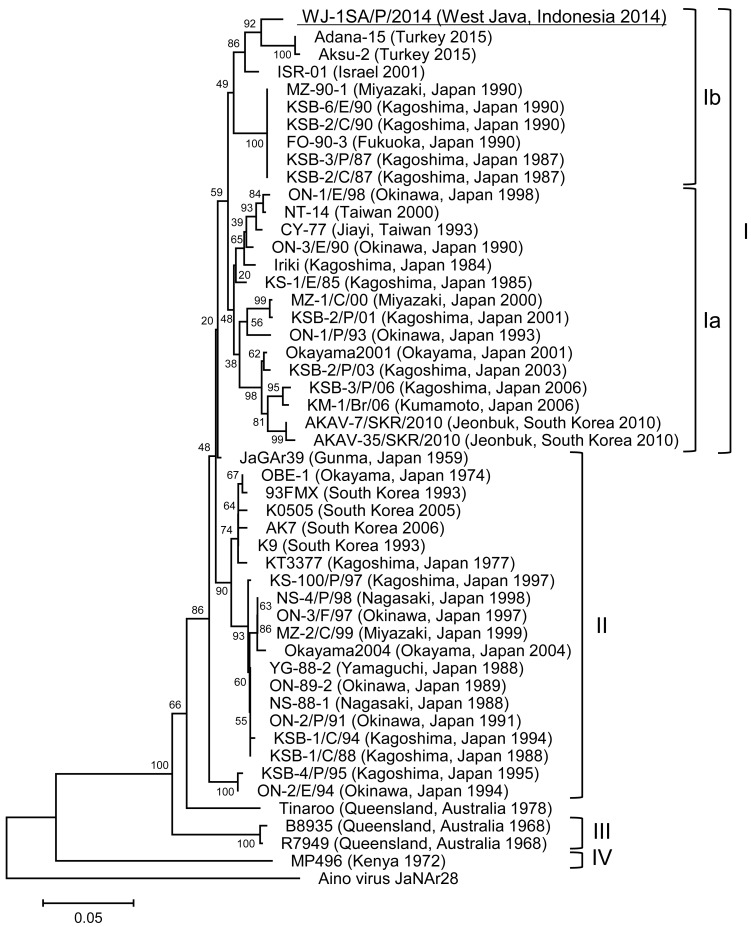
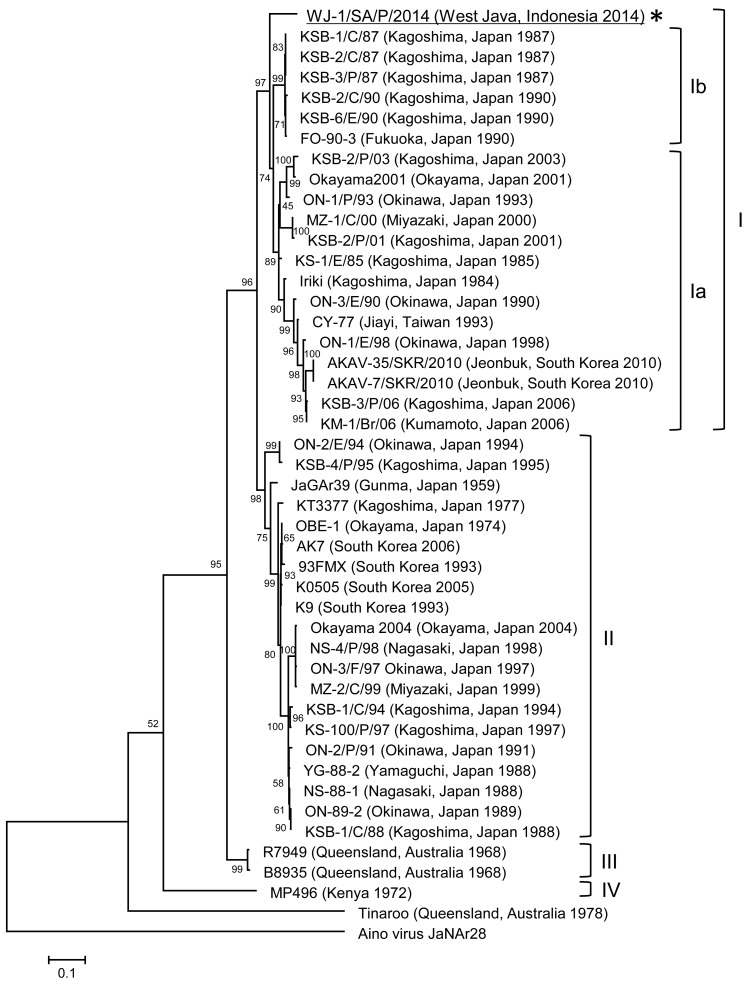
The PCR products were purified by using a QIAquick PCR Purification Kit (Qiagen) and then subjected to direct sequencing. The nucleotide sequence of the 443-nt fragment of the S segment was determined by using the primers, AKAI206F and SimbuS637-656 [11, 20], and the 616-nt fragment of the M segment was determined by using the primers, AKAVM-F and AKAVM-R [28]. To analyze the phylogenetic relationships among the isolate obtained in the present study and other isolates obtained elsewhere, the sequences were aligned by the Clustal W program [25]. Then, neighbor-joining (NJ) and maximum-likelihood (ML) comparisons were made by using MEGA5 [24]. The reliability of the branching orders was evaluated by the bootstrap test (n=1,000). The AKAV isolate in the present study, namely AKAV WJ-1SA/P/2014, was most closely related with two isolates from Israel and Turkey, ISR-01 and Adana-15, belonging to genogroup Ib in the phylogenetic trees of the 443-nt fragment of the S segment constructed by the NJ method (Fig. 2) and the ML method (data not shown), respectively. The identities of the WJ-1SA/P/2014 isolate to the ISR-01 and Adana-15 isolates were both 97.06% at nucleotide level. On the other hand, the AKAV WJ-1SA/P/2014 isolate was closely related with Japanese isolates belonging to genotypes Ia and Ib in the 616-nt fragment of the M segment constructed by the ML method (Fig. 3) and the NJ method (data not shown), respectively. The highest sequence identities between the AKAV WJ-1SA/P/2014 isolate and the isolates in genogroups Ia and Ib were 90.42% (KS-1/E/85) and 89.93% (FO-90-3), respectively, at nucleotide level. The AKAV WJ-1SA/P/2014 isolate was most closely related to the KS-1/E/85 isolate which was isolated in Kagoshima, Japan, in 1985, based on the partial sequence of the M segment. The nucleotide sequence data reported in this study were deposited in the DNA Data Bank of Japan under the accession numbers, LC113954 and LC113955.
Fig. 2.
Phylogenetic tree based on the 443-nt fragment of the genome segment S of AKAV, constructed using the neighbor-joining method. The AKAV WJ-1SA/P/2014 isolate (GenBank accession No. LC113954) is underlined. The percentage bootstrap values calculated from 1,000 replications are indicated around the internal nodes. Bar, 0.05% sequence divergence.
