Abstract
Human ribosomal genes (rDNA) are located in nucleolar organizer regions (NORs) on the short arms of acrocentric chromosomes. Metaphase NORs that were transcriptionally active in the previous cell cycle appear as prominent chromosomal features termed secondary constrictions that are achromatic in chromosome banding and positive in silver staining. The architectural RNA polymerase I (pol I) transcription factor UBF binds extensively across rDNA throughout the cell cycle. To determine if UBF binding underpins NOR structure, we integrated large arrays of heterologous UBF-binding sequences at ectopic sites on human chromosomes. These arrays efficiently recruit UBF even to sites outside the nucleolus and, during metaphase, form novel silver stainable secondary constrictions, termed pseudo-NORs, morphologically similar to NORs. We demonstrate for the first time that in addition to UBF the other components of the pol I machinery are found associated with sequences across the entire human rDNA repeat. Remarkably, a significant fraction of these same pol I factors are sequestered by pseudo-NORs independent of both transcription and nucleoli. Because of the heterologous nature of the sequence employed, we infer that sequestration is mediated primarily by protein–protein interactions with UBF. These results suggest that extensive binding of UBF is responsible for formation and maintenance of the secondary constriction at active NORs. Furthermore, we propose that UBF mediates recruitment of the pol I machinery to nucleoli independently of promoter elements.
Keywords: RNA polymerase I, ribosomal DNA, nucleolar organiser region, nucleolus, upstream-binding factor
In all eukaryotic organisms, the genes encoding the large ribosomal RNAs (rDNA) are transcribed by the dedicated transcription machinery of RNA polymerase I (pol I) in a specific nuclear location, the nucleolus. Determining how the pol I transcription machinery is selectively recruited by rDNA and how chromatin organization and nuclear localization of rDNA influences this process should provide more general insights into eukaryotic gene expression.
Approximately 400 copies of the 43-kb human ribosomal gene repeat are distributed among the short arms of the human acrocentric chromosomes (13, 14, 15, 21, and 22) (Henderson et al. 1972), each of which consists of three bands: p11, p12, and p13. Band p12 contains the ribosomal genes and is termed NOR (nucleolar organizer region), while p11 and p13 are comprised of satellite sequences packaged as heterochromatin. During metaphase, NORs that were transcriptionally active in the previous interphase form prominent chromosomal features termed secondary constrictions, which appear as achromatic gaps when stained with Giemsa or AT-specific fluorochromes such as DAPI (4′,6-diamidino-2-phenylindole) or quinacrine (Sumner 1982). Electron tomography revealed that chromatin within secondary constrictions is 10-fold less condensed than the rest of the chromosome (Heliot et al. 1997). Classically, secondary constrictions are positive in silver staining and often termed AgNORs (Goodpasture and Bloom 1975). Association of the pol I machinery with rDNA during mitosis correlates with the presence of a secondary constriction (Weisenberger and Scheer 1995; Roussel et al. 1996). It is generally considered, but not proven, that some component of the transcription machinery or indeed the act of transcription itself is responsible for the undercondensation of NORs during metaphase.
Efficient transcription of rDNA by pol I requires the formation of a preinitiation complex (PIC) on the promoter including upstream binding factor (UBF) and promoter selectivity factor (SL1) (Bell et al. 1988; Paule and White 2000; Moss and Stefanovsky 2002; Grummt 2003). SL1 is comprised of TATA-binding protein (TBP) and TBP-associated factorsI (TAFI110, TAFI63, and TAFI48) (Comai et al. 1992). SL1 interacts with promoter DNA in a highly sequence-specific manner as illustrated by the species-specific nature of rDNA transcription (Heix and Grummt 1995). PICs recruit an initiation competent subfraction of pol I, defined by the presence of TIF-IA/Rrn3 (Bodem et al. 2000; Miller et al. 2001). Although clearly involved in promoter function, mediated through its ability to interact with other components of the pol I machinery, highly abundant UBF (∼5×105 molecules per cell) has additional roles. It is a so-called architectural transcription factor and is a prime candidate for maintaining the “open” chromatin state of secondary constrictions. The remarkable ability of UBF to bend and loop DNA is mediated through its multiple HMG1-box motifs. Indeed, it can organize promoter DNA into a 360° loop (Bazett-Jones et al. 1994). UBF may also act to prevent or reverse the assembly of transcriptionally inactive chromatin structures catalyzed by linker histone H1 binding (Kermekchiev et al. 1997). A role for HMG1-box proteins in rDNA chromatin organization appears to have been conserved throughout evolution. Hmo1p, a nucleolar protein with a single HMG1-box, has some sequence similarity to UBF and strongly enhances rDNA transcription in yeast (Gadal et al. 2002). Both UBF and Hmo1 belong to the sequence nonspecific class of HMG1-box proteins (Thomas and Travers 2001). Consistent with this, UBF binding is not restricted to regulatory elements as it interacts in vivo with sequences across the entire rDNA repeat (O'Sullivan et al. 2002). Despite this diversity of binding sites, UBF binds selectively to rDNA as it is exclusively localized to nucleoli during interphase and at NORs during mitosis (Roussel et al. 1993). We have proposed a model in which rDNA repeats contain specialized high-affinity binding sequences that bind UBF in a cooperative manner and induce spreading of UBF onto adjacent lower-affinity binding sites across the rDNA repeat (O'Sullivan et al. 2002).
In order to determine if extensive binding of UBF is responsible for the undercondensation of NORs during metaphase, we have generated cell lines in which large arrays of a heterologous UBF-binding sequence have been integrated into ectopic sites on human chromosomes. These arrays bind UBF independent of nuclear location and adopt a morphology characteristic of NORs. Our results provide compelling support for a model in which UBF underpins NOR morphology.
A second major finding of the work presented here relates to the mechanism by which the pol I transcription machinery is recruited to nucleoli. An extensive chromatin immunoprecipitation (ChIP) analysis revealed that pol I and SL1 can be found at high levels associated with sequences throughout the 30-kb intergenic spacer of human rDNA. The inference being that this is mediated through protein–protein interactions with UBF. Support for this model is provided by the observation that ectopic UBF-binding site arrays sequester a significant fraction of every component of the pol I transcription machinery even to sites outside nucleoli and independent of both promoter sequences and transcription. These results force us to re-evaluate how the pol I machinery is recruited to nucleoli and rDNA.
Results
Generation and mapping of heterologous UBF-binding site arrays
Blocks of 60/81-bp repeats in the intergenic spacer of Xenopus ribosomal genes function as transcriptional enhancers (Labhart and Reeder 1984) and are among the best characterized UBF-binding sites. UBF binds cooperatively to these Xenopus Enhancer (XEn) elements (Pikaard et al. 1989; Putnam and Pikaard 1992) and is required for enhancer function (McStay et al. 1997). Despite the lack of sequence homology with human rDNA, human UBF can bind to these elements (Bell et al. 1989; McStay et al. 1997). It should be stressed that due to the species-specific nature of transcription by pol I, it is not considered likely that the other components of the human pol I transcription machinery would interact directly with these Xenopus sequences. We reasoned that by introducing large tandem arrays of these sequences into human chromosomes, we could determine, first, if UBF localization within the cell nucleus was solely a reflection of DNA binding specificity and, second, if extensive UBF binding was responsible for the morphology of NORs, i.e., secondary constriction and silver staining. To this end the plasmid pXEn8 was constructed, which contains eight blocks of the enhancer element or eighty 60/81-bp repeats. The pXEn8 insert was transfected into a human fibrosarcoma cell line, HT1080, together with a blasticidin resistance marker in a 200:1 ratio using calcium phosphate precipitation.
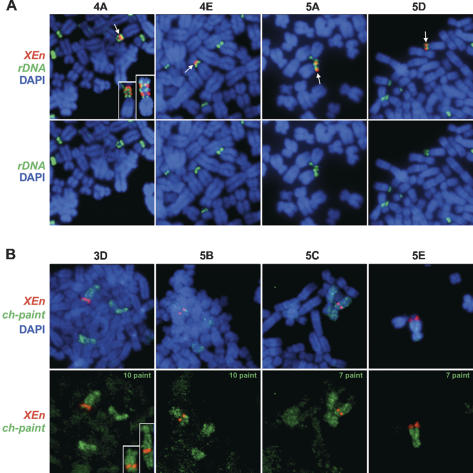
Eight stably transfected cell lines were chosen for further analysis. The XEn content of these cell lines was determined by Southern blotting and ranges from 105 kb to 2.1 Mb (Table 1). It should be pointed out that this represents an average cellular DNA content. Fluorescent in situ hybridization (FISH) was performed on metaphase spreads from each clone using spectrum red-labeled XEn DNA and a spectrum green-labeled human rDNA probe or chromosome paint to establish the site of integration of XEn sequences (Fig. 1). In four of the clones (4A, 4E, 5A, and 5D) XEn sequences have integrated into the p-arms of acrocentric chromosomes, the sites of human ribosomal gene clusters (Fig. 1A). In clones 5A and 5D, the XEn sequences appear to have inserted into the NOR present on 13p. In clone 4A XEn sequences again appear to have inserted into 13p, but in this case integration is associated with a high frequency of rearrangement such that there are alternating blocks of human rDNA and XEn present on this chromosome. The number of alternating blocks varies from cell to cell. In the examples shown there are one, two, or three blocks of each sequence type (see insets in Fig. 1A). We have observed as many as six alternating blocks in some metaphase spreads. Interestingly, a change in the size of the short arm of acrocentric chromosomes associated with duplication of the NOR is a naturally occurring variant observed in human chromosomes (Perez-Castillo et al. 1986). Human acrocentric chromosomes with as many as four NORs have been observed. In clone 4E, XEn sequences have integrated adjacent to or within the NOR present on acrocentric chromosome 21 or 22.
