Abstract
Introduction
Human studies report conflicting results on the predictive power of serum lipids on the progression of chronic kidney disease. We aimed to systematically identify the lipids that predict progression to end-stage kidney disease.
Methods
From the Chronic Renal Insufficiency Cohort, 79 patients with chronic kidney disease stages 2 to 3 who progressed to end-stage kidney disease over 6 years of follow-up were selected and frequency matched by age, sex, race, and diabetes with 121 nonprogressors with less than 25% decline in estimated glomerular filtration rate during the follow-up. The patients were randomly divided into training and test sets. We applied liquid chromatography-mass spectrometry-based lipidomics on visit year 1 samples.
Results
We identified 510 lipids, of which the top 10 coincided with false discovery threshold of 0.058 in the training set. From the top 10 lipids, the abundance of diacylglycerols and cholesteryl esters was lower, but that of phosphatidic acid 44:4 and monoacylglycerol 16:0 was significantly higher in progressors. Using logistic regression models, a multimarker panel consisting of diacylglycerols and monoacylglycerol independently predicted progression. The c-statistic of the multimarker panel added to the base model consisting of estimated glomerular filtration rate and urine protein-to-creatinine ratio as compared with that of the base model was 0.92 (95% confidence interval: 0.88–0.97) and 0.83 (95% confidence interval: 0.76–0.90, P < 0.01), respectively, an observation that was validated in the test subset.
Discussion
We conclude that a distinct panel of lipids may improve prediction of progression of chronic kidney disease beyond estimated glomerular filtration rate and urine protein-to-creatinine ratio when added to the base model.
Keywords: chronic kidney disease, lipids, proteinuria
According to the Center for Disease Control and Prevention, there are currently more than 20 million people above the age of 20 with chronic kidney disease (CKD) in the United States.1 In spite of its public health burden, the clinical care of the patients with CKD is largely dependent on the application of traditional biomarkers including serum creatinine, urine protein-to-creatinine ratio (UPCR), and estimated glomerular filtration rate (eGFR), which are significantly limited by their precision, accuracy, and prognostic values especially early in the course of disease.2, 3 In CKD, metabolic derangements start at early stages where these inherent deficiencies are most prominent. Such limitations necessitate a shift of paradigm from exclusive reliance on traditional biomarkers to systematic approaches for the identification of prognostic markers.
Lipids are diverse and abundant molecules with significant links to different metabolic pathways along with diverse cellular and biological functions.4, 5 In the past, lipid studies in CKD have largely been limited to studying the changes at class level of a limited number of lipids such as total cholesterol, triglycerides, low-density lipoprotein (LDL), and high-density lipoprotein with conflicting results in terms of the association between dyslipidemia and progression of CKD.6, 7, 8, 9, 10, 11 As a result of these limited approaches, the effect of diverse intraclass variation within these lipid classes as well as the alterations in various other classes of lipids on the progression of CKD has remained poorly understood. More recently, the use of conventional lipid measurements for the description of lipoprotein abnormalities in mild CKD has come into question.12 On the other hand, the application of the lipidomics and/or metabolomics approach in a number of diseases such as diabetes,13, 14 cardiovascular diseases,15 and other inflammatory processes16 has provided characteristic lipid signatures and mechanistic insights to disease processes.17 These studies provide proof-of-principle on the clinical applicability of the candidate metabolites for risk prediction, an approach that is rarely taken in CKD. In a recently published report, Reis et al.18 have compared the lipid signature of LDL in patients at the advanced stage of CKD (stages 4 and 5) with the control group using the liquid chromatography-mass spectrometry-based lipidomics approach. To our knowledge, there is no study in CKD aimed at the identification of lipid signature predictive of incident end-stage kidney disease (ESKD) at early stages of CKD. Therefore, this study examines the systematic identification of prognostic serum lipid metabolites at CKD stages 2 and 3 to predict progression to ESKD using liquid chromatography-mass spectrometry-based lipidomics in the Chronic Renal Insufficiency Cohort (CRIC) patient population.
Methods
Patients
This study is a case-control study nested in the core CRIC study. The design of CRIC is published previously.19, 20 CRIC is a multicenter cohort of patients with mild-to-moderate CKD, with recruitment starting in 2003 with the goals of examining risk factors for CKD and cardiovascular events, and developing predictive models that would identify high-risk subgroups. The core study has recruited 3939 subjects over a 5-year period through 2008. Inclusion criteria of the subcohort used for this study were eGFR ≥ 30 ml/min at visit year 1 and an age of 18 years or more with no racial or gender restriction. Cases were defined as patients who progressed to ESKD over the next 6 years of follow-up. ESKD is defined as needing chronic dialysis or having kidney transplantation. Controls were defined as patients who were frequency matched with cases by their baseline age, sex, race, and diabetes and had less than 25% decline in eGFR during the 6-year mean follow-up. One milliliter of fasting serum sample from visit year 1 as baseline was obtained from the selected subcohort. Demographic, clinical, and laboratory variables from baseline were retrieved from the corresponding patients. eGFR calculated by CKD Epidemiology Collaboration is used for multivariable adjustments.