Fig. 3.
Phylogenetic tree based on the 616-nt fragment of the genome segment M of AKAV, constructed using the maximum-likelihood method. The AKAV WJ-1SA/P/2014 isolate (GenBank accession No. LC113955) is underlined. It is clear that the AKAV WJ-1SA/P/2014 isolate belongs to genogroup I, but the isolate is closely related to both genogroups Ia and Ib in this phylogenetic tree (*). The percentage bootstrap values calculated from 1,000 replications are indicated around the internal nodes. Bar, 0.1% sequence divergence.
In this study, an AKAV isolate was obtained from the plasma of a cow showing no clinical symptoms in West Java, Indonesia. To the best of our knowledge, this is the first isolation of AKAV from Indonesia. Our data suggested that the isolate, WJ-1SA/P/2014, belongs to genogroup Ib, since it was most closely related to an Israeli isolate of AKAV genogroup Ib, ISR-01 [23], with 97.06% identity in the 443-nt fragment of the S segment.
The AKAV WJ-1SA/P/2014 isolate was closely related with AKAV isolates in genogroups Ia and Ib in the 616-nt fragment of the M segment. The highest sequence identities at nucleotide level between the AKAV WJ-1SA/P/2014 isolate and the isolates in genogroup Ia and Ib were 90.42 and 89.93%, respectively, while the identities to isolates in genogroups II (OBE-1), III (B8935) and IV (MP496) were 85.55, 81.33 and 66.55%, respectively. The identities to isolates in genogroups Ia and Ib described above were relatively low when compared with the identity to the AKAV ISR-01 and Adana-15 isolates in the 443-nt fragment of the S segment (97.06%). One of the possible reasons for the low identity was that GenBank sequence data were not available for the M segment of Israeli, Turkish or other Asian isolates. Another possible reason was that the 616-nt fragment of the M segment was too short to reflect the relationship among AKAV isolates correctly. Therefore, further analysis of full-length ORFs of the AKAV WJ-1SA/P/2014 isolate is needed to deepen our understanding of the genetic characteristics of the isolate, and comparison of the ORF sequence data with AKAV isolates from other countries, such as Israel, Turkey, Vietnam and China, would also be required to understand the molecular epidemiology of AKAV in Asia and the Middle East [2, 9, 23].