Table 1.
Size and location of XEn arrays
| Clone | Array size (kb) | Chromosomal location |
|---|---|---|
| 4A | 2100 | 13p (high frequency of duplication) |
| 4E | 250 | 21p or 22p |
| 5A | 250 | 13p |
| 5D | 250 | 13p |
| 3D | 1400 | 10q (close to telomere, rearrangements) |
| 5B | 175 | 10p (adjacent to centromere) |
| 5C | 105 | 7q (middle of arm) |
| 5E | 850 | 7q (adjacent to telomere) |
The size of XEn arrays present in each clone was determined by quantitative Southern blotting. Chromosomal location of XEn arrays was demonstrated by FISH on metaphase spreads (Fig. 1).
Figure 1.
Generation and mapping of UBF-binding site arrays. (A) XEn arrays map to the short arms of acrocentric chromosomes in clones 4A, 4E, 5A, and 5D. Human NORs were visualized using a spectrum green-labeled probe derived from the intergenic spacer of the human ribosomal gene repeat. XEn sequences were visualized with spectrum red-labeled insert of the plasmid pXEn8. Chromosome identity was determined using enhanced reverse DAPI banding (SmartCapture; data not shown). Merges of both probes and DAPI staining are shown in the top panels. Arrows indicate the XEn arrays. In the bottom panels, only human rDNA and DAPI signals are shown. (B) XEn arrays map to submetacentric chromosomes in clones 3D, 5B, 5C, and 5E. Chromosomes were initially identified using enhanced reverse DAPI banding and then confirmed using chromosome paints labeled with spectrum green. XEn arrays were visualized as above. Merges of both probes and DAPI staining are shown in the top panels. In the bottom panels, DAPI signal is omitted in order to more readily visualize the chromosome paint. The identity of the chromosome paint is shown in the top right corner of each panel.
In clones 3D, 5B, 5C, and 5E XEn sequences have inserted into nonacrocentric chromosomes (i.e., chromosomes that do not bear NORs and that are not associated with nucleoli) (Fig. 1B). In 3D, XEn sequences have inserted close to the telomere on the q-arm of chromosome 10. In a substantial number of cells in this clone, a rearrangement was observed in which additional chromosome 10 sequences have been added distal to the XEn array (see insets in Fig. 1B). In some cases the banding pattern suggests that the entire q-arm of 10 has been duplicated. In 5C and 5E XEn sequences are located in the middle of and adjacent to the telomere, respectively, of the q-arm of chromosome 7. In 5B XEn sequences are located close to the centromere on the p-arm of chromosome 10.
UBF binds to XEn arrays in vivo
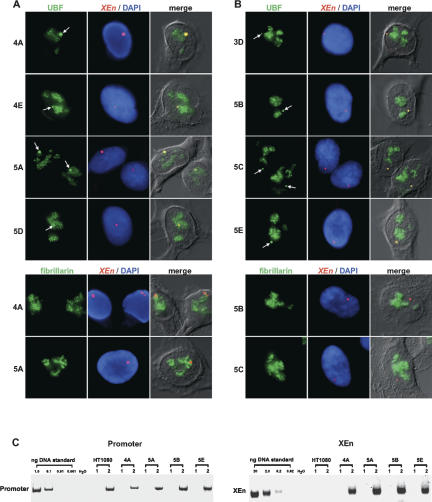
To determine the UBF loading status of the XEn arrays, we performed combined immuno-FISH on cells in which the three-dimensional structure has been maintained using paraformaldehyde (PFA) fixation (Fig. 2). XEn DNA was visualized as described above, and UBF was visualized with a FITC-labeled affinity-purified hUBF antibody. In every case UBF colocalizes with XEn arrays, whether associated with nucleoli or not. In clones 4A, 4E, 5A, and 5D, XEn arrays are associated with nucleoli in the majority of cells, as would be expected given their location on acrocentric chromosomes (Table 2). However, even when localized within nucleoli, UBF foci associated with XEn arrays can be clearly distinguished from “nucleolar” UBF due to their large size and high fluorescence intensity, especially apparent in clones 4A and 5A (Fig. 2A). In order to confirm that UBF-associated XEn arrays are distinct from nucleoli, we performed combined immuno-FISH using antibodies against fibrillarin, a nucleolar protein involved in processing of precursor ribosomal RNA. Whereas the overall nucleolar staining observed with UBF and fibrillarin antibodies is similar, no bright foci of fibrillarin staining are associated with XEn arrays. This point is more clearly illustrated in cells where XEn arrays are not associated with nucleoli (see below).
Figure 2.
UBF binds to XEn arrays in vivo. (A) Combined immuno-FISH on clones 4A, 4E, 5A, and 5D containing XEn arrays on the short arms of acrocentric chromosomes. (Top four rows) UBF was visualized using FITC-conjugated affinity-purified antibodies. (Bottom two rows) Fibrillarin was visualized in clones 4A and 5A using monoclonal antibody 72B9 combined with Cy2-labeled α-mouse secondary antibodies. XEn arrays were visualized with a spectrum red-labeled probe. For each clone, the row shows antibody staining (left), the XEn probe with DAPI counterstain (center), and the merge of antibody and XEn signals with a DIC image of the cell (right). Arrows indicate XEn arrays. (B) Combined immuno-FISH on clones 3D, 5B, 5C, and 5E containing XEn arrays on submetacentric chromosomes. Performed and illustrated as above. (C) Chromatin immunoprecipitation demonstrates that UBF is bound to XEn arrays. ChIP was performed on soluble chromatin fractions prepared from HT1080 parental cells and clones 4A, 5A, 5B, and 5E using control IgG (lanes labeled 1) or affinity-purified α-UBF antibodies (lanes labeled 2). PCR reactions were performed using primers specific for the human ribosomal gene promoter (left) or the XEn array (right). Serial dilutions of a cosmid comprising the entire human ribosomal repeat or genomic DNA isolated from clone 4A were used as standard templates in PCRs with promoter and XEn primers respectively.
Table 2.
Interphase location of XEn arrays
| Clone (chromosomal location) | XEn nonnucleolar |
|---|---|
| 4A (13p) | 17% (n = 54) |
| 4E (21p or 22p) | 38% (n = 37) |
| 5A (13p) | 5% (n = 63) |
| 5D (13p) | 4% (n = 48) |
| 3D (10q) | 83% (n = 35) |
| 5B (10p) | 71% (n = 34) |
| 5C (7q) | 84% (n = 44) |
| 5E (7q) | 83% (n = 54) |
Combined immuno-FISH experiments were performed on XEn array containing cell lines. The percentage of cells in which the XEn array (visualized with a spectrum red probe) and nucleoli (visualized with UBF, RPA43, or fibrillarin antibodies) were clearly separable was calculated. The number of cells analyzed (n) for each clone is also shown.
In clones 3D, 5B, 5C, and 5E, XEn arrays are present on nonacrocentric chromosomes. As would be expected, arrays present on these chromosomes are not associated with nucleoli (Fig. 2B). In 81% of the cells analyzed, XEn sequences are clearly distinct from the nucleolus (Table 2). Note that this figure is an underestimate since signals that abut nucleoli are not included. However, immuno-FISH clearly demonstrates that UBF still associates with these arrays. Furthermore, the extranucleolar foci of UBF staining are completely devoid of fibrillarin (Fig. 2B). The dissociation of nucleolar markers and extranucleolar UBF foci in these cell lines is further illustrated by combined immuno-FISH experiments that demonstrate that neither nucleolin or B23 (Nucleophosmin) is associated with XEn arrays in interphase nuclei (Supplementary Fig. 1).
Mammalian cells contain approximately equal amounts of two UBF forms as a result of splice site variation. UBF 2 differs from UBF 1 in missing 37 residues from the second HMG box (O'Mahony and Rothblum 1991). This deletion renders UBF 2 nonfunctional at the promoter (Kuhn et al. 1994). Although the antibodies used here cannot distinguish between UBF splice variants, we have shown in transfection experiments that both UBF 1 and 2 associate with XEn arrays (Supplementary Fig. 3A).
The binding of UBF to XEn arrays was further confirmed by ChIP in four of the clones (4A, 5A, 5B, and 5E) (Fig. 2C). In PCR with primers specific for the human rDNA promoter, product was obtained from α-UBF ChIPs in each cell line tested, including the parental HT1080 cell line. Using XEn primers product was obtained from each of the clones but not from HT1080 cells. Real-time PCR revealed that the XEn DNA content of α-UBF ChIPs was >100-fold higher than that of control ChIPs (data not shown). Thus, colocalization of UBF and XEn DNA reflects direct binding of UBF to the array.