Data Acquisition
Liquid chromatography-mass spectrometry-based shotgun lipidomics using a TripleTOF 5600 was applied for lipid identification (see the Supplementary Methods for details).
Statistical Analysis
After data acquisition, the missing values for lipids were imputed using the K nearest-neighbor method.17, 21 Then the data were log2 transformed followed by normalization using the cross-contribution compensating multiple internal standard normalization method.22 The cohort was randomly divided into the training and test sets with a 2:1 ratio in an attempt to develop the probabilistic predictive model of multimarker panel predictive of progression in the training set followed by its validation in the test set. The compound-by-compound t-test was applied to identify the top differentially regulated lipids that passed the nominal threshold P value of <0.05, followed by the Benjamini-Hochberg procedure for false discovery rate (FDR) correction23, 24 accounting for multiple comparisons. In parallel, the partial least square-discriminant analysis (PLS-DA)25, 26 and Random Forest (RF)27 classification methods were applied on the top lipids with nominal significance in the training set to generate the rank of the variable important in projection by each classification method separately (Figure 1). The rationale for using PLS-DA and RF classification methods besides the application of the Benjamini-Hochberg procedure for FDR correction was to assess concordance of the products of different classification methods and to compare if the proposed lipids by different methods differed. Then logistic regression models with and without adjusting for eGFR and UPCR as continuous variables were used to identify the independent predictors of progression from the ranking list of each classification method. The c-statistic,28 category-free continuous net reclassification improvement, and integrated discrimination improvement were calculated for the probabilistic model of the multimarker panels derived from logistic regression, and their improvement over the base model was tested in the training set and replicated in the test set. Linear regression analysis was applied to test the relationship between the mean ratio of lipid levels on the log2 scale in cases and controls with the number of carbons or double bonds within each lipid class. Over-representative enrichment analysis using the 2-sided Fisher exact test was applied to test the enrichment of lipid classes by taking into account the number of metabolites that have passed the FDR threshold and the number of metabolites that are detected within each class of lipids as compared with the rest of other lipids in the entire dataset. The lipid correlation network was built using Metscape.29 We applied a sparse graphical modeling algorithm based on the desparsified graphical lasso modeling procedure30 to calculate the Benjamini-Hochberg-adjusted partial correlations between each pair of lipids that displayed a significant difference between cases and controls using the “Correlation Calculator” tool (http://metscape.med.umich.edu/calculator.html).31 The study has 80% power at α = 0.05 to detect an increase in the area under receiver operating characteristics from 0.8 to 0.9 using a 2-sided z-test.32 MetaboAnalyst version 2.0,33, 34 R-Metabolomics version 0.1.3 (Melbourne, Australia),35 SPSS version 22 (Armonk, NY), and STATA version 10 (College Station, TX) were applied for the analysis.
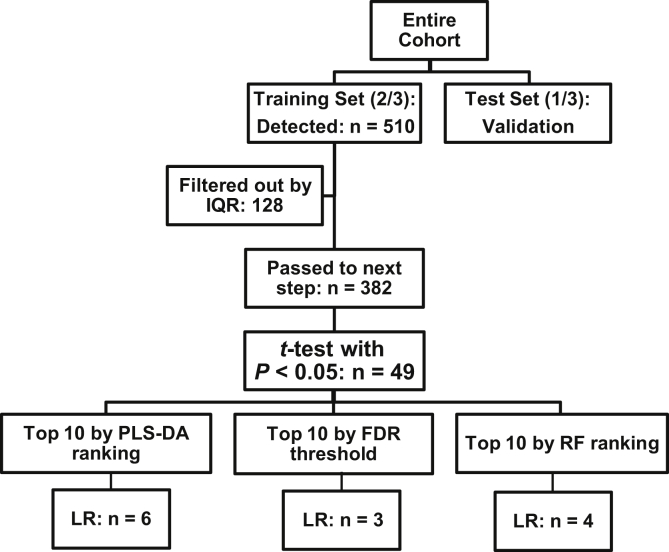
Figure 1.
Flow of identification and validation of the independent predictors of progression by different classification methods in the study subsets. FDR, false discovery rate; IQR, interquartile range; LR, logistic regression; PLS-DA, partial least square-discriminant analysis; RF, Random Forest.