The AKAV WJ-1SA/P/2014 isolate was then used for a neutralization test to understand anti-AKAV antibody prevalence in the study area where the AKAV isolate was obtained. Serum samples were collected from 115 heads of cattle in 19 herds in October 2015, and they were tested for anti-AKAV antibodies by the neutralization test as described previously [13]. As a result, 109 of the 115 samples (94.78%) tested positive. The antibodies were detected in 13 of the 14 samples (92.86%) collected from cattle aged 6−12 months and 96 of the 101 samples (95.05%) from cattle aged over 12 months, respectively. The anti-AKAV antibodies were detected in a large population of tested samples regardless of the ages of cattle, suggesting that initial infection of AKAV often occurs in young cattle and they produce the antibodies in the study area. Blood samples of cattle aged 5 months or less are not suitable for isolation of AKAV, since maternally derived antibodies against AKAV were estimated to be present until the age of 3.7−4.8 months old [27], and therefore, the high prevalence of antibodies against AKAV among cattle aged 6 months or more seems to make it difficult to isolate AKAV in the area.
Various pathogens can cause abortion, stillbirth, congenital abnormalities or encephalomyelitis in cattle, but AKAV is clearly one of the most prevalent, since it has caused many large-scale epidemics of Akabane disease in East Asia and Australia since 1972 [10, 19, 26, 32]. In Indonesia, although cattle with the above abnormalities are not generally tested for AKAV, several facts suggest that AKAV may be widespread and a major cause of disease in ruminants: (i) Neutralizing antibodies against AKAV were detected in bovine serum collected in Java and Bali in 1979 [16]; (ii) AKAV has been isolated in countries neighboring Indonesia, such as Australia and Vietnam [2, 5, 32]; (iii) Culicoides brevitarsis, which transmits several arboviruses, is present in Indonesia [1]; and (iv) An arbovirus that is transmitted by Culicoides biting midges, bluetongue virus (BTV), is widespread in ruminants in Indonesia [3]. Therefore, the spread of AKAV should be investigated in Indonesia as well as in neighboring countries [4, 6, 7], and bovine cases showing clinical signs and lesions similar to those of Akabane disease should also be tested for AKAV and anti-AKAV antibodies in these regions [21, 31]. Such efforts would be certainly beneficial for elucidating AKAV epidemiology and for improving control measures against bovine cases of abortion, stillbirth, congenital abnormalities and encephalomyelitis in Indonesia.
Acknowledgments
This work was supported by the JICA Project of Capacity Development of Animal Health Laboratory (project number 1000131). The authors gratefully acknowledge the staff at DIC Subang and KPSBU Lembang for their kind support in the sampling; Dr. Masato Kishima and Mr. Yasuyuki Maeda for their kind support in the arrangement and implementation of our study; and Mrs. Tera Tri Yusepi for her kind assistance with this work.