UBF binding to XEn arrays induces formation of novel silver staining secondary constrictions
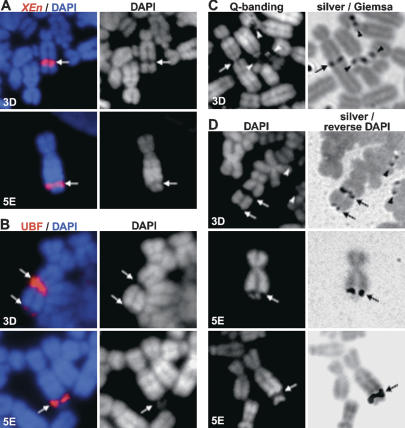
Human NORs contain on average 3 Mb of rDNA (Sakai et al. 1995). Although none of the XEn arrays described here are this large, we investigated the possibility that they would appear as secondary constrictions during metaphase. In these experiments we focused on clones 3D and 5E that contain large XEn arrays present on submetacentric chromosomes. DAPI staining of metaphase spreads prepared from clones 3D and 5E clearly demonstrated the presence of novel secondary constrictions (achromatic regions) that colocalized with XEn arrays visualized by FISH (Fig. 3A). Antibody staining of chromosomes confirmed that UBF remains associated with these novel secondary constrictions during mitosis (Fig. 3B). While achromatic regions coinciding with the presence of XEn DNA and UBF are particularly prominent in clones 3D and 5E, the achromatic region is less apparent in clones 5B and 5C, correlating with the smaller size of the XEn array (data not shown).
Figure 3.
XEn arrays display characteristics of NORs. (A) XEn arrays form secondary constrictions. Metaphase spreads prepared from clones 3D and 5E were hybridized with a spectrum red-labeled probe to visualize XEn arrays. Merges of probe and DAPI staining are shown in left panels. Grayscale images of DAPI staining are shown on the right. XEn arrays and secondary constrictions are denoted by arrows. (B) UBF remains associated with XEn arrays during metaphase. Chromosomes were stained with rhodamine-conjugated affinity-purified α-UBF antibodies and mounted in DAPI/Antifade (left). Grayscale images of DAPI staining are shown on the right. XEn arrays and secondary constrictions are denoted by arrows. (C) XEn arrays are positive in silver staining. (Left) Chromosomes prepared from clone 3D were Q-banded, and images were captured. (Right) Subsequently, chromosomes were stained with silver and counterstained with Giemsa, and images of the same fields were captured. Pseudo-NORs are indicated by arrows, NORs by arrowheads. (D) XEn arrays are positive in silver staining. Chromosomes prepared from clones 3D and 5E were stained with DAPI (left) and silver. The right panels show a merge of silver and reverse DAPI images. Note the presence of an additional UBF and silver-positive smaller XEn array at the telomere of the rearranged chromosome 10 in clone 3D. Pseudo-NORs are indicated by arrows and NORs by arrowheads.
A novel secondary constriction was also clearly visible after quinacrine staining (Q-banding) of chromosomes prepared from clone 3D (Fig. 3C). This locus was positive in subsequent silver staining. Chromosomes from clones 3D and 5E were also stained with silver followed by DAPI (Fig. 3D). Again silver deposits colocalized with the large secondary constrictions generated by the XEn array. We conclude from these experiments that recruitment of UBF to XEn arrays results in formation of a chromatin structure with all the primary characteristics of NORs. Henceforth, we will refer to these structures as pseudo-NORs since they do not organize formation of a nucleolus.
UBF does not associate with introduced LacO arrays
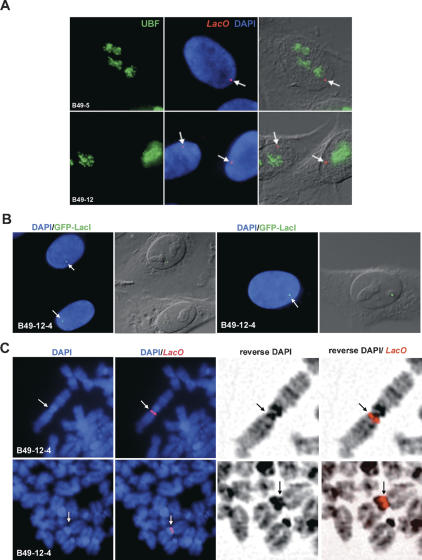
It is conceivable that association of UBF with XEn arrays could be a consequence of their highly repeated structure rather than the underlying DNA sequence. In order to demonstrate specificity of UBF binding to XEn arrays, we assessed the UBF loading status of extensive Lac operator (LacO) arrays introduced into HT1080 cells (Chubb et al. 2002). Combined immuno-FISH was performed on clones B49-12 and B49-5, which contain LacO arrays on chromosome 1 and 5, respectively (Fig. 4A). UBF was visualized as above, and the LacO arrays were visualized with a spectrum red-labeled probe. The results clearly demonstrate that UBF does not associate with LacO arrays, thus illustrating the specificity of UBF binding to XEn arrays.
Figure 4.
HT1080 cells with arrays of lac repressor-binding sites (LacO). (A) UBF is not targeted to LacO arrays. Combined immuno-FISH was performed on HT1080 cells containing arrays of lac operators on chromosome 1 (clone B49-12) and 5 (clone B49-5). UBF was detected using FITC-conjugated affinity-purified antibodies (left); the LacO arrays were visualized with a spectrum red-labeled probe (center, merged with DAPI). (Right) Merged fluorescence and DIC images of the cells. Arrows depict LacO arrays. (B) Clone B49-12-4 is a derivative of B49-12 (above, in A) in which GFP-LacI is stably expressed. Panels show GFP-LacI visualized in interphase cells. The left panel in each case shows a merge of GFP and DAPI signals, the right shows a merge of GFP and DIC images. (C) Binding of GFP-Lac repressor fusion protein (GFP-LacI) to LacO arrays does not induce formation of a secondary constriction. Metaphase spreads prepared from B49-12-4 cells were stained with DAPI, and LacO arrays were visualized with a spectrum red-labeled probe. Panels on the left show DAPI staining alone and merged with the spectrum red signal. Relevant sections of metaphase spreads are shown enlarged in the right panels. DAPI staining has been reversed to enhance chromosome banding.
Lac repressor binding does not induce formation of a secondary constriction over LacO arrays
It is possible that formation of pseudo-NORs is a consequence of binding of any abundant sequence-specific factor to extensive arrays of its target site. To address this issue, we have determined if expression of Lac repressor can induce formation of a secondary constriction over LacO arrays. In cell line B49-12-4 (derived from B49-12), a Lac repressor GFP fusion protein (GFP-LacI) is stably expressed (Chubb et al. 2002). Binding of GFP-LacI to the LacO array present at 1q11 is demonstrated by a focus of GFP signal in interphase nuclei (Fig. 4B). Metaphase chromosomes were then prepared from this cell line, and the LacO array was revealed using a spectrum red-labeled probe (Fig. 4C). Clearly, GFP-LacI does not induce formation of a novel secondary constriction associated with the LacO array. This is particularly evident in the reversed DAPI image shown in the right panels. In a previous study (Strukov et al. 2003) with cell lines containing larger LacO arrays (∼2 Mb), binding of GFP-LacI was shown not to result in an altered metaphase chromosome structure as determined both by DAPI staining and scanning electron microscopy. These results highlight the specialized role that UBF has in specifying NOR structure.
The pol I transcription machinery interacts with sequences across the entire human rDNA repeat.
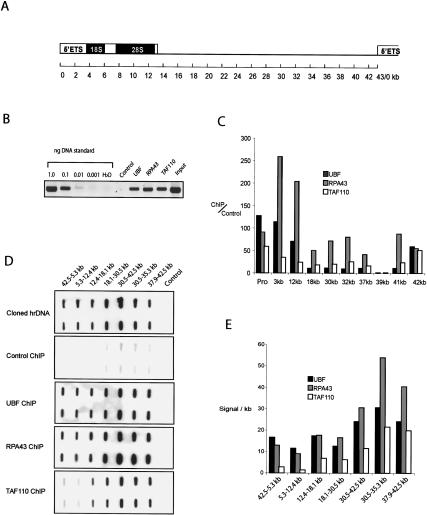
To date it has been assumed that the pol I transcription machinery is recruited directly to the rDNA promoter, mediated by interactions between promoter-bound UBF and other components of the pol I transcription machinery (Beckmann et al. 1995; Hanada et al. 1996). Indeed, nucleolar localization of the pol I machinery is considered to be entirely due to promoter interaction and engagement in transcription (Dundr et al. 2002). Our recent finding that UBF binds extensively across the rDNA repeat (O'Sullivan et al. 2002) already suggested an alternative model. Specifically, nonpromoter-bound UBF could greatly enhance the ability of rDNA to recruit the pol I machinery. A prediction of this model is that components of the pol I machinery could be found associated with sequences across the entire rDNA repeat. In our previous work, a limited series of ChIP experiments with pol I and SL1 antibodies suggested that this was not the case (O'Sullivan et al. 2002). Here we have reinvestigated this question using improved antibodies and more quantitative methodologies. ChIP was performed on nucleolar chromatin prepared from HT1080 cells with affinity-purified UBF, RPA43, and TAFI110 antibodies. Initially, we performed PCR using primers specific for the human rDNA promoter (Fig. 5B). As expected, each of these factors is present at the promoter. Real-time PCR was then performed using primer pairs specific to the promoter and primer pairs specific for a further nine sites distributed across the 43-kb rDNA repeat (Fig. 5C). The amount of target sequence present in UBF, RPA43 and TAFI110 ChIPs was calculated relative to that in a control ChIP. From these data it is clear that each of these factors can be found associated with sequences across the rDNA repeat. To obtain a more global and quantitative description of the distribution of these factors across the rDNA repeat, we radiolabeled DNA recovered from UBF, RPA43, TAFI110, and control ChIPs. Direct labeling was possible since real-time PCR data indicated that we recovered between 5 and 10 ng of rDNA from each ChIP. Labeled DNA was then used as a probe against slot blots loaded with a panel of cloned rDNA subfragments that covers the entire rDNA repeat (Fig. 5D). Radiolabeled cosmid DNA from a clone comprising the entire rDNA repeat was used as a hybridization control. Hybridization signals were quantified using a PhosphorImager. Typically the signal observed over a given rDNA subfragment with labeled UBF, RPA43 and TAFI110 ChIPs was 20- to 40-fold above that observed with the control ChIP. Ratios of signals with radiolabeled cosmid closely reflected that predicted on the basis of the size of the rDNA subclone loaded in each slot. Consequently, we expressed the results as signal per kilobase over each subfragment of the rDNA repeat (Fig. 5E). As previously shown, results presented here confirm that UBF is found associated with sequences across the entire rDNA repeat. The pol I subunit RPA43 is similarly distributed. Levels of both UBF and RPA43 vary by at most two- to threefold across the repeat. In contrast TAFI110 is found associated with sequences across the intergenic spacer at levels that are 10- to 15-fold above that observed over transcribed sequences. The somewhat surprising conclusion from these experiments is that higher levels of pol I and SL1 are associated with the intergenic spacer than with the promoter and transcribed sequences. Moreover, the majority of the pol I transcription machinery that is associated with rDNA is not actively engaged in the transcription process.