Results
Baseline
From patients aged ≥18 years with baseline eGFR ≥ 30 ml/min, 79 patients who progressed to ESKD over 6 years of follow-up were selected and frequency matched with 121 nonprogressors (<25% decline in eGFR during follow-up) by age, sex, race, and diabetes. Mean age was 59 years (SD = 10). There were 112 males (56%), 100 patients with diabetes (50%), and equal numbers of African American and Caucasians (100 in each group). The distribution of baseline demographic characteristics, medications, and comorbidities in cases and controls is presented for the entire cohort (Table 1) as well as in the training and the test subsets (Supplementary Table S1). Accordingly, the progressors are reasonably matched with nonprogressors as is evident by the lack of clinical and statistical differences in most of the baseline variables. However, the mean eGFR was lower by 10 ml/min, and the median UPCR was higher by 0.7 in progressors as compared with nonprogressors at baseline (P < 0.001).
Table 1.
Comparison of baseline characteristics of the progressors and nonprogressors
| Variable | Nonprogressors | Progressors | P value |
|---|---|---|---|
| N | 121 | 79 | |
| Age (yr) | 59 ± 10 | 59 ± 10 | 0.705 |
| Male gender (%) | 68 (56.2) | 44 (55.7) | 0.944 |
| Race | 0.885 | ||
| White (%) | 61 (50.4) | 39 (49.4) | |
| Black (%) | 60 (49.6) | 40 (50.6) | |
| Current smoking (%) | 23 (19.0) | 16 (20.3) | 0.828 |
| Medications | |||
| ACEI (%) | 61 (50.4) | 41 (52.6) | 0.767 |
| ARB (%) | 33 (27.3) | 23 (29.5) | 0.735 |
| Beta blocker (%) | 56 (46.3) | 44 (56.4) | 0.163 |
| Ca channel blocker (%) | 48 (39.7) | 42 (53.8) | 0.050 |
| Diuretics (%) | 68 (56.2) | 48 (61.5) | 0.456 |
| Statins (%) | 73 (60.6) | 51 (65.4) | 0.473 |
| Other lipid-lowering agents (%) | 15 (12.4) | 8 (10.3) | 0.645 |
| Steroids (%) | 80 (66.1) | 53 (67.1) | 0.887 |
| Aspirin (%) | 66 (54.5) | 39 (50.0) | 0.531 |
| Antiplatelets (%) | 67 (55.4) | 42 (53.8) | 0.833 |
| Comorbidities (history) | |||
| Diabetes (%) | 57 (47.1) | 43 (54.4) | 0.311 |
| Hypertension (%) | 103 (85.1) | 73 (92.4) | 0.121 |
| PVD (%) | 9 (7.4) | 7 (9.0) | 0.717 |
| CHF (%) | 9 (7.4) | 14 (17.7) | 0.026 |
| Stroke (%) | 6 (5.0) | 9 (11.4) | 0.091 |
| A-fib (%) | 19 (15.7) | 22 (27.8) | 0.038 |
| Height (m) | 1.7 ± 0.1 | 1.7 ± 0.1 | 0.492 |
| Weight (kg) | 93 ± 21 | 97 ± 28 | 0.229 |
| BMI (kg/m2) | 32.0 ± 7.1 | 33.7 ± 8.9 | 0.139 |
| Waist (m) | 1.1 ± 0.2 | 1.1 ± 0.2 | 0.203 |
| Systolic BP (mm Hg) | 128 ± 22 | 132 ± 19 | 0.162 |
| Diastolic BP (mm Hg) | 71 ± 14 | 71 ± 13 | 0.774 |
| Pulse (per min) | 67 ± 11 | 69 ± 11 | 0.205 |
| HbA1c (%)a | 7.8 ± 1.8 | 8.2 ± 1.8 | 0.080 |
| Sodium (mmol/l) | 140 ± 2.3 | 140 ± 2.9 | 0.455 |
| CO2 (mmol/l) | 24.8 ± 2.7 | 24.1 ± 2.8 | 0.110 |
| Chloride (mmol/l) | 104 ± 3 | 105 ± 4 | 0.093 |
| ALT (IU/l) | 35 ± 21 | 32 ± 13 | 0.249 |
| AST (IU/l) | 26 ± 12 | 26 ± 13 | 0.695 |
| TAG (mg/dl) | 149 ± 104 | 154 ± 77 | 0.699 |
| Total cholesterol (mg/dl) | 182 ± 47 | 179 ± 47 | 0.648 |
| HDL (mg/dl) | 49 ± 14 | 47 ± 15 | 0.409 |
| LDL (mg/dl) | 103 ± 38 | 96 ± 31 | 0.226 |
| eGFR (ml/min) | 48 ± 13 | 38 ± 8 | <0.001 |
| UPCRb | 0.1 [0.1–0.4] | 1.8 [0.2–2.1] | <0.001 |
ACEI, angiotensin-converting-enzyme inhibitor; ALT, alanine aminotransferase; ARB, angiotensin receptor blocker; AST, aspartate aminotransferase; BMI, body mass index; BP, blood pressure; CHF, congestive heart failure; eGFR, estimated glomerular filtration rate; HDL, high-density lipoprotein; LDL, low-density lipoprotein; PVD, peripheral vascular disease; TAG, triacylglycerol; UPCR, urine protein-to-creatinine ratio.