REFERENCES
- 1.Bellis G., Dyce A., Gopurenko D., Yanase T., Garros C., Labuschagne K., Mitchell A.2014. Revision of the Culicoides (Avaritia) Imicola complex Khamala & Kettle (Diptera: Ceratopogonidae) from the Australasian region. Zootaxa 3768: 401–427. doi: 10.11646/zootaxa.3768.4.1 [DOI] [PubMed] [Google Scholar]
- 2.Bryant J. E., Crabtree M. B., Nam V. S., Yen N. T., Duc H. M., Miller B. R.2005. Isolation of arboviruses from mosquitoes collected in northern Vietnam. Am. J. Trop. Med. Hyg. 73: 470–473. [PubMed] [Google Scholar]
- 3.Daniels P., Lunt R., Pritchard I.2009. Bluetongue viruses in Australasia and East Asia. pp. 223−234. In: Bluetongue (Biology of Animal Infections) (Mellor, P., Matthew, B. and Mertens, P. eds.), Elsevier, London. [Google Scholar]
- 4.Daniels P. W., Sendow I., Pritchard L. I., Sukarsih, Eaton B. T.2004. Regional overview of bluetongue viruses in South-East Asia: viruses, vectors and surveillance. Vet. Ital. 40: 94–100. [PubMed] [Google Scholar]
- 5.Della-Porta A. J., O’Halloran M. L., Parsonson I. M., Snowdon W. A., Murray M. D., Hartley W. J., Haughey K. J.1977. Akabane disease: isolation of the virus from naturally infected ovine foetuses. Aust. Vet. J. 53: 51–52. doi: 10.1111/j.1751-0813.1977.tb15825.x [DOI] [PubMed] [Google Scholar]
- 6.Eagles D., Melville L., Weir R., Davis S., Bellis G., Zalucki M. P., Walker P. J., Durr P. A.2014. Long-distance aerial dispersal modelling of Culicoides biting midges: case studies of incursions into Australia. BMC Vet. Res. 10: 135. doi: 10.1186/1746-6148-10-135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eagles D., Walker P. J., Zalucki M. P., Durr P. A.2013. Modelling spatio-temporal patterns of long-distance Culicoides dispersal into northern Australia. Prev. Vet. Med. 110: 312–322. doi: 10.1016/j.prevetmed.2013.02.022 [DOI] [PubMed] [Google Scholar]
- 8.Elliott R. M., Blakqori G.2011. Molecular biology of Orthobunyaviruses. pp. 1−39. In: Bunyaviridae: Molecular and Cellular Biology (Plyusnin, A. and Elliott, R. M. eds.), Caister Academic Press, Norfolk. [Google Scholar]
- 9.Feng Y., He B., Fu S., Yang W., Zhang Y., Tu C., Liang G., Zhang H.2015. [Isolation and identification of the Akabane virus from mosquitoes in Yunnan Province, China]. Bing Du Xue Bao 31: 51–57[Article in Chinese]. [PubMed] [Google Scholar]
- 10.Forman S., Hungerford N., Yamakawa M., Yanase T., Tsai H. J., Joo Y. S., Yang D. K., Nha J. J.2008. Climate change impacts and risks for animal health in Asia. Rev. - Off. Int. Epizoot. 27: 581–597. doi: 10.20506/rst.27.2.1814 [DOI] [PubMed] [Google Scholar]
- 11.Kato T., Matsumoto H., Hirashima Y., Shirafuji H., Yamakawa M., Yanase T.2013. Improvement of RT-PCR assay for detection of orthobunyaviruses isolated in Japan. Bull. Natl. Inst. Anim. Hlth. 119: 47–52[Article in Japanese with English abstract]. [Google Scholar]
- 12.Kobayashi T., Yanase T., Yamakawa M., Kato T., Yoshida K., Tsuda T.2007. Genetic diversity and reassortments among Akabane virus field isolates. Virus Res. 130: 162–171. doi: 10.1016/j.virusres.2007.06.007 [DOI] [PubMed] [Google Scholar]
- 13.Kono R., Hirata M., Kaji M., Goto Y., Ikeda S., Yanase T., Kato T., Tanaka S., Tsutsui T., Imada T., Yamakawa M.2008. Bovine epizootic encephalomyelitis caused by Akabane virus in southern Japan. BMC Vet. Res. 4: 20. doi: 10.1186/1746-6148-4-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurogi H., Akiba K., Inaba Y., Matumoto M.1987. Isolation of Akabane virus from the biting midge Culicoides oxystoma in Japan. Vet. Microbiol. 15: 243–248. doi: 10.1016/0378-1135(87)90078-2 [DOI] [PubMed] [Google Scholar]
- 15.Liao Y. K., Lu Y. S., Goto Y., Inaba Y.1996. The isolation of Akabane virus (Iriki strain) from calves in Taiwan. J. Basic Microbiol. 36: 33–39. doi: 10.1002/jobm.3620360108 [DOI] [PubMed] [Google Scholar]