Figure 5.
RNA pol I transcription machinery interacts with DNA sequences across the entire rDNA repeat. (A) The 43-kb human rDNA repeat derived from GenBank accession number U13369 is shown in cartoon form; 0 kb defines the origin of the 13-kb 47S pre rRNA transcript. (B) Nucleolar ChIP. ChIP was performed on nucleolar chromatin prepared from HT1080 cells with affinity-purified UBF, RPA43, and TAF110 antibodies. PCR reactions were performed on one-fortieth of the DNA recovered from each ChIP using primers specific for the human ribosomal gene promoter. A serial dilution of a cosmid comprising the entire human ribosomal repeat was used as a standard template. In a control ChIP, antibody was omitted. (C) Real-time PCR. Real-time PCR was performed on the above ChIPs and DNA standards with promoter specific primers and primers located at a further nine sites across the human rDNA repeat. The amount of target sequence in antibody ChIPs versus control ChIPs was calculated and plotted for each factor and for each primer set. The location of primer pairs is shown below each data set as appropriate. (D) Hybridization with radiolabeled ChIPs. Half of the DNA recovered from antibody and control ChIPs was radiolabeled and used to probe slot blots that were loaded in duplicate with cloned subfragments of the rDNA repeat and vector DNA. To demonstrate uniformity of hybridization across the repeat, the array was also probed with a cosmid clone that comprises the entire 43 kb of human rDNA. The identity of the probe is shown on the left of each panel, and the location of each rDNA subfragment is shown above the appropriate slots. Note that all hybridizations were performed and exposed simultaneously. (E) Quantitation of hybridization. Hybridization signals were quantified by using a Molecular Imager (Bio-Rad). Hybridization signal per kilobase (in arbitrary units) was plotted for each rDNA subfragment and each probe.
Pseudo-NORs sequester the pol I transcription machinery
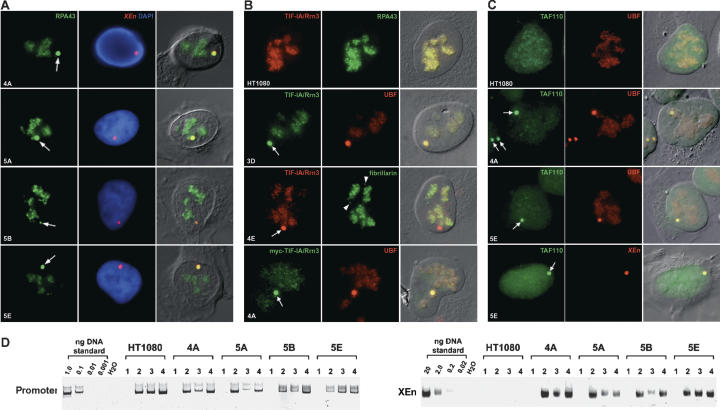
The most reasonable interpretation of the above data is that the pol I machinery is distributed across the entire rDNA repeat as a consequence of interactions with UBF rather than directly with DNA. The pseudo-NORs described here provide a unique opportunity to look at recruitment of the pol I transcription machinery to UBF loaded sequences uncoupled from promoters, transcription, and the nucleolar environment. Initially we looked at RPA43. Combined immuno-FISH clearly demonstrated colocalization of a significant fraction of total RPA43 with XEn arrays (Fig. 6A). Pol I subunits, RPA195, RPA135, and PAF53 are also associated with UBF on these arrays (Supplementary Figs. 2, 3B). Thus, at least four pol I subunits are sequestered by pseudo-NORs.
Figure 6.
Pol I transcription machinery targets to pseudo-NORs. (A) Colocalization of RPA43 and XEn sequences. Combined immuno-FISH was performed on clones 4A, 5A, 5B, and 5E. XEn arrays were visualized with a spectrum red-labeled probe and RPA43 with FITC-conjugated affinity-purified antibodies. Left and center panels show antibody staining and XEn probe with DAPI counterstain, respectively. (B, left panels) TIF-IA/Rrn3 was visualized in HT1080 cells and clones 3D and 4E with affinity-purified antibodies conjugated with FITC or rhodamine. (Center panels) Cells were counterstained with α-RPA43, α-UBF or α-fibrillarin antibodies. (Bottom panels) Clone 4A was transfected with a myc-tagged TIF-IA/Rrn3 expression construct. (Left panel) Exogenous TIF-IA/Rrn3 was visualized with α-myc monoclonal antibody 9E10. (Center panel) Cells were also stained with α-UBF antibodies. (C, left panels) TAFI110 was visualized in HT1080 cells and clones 4A and 5E with affinity-purified antibodies conjugated with FITC. (Center panels) Cells were counterstained with rhodamine-conjugated α-UBF antibodies. (Bottom panels) Combined immuno-FISH was performed on clone 4A. TAFI110 was visualized as above (left panel) and XEn arrays with a spectrum red-labeled probe (center panel). In A–C, merges of fluorescence signals with a DIC image of the cell are shown in the right panels. Pseudo-NORs and Cajal bodies are indicated by arrows and arrowheads, respectively. (D) ChIP confirms that pol I, SL1, and TIF-IA/Rrn3 are associated with XEn Arrays. ChIP was performed on soluble chromatin fractions prepared from HT1080 parental cells and clones 4A, 5A, 5B, and 5E with control IgG (lane 1), α-RPA43 (lane 2), α-TIF-IA/Rrn3 (lane 3), or α-TAFI110 affinity-purified antibodies (lane 4). PCR reactions were performed using primers specific for the human ribosomal gene promoter (left) or the XEn array (right). Standards were as in Figure 2.
The presence of TIF-IA/Rrn3 at pseudo-NORs could render the associated pol I competent for transcription initiation. In each of the clones but not the parental HT1080 cells, we observed a characteristic focus of TIF-IA/Rrn3 staining that colocalizes with UBF but is not associated with fibrillarin (Fig. 6B). Additionally, myc epitope tagged TIF-IA/Rrn3, introduced into cells by transient transfection, colocalizes with UBF at pseudo-NORs.
Next, we examined the possibility that pseudo-NORs could sequester SL1. By using TAFI110 antibodies, we generally observed a weak overall nuclear staining in the parental HT1080 cells, with a slight preference for the nucleolus (Fig. 6C). Remarkably, in XEn containing clones a large fraction of cellular TAFI110 colocalized with UBF in the characteristic bright foci of pseudo-NORs. Immuno-FISH confirmed association of TAFI110 with XEn sequences (Fig. 6C). We have also demonstrated that TBP targets to pseudo-NORs (Supplementary Fig. 3C). These results show that at least two components of SL1 are sequestered by pseudo-NORs.
It is most likely that pol I, TIF-IA/Rrn3, and SL1 are associated with pseudo-NORs through contacts with UBF rather than directly with XEn sequences. This close association was confirmed by ChIP experiments. Affinity-purified antibodies against RPA43, TIF-IA/Rrn3, TAFI110, and control IgGs were used in ChIPs of chromatin prepared from clones 4A, 5A, 5B, 5E and the parental HT1080 cell line (Fig. 6D). As before, PCR was performed on DNA recovered from ChIPs with primers either specific for XEn sequences or the human ribosomal gene promoter. In PCR with primers specific for the human rDNA promoter, product was obtained from ChIPs with RPA43, TIF-IA/Rrn3, and TAFI110 antibodies but not control IgGs in each cell line tested, including the parental HT1080 cell line. Using XEn primers product was obtained from each of the clones but not from parental HT1080 cells. Thus, colocalization of RPA43, TIF-IA/Rrn3, and TAFI110 with XEn arrays reflects intimate association of these components of the pol I transcription machinery with the XEn array in all likelihood through UBF.
One might imagine that the open chromatin structure of pseudo-NORs, specified by UBF, might provide a sink not just for the pol I transcription machinery but for that of pol II or III. We have demonstrated that pol II can be ChIPed onto U1snRNA genes (transcribed by pol II) but not pseudo-NORs or rDNA. Conversely, pol I can be ChIPed onto pseudo-NORs and rDNA but not U1snRNA genes (Supplementary Fig. 4). This result illustrates the specificity with which the pol I transcription machinery interacts with pseudo-NORs.
Pseudo-NORs are transcriptionally silent
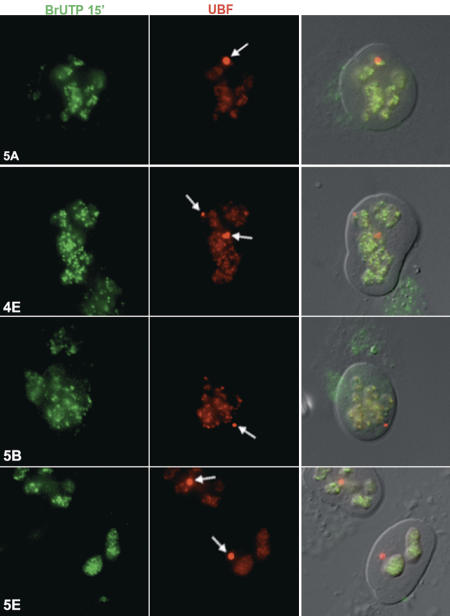
Although pol I would not be expected to transcribe XEn arrays due to the lack of promoter sequences, the presence of the entire pol I machinery prompted us to formally address this issue. BrUTP incorporation experiments were performed in permeabilized cells in the presence of α-amanitin to suppress pol II and III transcription (Fig. 7). Incorporated BrUTP was visualized with anti-BrdUTP antibodies (Roche) combined with FITC-conjugated secondary antibodies. Nucleoli and pseudo-NORs were visualized with UBF antibodies. Within nuclei, BrUTP incorporation is restricted to nucleoli. No BrUTP incorporation is detected at pseudo-NORs, whether associated with nucleoli (clones 5A and 4E) or independent of nucleoli (clones 5B and 5E). Thus, despite the fact that the entire pol I transcription machinery appears to be associated with pseudo-NORs, it is not engaged in transcription.
Figure 7.
Pseudo-NORs are transcriptionally inactive. Ongoing pol I-mediated transcription was visualized in clones 5A, 4E, 5B, and 5E by incorporation of BrUTP into nascent transcripts. (Left panels) BrUTP was visualized with anti-BrdUTP antibodies (Roche) combined with FITC-conjugated secondary antibodies. (Center panels) UBF was detected using affinity-purified antibodies conjugated to rhodamine. Right panels show merged fluorescence and DIC images of the cells. Arrows depict pseudo-NORs.
Finally, an RNA analysis revealed that the presence of pseudo-NORs has no detectable effect on the levels of pre-rRNA synthesis (Supplementary Fig. 5).
Discussion
One of the remarkable features of this work is that without exception XEn arrays are bound by UBF in vivo. This occurs independently of integration site and position within the interphase nucleus. The first obvious implication of this result is that UBF localization within the nucleus is entirely driven by rDNA binding specificity. Moreover, the strictly nucleolar localization of UBF in normal cells reflects the presence of rDNA. Nucleolar targeting of UBF independently of its DNA binding activity appears to have little or no contribution to its subnuclear localization (Maeda et al. 1992). XEn sequences were originally chosen for experimental reasons. These included their high affinity for UBF and lack of homology with the human ribosomal gene repeat. An important feature is their ability to bind UBF in a cooperative manner (Putnam and Pikaard 1992), a fact we believe was critical to the success of these experiments. We envisage that murine enhancer elements (Kuhn et al. 1990) and as yet unidentified elements in the human ribosomal gene repeat bind UBF in a similar manner to XEn sequences. A remaining issue is how UBF spreads from high-affinity binding sites to lower-affinity sites across the rDNA repeat. In the future it will be of interest to include non-UBF-binding sequences in XEn arrays to study this spreading process.
At the chromosomal level, the results presented here provide compelling evidence for the involvement of UBF in specifying the morphology of NORs. XEn arrays are sufficient to induce formation of structures apparently identical to true NORs as evidenced by positive silver staining and the lack of DAPI staining or Q-banding. During interphase the major AgNOR proteins are nucleolin and B23/nucleophosmin, and during mitosis the AgNOR proteins are the pol I large subunit RPA195 and 135, UBF, and a 50-kDa protein (Roussel and Hernandez-Verdun 1994). Nucleolin and B23/nucleophosmin do not associate with pseudo-NORs during interphase. Both pol I and UBF associate with mitotic pseudo-NORs and presumably are responsible for silver staining.
The precise role of UBF in forming secondary constrictions is not yet clear to us, but we can now make a number of conclusions. First, as pseudo-NORs are inactive, transcription in the previous interphase is not a prerequisite. Second, the ability to form secondary constrictions is intrinsic to rDNA. The de novo formation of pseudo-NORs at ectopic sites demonstrates that sequences distal and proximal to NORs are not required. Finally, as UBF-binding sites are sufficient to specify this structure, binding sites for other pol I factors are not required. A remaining issue is whether core histones are associated with pseudo-NORs. Initial ChIP analysis has revealed the presence of core histones on XEn sequences (data not shown). However, in the absence of sequential ChIP data, we cannot confirm precise colocalization of UBF and nucleosomes. In this regard it is worth pointing out that Pikaard and colleagues (Kermekchiev et al. 1997) have demonstrated that UBF can bind to nucleosomes with high affinity and displace linker histone H1. This observation may be relevant to the undercondensation observed at NORs and pseudo-NORs during metaphase.
At this point it is worth reviewing recent work concerning the activation and repression of rDNA in a chromatin and chromosomal context. The presence of a binding site (T0) for the pol I transcription termination factor TTF1 immediately upstream of the gene promoter appears to be a conserved feature of vertebrate rDNA. Early evidence suggested that in addition to its role in transcription termination this element also functioned as a promoter element (McStay and Reeder 1990). More recently, it has been demonstrated that in vitro transcription of plasmid templates, packaged as chromatin, is entirely dependent on the presence of TTF1 and the T0-binding site (Langst et al. 1997). It was proposed that TTF1 mediated the recruitment of an activating chromatin remodeling complex to rDNA (Langst et al. 1998). Curiously, the only TTF1-associated remodeling complex described thus far functions in silencing of rDNA through the action of histone deacetylases and DNA methyltransferases (Santoro et al. 2002). The process of reactivating a silent NOR may require TTF1 and activating remodeling complexes. However, our observation that pseudo-NORs adopt the morphology of active NORs, including recruitment of the pol I machinery, all without the presence of TTF1-binding sites, argues strongly that the presence of high-affinity UBF-binding sites is sufficient for maintaining the open chromatin state of NORs once activated.
UBF is only found in vertebrates, yet secondary constrictions occur in all eukaryotes. In yeast, Hmo1p is thought to be the functional homolog of UBF (Gadal et al. 2002). It is tempting to speculate that other nonvertebrate eukaryotic organisms will utilize an abundant, nucleolar Hmo1-like HMG protein to specify NOR structure.
We have previously demonstrated that human acrocentric chromosomes with transcriptionally silent NORs can nonetheless associate with nucleoli (Sullivan et al. 2001). This occurred independently of their UBF loading status. We interpreted this as evidence for the presence of elements on human acrocentric chromosomes out with the NOR that are responsible for this localization. Our observation that pseudo-NORs at ectopic chromosomal locations seldom associate with nucleoli supports this conclusion. However, it is also conceivable that when located on nonacrocentric chromosomes, pseudo-NORs come under the influence of more dominant positional cues.
The other major finding of this work has profound implications for our understanding of how the pol I transcription machinery is recruited to rDNA. We have demonstrated that a significant amount of the nucleolar pol I transcription machinery is associated with sequences present in the 30-kb intergenic spacer of the human rDNA repeat. Polymerase associated with the transcribed region of the repeat is presumably engaged in transcription, whereas that associated with the intergenic spacer is not. This point is not formally proven here, but our results with pseudo-NORs support this conclusion. SL1 appears to be more restricted to the intergenic spacer; possibly it is excluded from actively transcribing polymerases. The most reasonable assumption is that these factors are recruited to sites in the intergenic spacer through protein–protein contacts with UBF. Strong support for this contention comes from our observation that pseudo-NORs, generated using a heterologous sequence, sequester pol I, SL1, and TIF-IA/Rrn3 independent of promoters, transcription, and nucleoli. SL1 subunits, TAFI48, and TBP have been shown individually to interact with UBF (Kwon and Green 1994; Beckmann et al. 1995). The pol I subunit PAF53 has been shown to contact UBF directly (Hanada et al. 1996). The yeast homolog of PAF53, RPA49, interacts with the presumptive yeast homolog of UBF, hmo1p (Gadal et al. 2002). Direct contacts between TIF-IA/Rrn3 and UBF have not been described. Presumably TIF-IA/Rrn3 is recruited to pseudo-NORs by interactions with pol I or SL1 (Miller et al. 2001; Yuan et al. 2002). Although promoters may contribute to sequestration of the pol I machinery by rDNA in nucleoli, the efficiency of targeting to pseudo-NORs suggests that UBF bound to nonpromoter sequences provides the major contributing factor. The observation that the presence of pseudo-NORs does not result in down-regulation of rDNA transcription provides further evidence for the excess of nucleolar pol I transcription factors.