- 16.Miura Y., Inaba Y., Tsuda T., Tokuhisa S., Sato K., Akashi H., Matumoto M.1982. A survey of antibodies to arthropod-borne viruses in Indonesian cattle. Jpn. J. Vet. Sci. 44: 857–863. doi: 10.1292/jvms1939.44.857 [DOI] [PubMed] [Google Scholar]
- 17.Miyazato S., Miura Y., Hase M., Kubo M., Goto Y., Kono Y.1989. Encephalitis of cattle caused by Iriki isolate, a new strain belonging to Akabane virus. Jpn. J. Vet. Sci. 51: 128–136. doi: 10.1292/jvms1939.51.128 [DOI] [PubMed] [Google Scholar]
- 18.Niwa T., Shirafuji H., Ikemiyagi K., Nitta Y., Suzuki M., Kato T., Yanase T.2015. Occurrence of bovine ephemeral fever in Okinawa Prefecture, Japan, in 2012 and development of a reverse-transcription polymerase chain reaction assay to detect bovine ephemeral fever virus gene. J. Vet. Med. Sci. 77: 455–460. doi: 10.1292/jvms.14-0492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oem J. K., Lee K. H., Kim H. R., Bae Y. C., Chung J. Y., Lee O. S., Roh I. S.2012. Bovine epizootic encephalomyelitis caused by Akabane virus infection in Korea. J. Comp. Pathol. 147: 101–105. doi: 10.1016/j.jcpa.2012.01.013 [DOI] [PubMed] [Google Scholar]
- 20.Ohashi S., Yoshida K., Yanase T., Kato T., Tsuda T.2004. Simultaneous detection of bovine arboviruses using single-tube multiplex reverse transcription-polymerase chain reaction. J. Virol. Methods 120: 79–85. doi: 10.1016/j.jviromet.2004.04.006 [DOI] [PubMed] [Google Scholar]
- 21.OIE 2014. Bunyaviral diseases of animals. pp. 1−16. In: Manuals of Diagnostic Tests and Vaccines for Terrestrial Animals 2014 (OIE ed.). Office International des Epizooties, Paris. [Google Scholar]
- 22.Oya A., Okuno T., Ogata T., Kobayashii, Matsuyama T.1961. Akabane, a new arbor virus isolated in Japan. Jpn. J. Med. Sci. Biol. 14: 101–108. doi: 10.7883/yoken1952.14.101 [DOI] [PubMed] [Google Scholar]
- 23.Stram Y., Brenner J., Braverman Y., Banet-Noach C., Kuznetzova L., Ginni M.2004. Akabane virus in Israel: a new virus lineage. Virus Res. 104: 93–97. doi: 10.1016/j.virusres.2004.03.004 [DOI] [PubMed] [Google Scholar]
- 24.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S.2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. doi: 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson J. D., Higgins D. G., Gibson T. J.1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. doi: 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsuda T.2000. Congenital abnormalities of cattle caused by the arboviral infection. Yamaguchi J. Vet. Med. 27: 1–18[Article in Japanese with English abstract]. [Google Scholar]
- 27.Tsutsui T., Yamamoto T., Hayama Y., Akiba Y., Nishiguchi A., Kobayashi S., Yamakawa M.2009. Duration of maternally derived antibodies against Akabane virus in calves: survival analysis. J. Vet. Med. Sci. 71: 913–918. doi: 10.1292/jvms.71.913 [DOI] [PubMed] [Google Scholar]
- 28.Yamakawa M., Shirafuji H.2012. Genetic testing for diagnosis of arbovirus infections in livestock animals. Bull. Kagoshima Vet. Med. Assoc. 49: 8–9[Article in Japanese]. [Google Scholar]
- 29.Yamakawa M., Yanase T., Kato T., Tsuda T.2006. Chronological and geographical variations in the small RNA segment of the teratogenic Akabane virus. Virus Res. 121: 84–92. doi: 10.1016/j.virusres.2006.04.007 [DOI] [PubMed] [Google Scholar]
- 30.Yanase T., Kato T., Kubo T., Yoshida K., Ohashi S., Yamakawa M., Miura Y., Tsuda T.2005. Isolation of bovine arboviruses from Culicoides biting midges (Diptera: Ceratopogonidae) in southern Japan: 1985--2002. J. Med. Entomol. 42: 63–67. doi: 10.1093/jmedent/42.1.63 [DOI] [PubMed] [Google Scholar]
- 31.Yoshida K., Tsuda T.1998. Rapid detection of antigenic diversity of Akabane virus isolates by dot immunobinding assay using neutralizing monoclonal antibodies. Clin. Diagn. Lab. Immunol. 5: 192–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeller H., Bouloy M.2000. Infections by viruses of the families Bunyaviridae and Filoviridae. Rev. - Off. Int. Epizoot. 19: 79–91. doi: 10.20506/rst.19.1.1208 [DOI] [PubMed] [Google Scholar]