Until recently, it was generally considered that rDNA transcription was mediated by stable PICs that supported multiple rounds of transcription initiation. The appeal of this model was that it explained the rapid reinitiation rate on ribosomal gene promoters evidenced by the high density of polymerase molecules observed on active genes (Miller and Bakken 1972; Puvion-Dutilleul 1983; Scheer and Benavente 1990). Recently, however, this view of transcription has been challenged. In yeast, there is now strong evidence that PICs are recycled after each round of transcription (Aprikian et al. 2001). Microscopy of live mammalian cells expressing GFP-tagged transcription factors and pol I subunits has revealed the dynamic nature of the eukaryotic nucleus. Photobleaching experiments have revealed that UBF and SL1 dwell on ribosomal DNA only for a few seconds (Dundr et al. 2002). Consequently, it was calculated that PICs support a single or at most a few rounds of initiation. Experiments with GFP-tagged pol I subunits led to the more startling conclusion that components of pol I itself are brought to ribosomal genes as distinct subunits and assemble via metastable intermediates into a functional polymerase that then breaks apart following transcription. Inverse FRAP demonstrated that the majority of each pol I subunit associated with ribosomal genes is not engaged in transcription and rapidly dissociates from rDNA. Only 10% of nucleolar pol I is engaged in transcription elongation, as evidenced by its slower dissociation kinetics. Each ribosomal gene is transcribed by ∼100 pol I molecules at any given time (Miller and Bakken 1972; Puvion-Dutilleul 1983; Scheer and Benavente 1990). Consequently, significantly >100 pol I complexes must be associated with each repeat but not engaged in transcription. Clearly this is more than can be accounted for by interaction with the promoter. We believe that this is the fraction of pol I we observe by ChIP to be associated with the human intergenic spacer sequences. Our results with pseudo-NORs support this model.
ChIP data with endogenous rDNA and observations with pseudo-NORs similarly lead us to conclude that the vast majority of SL1 although associated with rDNA is not engaged in PICs. Live cell imaging experiments with GFP-tagged SL1 components cannot specifically identify the small fraction of SL1 engaged in PICs and consequently cannot be used as evidence for PIC recycling. Despite this re-evaluation of live cell imaging data, we still favor a model in which PICs are recycled after each round of transcription initiation, as it provides a role for the high concentration of unengaged pol I transcription factors associated with rDNA. The unique morphology of NORs, mediated by UBF binding, creates a local environment rich in pol I transcription factors, which may support rapid PIC formation and/or recycling.
Materials and methods
Antibodies
Antibodies against human UBF, RPA43, PAF53, and TIF-IA/Rrn3 were raised in sheep immunized with full-length recombinant proteins produced using baculovirus or Escherichia coli expression systems. Antibodies against human TAFI110 were raised in sheep immunized with peptides produced in E. coli that encoded the C-terminal 184 amino acids of TAFI110. All these antibodies were affinity purified against the appropriate antigen coupled onto 1 mL HiTrap NHS-activated HP columns (Amersham Biosciences). Antibody was bound and eluted using ImmunoPure Gentle Binding and Elution buffers (Pierce). For immunolocalization, affinity-purified antibodies were conjugated with FITC or Rhodamine (Sigma). Monoclonal antibody 72B9 (provided by Ulrich Scheer, Würzburg) combined with Cy2-labeled α-mouse secondary antibody (Jackson ImmunoResearch) was used to detect fibrillarin. Human pol I subunit RPA195 was visualized using autoimmune serum 57299 (generously provided by Ulrich Scheer, Würzburg) combined with α-human Cy2-labeled secondary antibody (Jackson ImmunoResearch). Myc epitope tags were detected using the monoclonal antibody 9E10 combined with Cy2- or rhodamine-labeled α-mouse secondary antibody.
Plasmids
The plasmid pXEn8 contains a tandem array of eight copies of the 625-bp X. laevis enhancer cloned as a SalI–XhoI fragment in a modified pGEM3 vector (Promega). For transfections and probe preparation, the insert was released by double digestion and purified from preparative agarose gels. pJRC41, a gift from Jonathan Chubb (MRC Human Genetics Unit), contains a blasticidin resistance marker driven by the SV40 early promoter (Chubb et al. 2002). Open reading frames for human UBF1/2, human pol I subunits (RPA195, RPA135, PAF53, and RPA43), and human SL1 components (TBP and TAFI110) were introduced into Gateway Entry vectors (Invitrogen) and subsequently recombined into Cyan Fluorescent Protein mammalian expression vector pdECFP-amp (Simpson et al. 2000). CFP is fused to the N terminus of open reading frames.
Cell culture
HT1080 cells were grown in Dulbecco's MEM (+Glutamax, sodium pyruvate, and 4.5 g/L glucose; GIBCO) supplemented with 10% fetal bovine serum and antibiotics. Transfections were performed using a standard calcium phosphate protocol. One hundred-micrometer plates were transfected with 20 μg of pXEn8 insert DNA and 100 ng pJRC41. Selection was carried out in the above medium containing 5 μg/mL blasticidin (GIBCO). Colonies were picked after 10 d and maintained in blasticidin-containing medium. LacO and GFP-LacI containing HT1080 derivatives were maintained as described previously (Chubb et al. 2002).
Immunofluorescent staining
Cells grown on coverslips were rinsed in PBS, fixed for 10 min at room temperature in 2% PFA in PBS, rinsed again in PBS, and subsequently permeabilized with 0.1% Triton X-100/PBS for 10 min at room temperature. All antibody incubations were performed for 30–45 min at room temperature in a humidified chamber, followed by rinsing in PBS. Hoechst 55328 stain for visualization of DNA was added to the coverslips during the last antibody incubation step at a concentration of 5 μg/mL for 10 min. In double-labeling experiments the antibodies were applied consecutively, starting with unconjugated primary, followed by conjugated secondary and finally conjugated primary antibodies. Coverslips were mounted in DABCO solution (0.2M Tris-HCl at pH 8.5/glycerol, 1:9, containing 1.5% DABCO).
FISH and silver staining on metaphase spreads
FISH on metaphase spreads was performed essentially as described previously (Sullivan et al. 2001). Cells were treated with colcemid (10 μg/mL) for 1 h prior to harvesting by mitotic shake off. Cells were recovered by centrifugation, washed in PBS, repelleted, and hypotonically shocked by resuspension in 75 mM KCl. After fixation in methanol:acetic acid (3:1) at room temperature, cells were dropped onto slides and air dried. To denature chromosomal DNA, slides were incubated in 70% formamide/2× SSC (pH 7.0) for 1 min at 70°C and then dehydrated through an ethanol series. Seven microliters of hybridization buffer (Hybrisol VII, Qbiogene) containing probe(s) and competitor herring sperm DNA were applied to the slide; a coverslip was sealed with rubber cement. Slides were hybridized overnight at 37°C in a humidified chamber and then washed in 0.4× SSC, 0.3% NP-40 for 2 min at 74°C, followed by 2× SSC, 0.1% NP-40 for 1 min at room temperature. Slides were mounted in DAPI/Antifade (Qbiogene). Probes were labeled with spectrum red or green by nick translation (Vysis). XEn sequences were visualized with spectrum red-labeled pXEn8 insert DNA. Human NORs were visualized with an 11.9-kb EcoRI restriction fragment that contains human intergenic spacer sequences immediately upstream of the gene promoter, labeled with spectrum green. Spectrum green-labeled individual chromosome paints were prepared by nick translation of DOP–PCR products from flow-sorted chromosomes. For silver staining of NORs, air-dried metaphase spreads were aged for 2–3 d at room temperature. A drop of a 2% gelatine solution supplemented with 0.1% formic acid was added to the slide, followed by a drop of a 50% silver nitrate solution. After applying a coverslip, the slides were incubated for 2–4 min at 60°C–65°C until the solution turned golden brown. The slides were rinsed with water, air dried, and mounted in DAPI/Antifade (Qbiogene). For Q-banding of chromosomes, aged metaphase spreads were rehydrated through a decreasing ethanol series followed by two 30-s incubations in MacIlvaine's buffer (0.1 M citric acid, 0.2 M Na2HPO4 at pH 7.0). Staining was performed for 20 min at room temperature in MacIlvaine's buffer containing 50 μg/mL Quinacrine mustard (Sigma). Slides were rinsed twice in MacIlvaine's buffer and mounted in saturated sucrose solution. After image capture the slides were subjected to silver staining as described and remounted in saturated sucrose.
Combined immunofluorescence and FISH on three-dimensionally preserved nuclei
Combined immunofluorescence and FISH was performed essentially as described previously (Bridger and Lichter 1999). HT1080 cells and XEn-containing clones were grown on Superfrost Plus microscope slides (Scientific Laboratory Supplies) and fixed for FISH and indirect immunofluorescence by a 10-min incubation in 4% PFA (w/v) in PBS, rinsed three times in PBS, and permeabilized by incubation for 10 min in 0.5% saponin (w/v) and 0.5% Triton-X (v/v) in PBS. After rinsing in PBS, cells were incubated in 20% glycerol/PBS for 2 h, snap-frozen in liquid nitrogen, and stored at –80°C. The hybridization probe used in these experiments was spectrum red (Vysis)-labeled XEn. Immediately prior to hybridization, slides were thawed, washed in PBS, depurinated in 0.1 N HCl for 10 min at room temperature, and washed twice in PBS. For denaturation, cells were placed in 70% formamide/2× SSC for 3 min at 73°C followed by 1 min in 50% formamide/2× SSC at 73°C. Denatured probe (50 ng/slide) in 10 μL/slide Hybrisol VII (Qbiogene) was then added to the cells and allowed to hybridize for at least 18 h at 37°C in a humidified chamber. Post-hybridization washes were 3 × 5 min in 50% formamide/2× SSC at 42°C and 3 × 5 min in 0.1× SSC at 60°C. Slides were washed in PBS and then subjected to antibody staining. UBF and RPA43 were visualized using FITC-labeled affinity-purified antibodies (see above). Fibrillarin and TAFI110 were visualized prior to depurination and hybridization using the primary and secondary antibodies described above. Bound antibodies were cross-linked with 4% PFA in PBS. Slides were mounted in DAPI/Antifade (Qbiogene).
Imaging
Fluorescent and DIC images were captured and merged using a Photometric Coolsnap HQ camera and Digital Scientific SmartCapture extensions (supplied by Applied Imaging) with 100× Plan Neofluar (metaphase spreads) and 63× Plan Apochromat (cells) Zeiss objectives mounted on a Zeiss Axioplan2 imaging microscope.
Nuclear ChIP
Cells (1 × 107 to 2 × 107) were cross-linked for 10 min by the addition of formaldehyde directly to the tissue culture media to a final concentration of 1%. Cross-linking was stopped by the addition of glycine to a final concentration of 0.125 M and incubation for 5 min. Cells were washed twice in PBS, harvested by combined trypsinization and scraping, and washed in PBS, and the cell pellet was finally resuspended in 2 mL ice-cold cell lysis buffer (5 mM PIPES at pH 8.0, 85 mM KCl, 0.5% NP40 and protease inhibitors). After incubation on ice for 15 min, nuclei were recovered by centrifugation and resuspended in 1 mL nuclear lysis buffer (50 mM Tris-HCl at pH 8.1, 10 mM EDTA, 1% SDS and protease inhibitors) at RT. Nuclear lysate was sonicated to generate chromatin fragments with an average length of 500 bp. The lysate was microfuged at 14,000 rpm for 5 min and the supernatant then snap frozen in liquid nitrogen and stored at –80°C. For immunoprecipitations, 5-μL aliquots of Protein G Dynabeads (10 mg/mL, Dynal), were pre-equilibrated in 20 μL PBS containing 0.1% IgG free BSA and 100 μg/mL E. coli RNA. Beads were recovered magnetically and incubated with 6 μg of the appropriate affinity-purified antibody or 10 μL of pre-immune serum as an IgG control. IgG loaded Dynabeads were incubated with 20 μL of the above chromatin fraction that had been premixed with 130 μL of IP dilution buffer (14.5 mM Tris-HCl at pH 7.6, 0.75% Triton X-100, 182.5 mM NaCl). Following overnight incubation at 4°C with mixing, beads were recovered, washed twice with 150 μL IP wash buffer 1 (20 mM Tris-HCl at pH 8.0, 2 mM EDTA, 0.2% SDS, 0.5% Triton X-100, and 150 mM NaCl) and twice with 150 μL IP wash buffer 2 (20 mM Tris-HCl at pH 8.0, 2 mM EDTA, 0.2% SDS, 0.5% Triton X-100, and 500 mM NaCl). Immunoprecipitated material was eluted from the beads, cross-links were reversed, and DNA was recovered by ethanol precipitation as previously described (O'Sullivan et al. 2002). DNA pellets were resuspended in 100 μL H2O.
Immunoprecipitated DNA was analyzed by PCR using either primers specific for human ribosomal gene promoter or the Xenopus enhancer sequences. Human promoter primers (5′-CG CTGCTCCCGCGTGTGTCC-3′ and 5′-CAGCGACAGGTCG CCAGAGG-3′) were used to amplify sequences from –198 to 33 of the ribosomal gene promoter. Xenopus enhancer primers (5′GACCGGGAGTTCCAGGAG-3′ and 5′-CAGGGCAGGGGG ACGAG-3′) were used to amplify 54 bp present in each 60/81-bp element of the Xenopus enhancer. PCR reactions were performed using an iCycler IQ real-time PCR system (Bio-Rad). Reactions (20 μL) contained 10 μL of a 2× hot-start mastermix (Biogene), 5 μL of IP DNA, and a 1/28500 dilution of SYBR Green (Biogene). Specificity of PCR products was determined using a combination of melting curve profiles and gel electrophoresis. Standard curves for quantitation were generated using serially diluted genomic DNA from clone 4A (XEn primers) or a cosmid clone that contains the entire 43-kb human ribosomal gene repeat (promoter primers). Reactions included an initial step of 10 min at 95°C followed by 27–40 cycles of 20 sec at 95°C, 20 sec at 57.8°C, 10 sec at 72°C, and 10 sec at 87°C for XEn primers or 28–40 cycles of 20 sec at 95°C, 20 sec at 60°C, and 10 sec at 72°C for promoter primers. For analysis by gel electrophoresis, 27–30 cycles of amplification were performed. Products were run on 8% polyacrylamide gels in TBE buffer. Gels were stained with ethidium bromide and images obtained using a Molecular Imager (Bio-Rad). For real-time quantitation, 40 cycles of amplification were performed. The amount of target present in each IP was calculated relative to that in control IPs.
Nucleolar ChIP
Nucleolar chromatin was prepared as described previously (O'Sullivan et al. 2002). Nucleolar chromatin (150 μL) was diluted 10-fold into 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl (pH 8.1), and 167 mM NaCl. Diluted chromatin was precleared with Protein A/G Sepharose beads (50 μL of a 50% slurry pre-equilibrated in E. coli RNA and BSA). Following addition of affinity-purified antibodies (∼5 μg) and immunoprecipitation with Protein A/G Sepharose beads (50 μL as above), immune complexes were washed successively with low-salt wash (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl at pH 8.1, 150 mM NaCl), high salt wash (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl at pH 8.1, 500 mM NaCl), LiCl wash (0.25 M LiCl, 1% IGEPAL-CA630, 1% deoxycholic acid [sodium salt], 1 mM EDTA, 10 mM Tris at pH 8.1) and TE buffer (10 mM Tris-Hcl at pH 8.0, 1 mM EDTA,). Complexes were eluted in TE buffer plus 2% SDS. PCR was performed on one-fortieth of the DNA recovered from each ChIP as described above. Sequences of primers and PCR conditions are available on request. Radiolabeling of DNA from ChIPs by random prime labeling, production of slot blots and hybridization were as described previously (O'Sullivan et al. 2002). A full description of the subcloned rDNA subfragments is available on request.
BrUTP incorporation in permeabilized cells
After permeabilization of cells with 0.05% Triton X-100 for 2 min at room temperature, run-on transcription was performed for 15 min in transcription buffer (100 mM KCl, 50 mM Tris-HCl at pH 7.4, 5 mM MgCl2, 0.5 mM EGTA, 5 U/mL RNasin [Promega] supplemented with 0.5 mM ATP, GTP, CTP, 0.2 mM BrUTP [Sigma], and α-amanitin [50 μg/mL]). After run-on transcription, cells were fixed with 2% PFA and permeabilised with 0.1% Triton X-100. BrUTP was visualized with anti-BrdUTP antibodies (Roche) combined with FITC-conjugated secondary antibodies.
Acknowledgments
We thank J. Chubb and W. Bickmore for plasmid pJRC41 and Lac operator containing cell lines, U. Scheer for pol I and fibrillarin antibodies, and S. Wiemann for plasmid pdECFP-amp. We also thank S. Keyse for critically reading the manuscript. This work was supported by a program grant from the MRC UK (B.M.). C.M. was funded by a project grant from the Wellcome Trust (B.M.).
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.310705.
References
- Aprikian P., Moorefield, B., and Reeder, R.H. 2001. New model for the yeast RNA polymerase I transcription cycle. Mol. Cell. Biol. 21: 4847–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazett-Jones D.P., Leblanc, B., Herfort, M., and Moss, T. 1994. Short-range DNA looping by the Xenopus HMG-box transcription factor, xUBF. Science 264: 1134–1137. [DOI] [PubMed] [Google Scholar]
- Beckmann H., Chen, J.L., O'Brien, T., and Tjian, R. 1995. Co-activator and promoter-selective properties of RNA polymerase I TAFs. Science 270: 1506–1509. [DOI] [PubMed] [Google Scholar]
- Bell S.P., Learned, R.M., Jantzen, H.M., and Tjian, R. 1988. Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science 241: 1192–1197. [DOI] [PubMed] [Google Scholar]
- Bell S.P., Pikaard, C.S., Reeder, R.H., and Tjian, R. 1989. Molecular mechanisms governing species-specific transcription of ribosomal RNA. Cell 59: 489–497. [DOI] [PubMed] [Google Scholar]
- Bodem J., Dobreva, G., Hoffmann-Rohrer, U., Iben, S., Zentgraf, H., Delius, H., Vingron, M., and Grummt, I. 2000. TIF-IA, the factor mediating growth-dependent control of ribosomal RNA synthesis, is the mammalian homolog of yeast Rrn3p. EMBO Rep. 1: 171–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridger J.M. and Lichter, P. 1999. Analysis of mammalian interphase chromosomes by FISH and immunofluorescence. In Chromosome structural analysis (ed. W.A. Bickmore), pp. 103–121. Oxford University Press, Oxford.
- Chubb J.R., Boyle, S., Perry, P., and Bickmore, W.A. 2002. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr. Biol. 12: 439–445. [DOI] [PubMed] [Google Scholar]
- Comai L., Tanese, N., and Tjian, R. 1992. The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell 68: 965–976. [DOI] [PubMed] [Google Scholar]
- Dundr M., Hoffmann-Rohrer, U., Hu, Q., Grummt, I., Rothblum, L.I., Phair, R.D., and Misteli, T. 2002. A kinetic framework for a mammalian RNA polymerase in vivo. Science 298: 1623–1626. [DOI] [PubMed] [Google Scholar]
- Gadal O., Labarre, S., Boschiero, C., and Thuriaux, P. 2002. Hmo1, an HMG-box protein, belongs to the yeast ribosomal DNA transcription system. EMBO J. 21: 5498–5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodpasture C. and Bloom, S.E. 1975. Visualization of nucleolar organizer regions in mammalian chromosomes using silver staining. Chromosoma 53: 37–50. [DOI] [PubMed] [Google Scholar]
- Grummt I. 2003. Life on a planet of its own: Regulation of RNA polymerase I transcription in the nucleolus. Genes & Dev. 17: 1691–1702. [DOI] [PubMed] [Google Scholar]
- Hanada K., Song, C.Z., Yamamoto, K., Yano, K., Maeda, Y., Yamaguchi, K., and Muramatsu, M. 1996. RNA polymerase I associated factor 53 binds to the nucleolar transcription factor UBF and functions in specific rDNA transcription. EMBO J. 15: 2217–2226. [PMC free article] [PubMed] [Google Scholar]
- Heix J. and Grummt, I. 1995. Species specificity of transcription by RNA polymerase I. Curr. Opin. Genet. Dev. 5: 652–656. [DOI] [PubMed] [Google Scholar]
- Heliot L., Kaplan, H., Lucas, L., Klein, C., Beorchia, A., Doco-Fenzy, M., Menager, M., Thiry, M., O'Donohue, M.F., and Ploton, D. 1997. Electron tomography of metaphase nucleolar organizer regions: Evidence for a twisted-loop organization. Mol. Biol. Cell. 8: 2199–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson A.S., Warburton, D., and Atwood, K.C. 1972. Location of ribosomal DNA in the human chromosome complement. Proc. Natl. Acad. Sci. 69: 3394–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermekchiev M., Workman, J.L., and Pikaard, C.S. 1997. Nucleosome binding by the polymerase I transactivator upstream binding factor displaces linker histone H1. Mol. Cell. Biol. 17: 5833–5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A., Deppert, U., and Grummt, I. 1990. A 140-base-pair repetitive sequence element in the mouse rRNA gene spacer enhances transcription by RNA polymerase I in a cell-free system. Proc. Natl. Acad. Sci. 87: 7527–7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A., Voit, R., Stefanovsky, V., Evers, R., Bianchi, M., and Grummt, I. 1994. Functional differences between the two splice variants of the nucleolar transcription factor UBF: The second HMG box determines specificity of DNA binding and transcriptional activity. EMBO J. 13: 416–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H. and Green, M.R. 1994. The RNA polymerase I transcription factor, upstream binding factor, interacts directly with the TATA box-binding protein. J. Biol. Chem. 269: 30140–30146. [PubMed] [Google Scholar]
- Labhart P. and Reeder, R.H. 1984. Enhancer-like properties of the 60/81 bp elements in the ribosomal gene spacer of Xenopus laevis. Cell 37: 285–289. [DOI] [PubMed] [Google Scholar]
- Langst G., Blank, T.A., Becker, P.B., and Grummt, I. 1997. RNA polymerase I transcription on nucleosomal templates: The transcription termination factor TTF-I induces chromatin remodeling and relieves transcriptional repression. EMBO J. 16: 760–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langst G., Becker, P.B., and Grummt, I. 1998. TTF-I determines the chromatin architecture of the active rDNA promoter. EMBO J. 17: 3135–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y., Hisatake, K., Kondo, T., Hanada, K., Song, C.Z., Nishimura, T., and Muramatsu, M. 1992. Mouse rRNA gene transcription factor mUBF requires both HMG-box1 and an acidic tail for nucleolar accumulation: Molecular analysis of the nucleolar targeting mechanism. EMBO J. 11: 3695–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay B. and Reeder. R.H. 1990. An RNA polymerase I termination site can stimulate the adjacent ribosomal gene promoter by two distinct mechanisms in Xenopus laevis. Genes & Dev. 4: 1240–1251. [DOI] [PubMed] [Google Scholar]
- McStay B., Sullivan, G.J., and Cairns, C. 1997. The Xenopus RNA polymerase I transcription factor, UBF, has a role in transcriptional enhancement distinct from that at the promoter. EMBO J. 16: 396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller O.L. and Bakken, A.H. 1972. Morphological studies of transcription. Acta Endocrinol. Suppl. (Copenh.) 168: 155–177. [DOI] [PubMed] [Google Scholar]
- Miller G., Panov, K.I., Friedrich, J.K., Trinkle-Mulcahy, L., Lamond, A.I., and Zomerdijk, J.C. 2001. hRRN3 is essential in the SL1-mediated recruitment of RNA Polymerase I to rRNA gene promoters. EMBO J. 20: 1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T. and Stefanovsky, V.Y. 2002. At the center of eukaryotic life. Cell 109: 545–548. [DOI] [PubMed] [Google Scholar]
- O'Mahony D.J. and Rothblum, L.I. 1991. Identification of two forms of the RNA polymerase I transcription factor UBF. Proc. Natl. Acad. Sci. 88: 3180–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan A.C., Sullivan, G.J., and McStay, B. 2002. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol. Cell. Biol. 22: 657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paule M.R. and White, R.J. 2000. Survey and summary: Transcription by RNA polymerases I and III. Nucleic Acids Res. 28: 1283–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Castillo A., Martin-Lucas, M.A., and Abrisqueta, J.A. 1986. New insights into the effects of extra nucleolus organizer regions. Hum. Genet. 72: 80–82. [DOI] [PubMed] [Google Scholar]
- Pikaard C.S., McStay, B., Schultz, M.C., Bell, S.P., and Reeder, R.H. 1989. The Xenopus ribosomal gene enhancers bind an essential polymerase I transcription factor, xUBF. Genes & Dev. 3: 1779–1788. [DOI] [PubMed] [Google Scholar]
- Putnam C.D. and Pikaard, C.S. 1992. Cooperative binding of the Xenopus RNA polymerase I transcription factor xUBF to repetitive ribosomal gene enhancers. Mol. Cell. Biol. 12: 4970–4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puvion-Dutilleul F. 1983. Morphology of transcription at cellular and molecular levels. Int. Rev. Cytol. 84: 57–101. [DOI] [PubMed] [Google Scholar]
- Roussel P. and Hernandez-Verdun, D. 1994. Identification of Ag-NOR proteins, markers of proliferation related to ribosomal gene activity. Exp. Cell. Res. 214: 465–472. [DOI] [PubMed] [Google Scholar]
- Roussel P., Andre, C., Masson, C., Geraud, G., and Hernandez, V.D. 1993. Localization of the RNA polymerase I transcription factor hUBF during the cell cycle. J. Cell. Sci. 104: 327–337. [DOI] [PubMed] [Google Scholar]
- Roussel P., Andre, C., Comai, L., and Hernandez-Verdun, D. 1996. The rDNA transcription machinery is assembled during mitosis in active NORs and absent in inactive NORs. J. Cell. Biol. 133: 235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai K., Ohta, T., Minoshima, S., Kudoh, J., Wang, Y., de Jong, P.J., and Shimizu, N. 1995. Human ribosomal RNA gene cluster: Identification of the proximal end containing a novel tandem repeat sequence. Genomics 26: 521–526. [DOI] [PubMed] [Google Scholar]
- Santoro R., Li, J., and Grummt, I. 2002. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat. Genet. 32: 393–396. [DOI] [PubMed] [Google Scholar]
- Scheer U. and Benavente, R. 1990. Functional and dynamic aspects of the mammalian nucleolus. Bioessays 12: 14–21. [DOI] [PubMed] [Google Scholar]
- Simpson J.C., Wellenreuther, R., Poustka, A., Pepperkok, R., and Wiemann, S. 2000. Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing. EMBO Rep. 1: 287–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strukov Y.G., Wang, Y., and Belmont, A.S. 2003. Engineered chromosome regions with altered sequence composition demonstrate hierarchical large-scale folding within metaphase chromosomes. J. Cell. Biol. 162: 23–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan G.J., Bridger, J.M., Cuthbert, A.P., Newbold, R.F., Bickmore, W.A., and McStay, B. 2001. Human acrocentric chromosomes with transcriptionally silent nucleolar organizer regions associate with nucleoli. EMBO J. 20: 2867–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner A.T. 1982. The nature and mechanisms of chromosome banding. Cancer Genet. Cytogenet. 6: 59–87. [DOI] [PubMed] [Google Scholar]
- Thomas J.O. and Travers, A.A. 2001. HMG1 and 2, and related “architectural” DNA-binding proteins. Trends Biochem. Sci. 26: 167–174. [DOI] [PubMed] [Google Scholar]
- Weisenberger D. and Scheer, U. 1995. A possible mechanism for the inhibition of ribosomal RNA gene transcription during mitosis. J. Cell. Biol. 129: 561–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Zhao, J., Zentgraf, H., Hoffmann-Rohrer, U., and Grummt, I. 2002. Multiple interactions between RNA polymerase I, TIF-IA and TAFI subunits regulate preinitiation complex assembly at the ribosomal gene promoter. EMBO Rep. 3: 1082–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]