N for HbA1c is 54 nonprogressors and 43 progressors all in diabetic patients.
Values are median and interquartile range.
Analytic Flow of Narrowing Down to Independent Predictors of Progression
Figure 1 illustrates the flow of identification and validation of the independent candidates for predicting progression of CKD. First, the entire cohort was randomly divided into the discovery or training set (77 nonprogressors and 57 progressors) and the validation or test set (44 nonprogressors and 22 progressors) with a 2:1 ratio. Using the training set, from the 510 identified known lipids, 128 lipids that were unlikely to be used in the downstream analyses were filtered out using the interquartile range filtering protocol36 that left 382 lipids of which 49 passed the nominal significance by a t-test (P < 0.05). From the top 49 lipids, we also used PLS-DA and RF besides using the Benjamini-Hochberg procedure for FDR correction to explore if different classification methods nominate different candidates. Figure 2 illustrates the distribution of statistical significance by the log2 mean fold change of the identified lipids in progressors versus nonprogressors in the training set, suggesting lower abundance of differentially regulated diacylglycerols (DAGs) and cholesterol esters(CEs) in progressors. Supplementary Table S3 shows the compound-by-compound comparison of identified lipids by status of progression using a t-test, as well as the corresponding unadjusted and adjusted logistic regression models. Accordingly, from the FDR-projected list, the top 10 lipids coincide with q ≤ 0.058. We then identified the top lipid candidates by each classification method followed by internal validation in the test set.
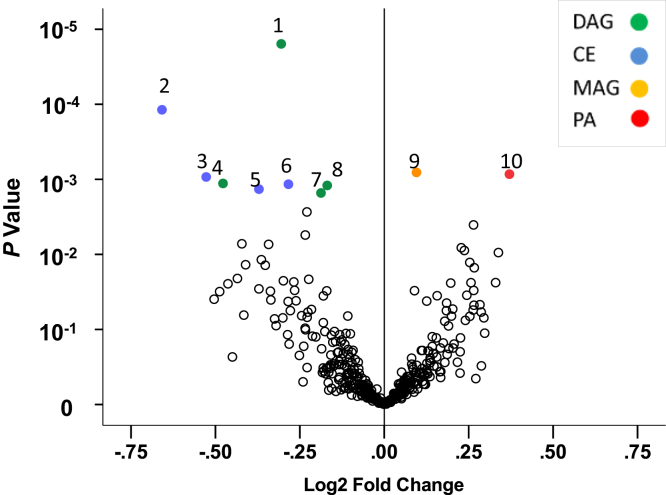
Figure 2.
Volcano plot in the training set illustrating the statistical significance of the fold change of the mean values of detected features in progressors to end-stage kidney disease versus nonprogressors derived from the compound-by-compound t-test. 1 = DAG 36:0; 2 = CE 20:5; 3 = CE 22:5; 4 = DAG 34:5; 5 = CE 20:3; 6 = CE 18:2; 7 = DAG 34:0; 8 = DAG 32:0; 9 = MAG 16:0; 10 = PA 44:4; lipids with a nominal P value ≤ 0.0027 are shown in color. CE, cholesterol esters; DAG, diacylglycerol; MAG, monoacylglycerol; PA, phosphatidic acid.
Top Lipid Candidates Nominated by Different Classification Methods
In the next step, we compared the products of different classification methods. Supplementary Table S4 suggests a high concordance between the top 10 lipids ranked by the 3 classification methods particularly for the DAGs. We then used logistic regression models on the top 10 lipids of each classification method to further narrow down to the independent predictors of progression after adjusting for eGFR, UPCR, age, sex, race, diabetes, hypertension, and congestive heart failure. As a result, the independent lipids projected by the FDR list were DAG 36:0, DAG 32:0, and monoacylglycerol (MAG) 16:0 (most conservative model). The independent lipids projected from the PLS-DA were DAG 36:0, DAG 32:0, MAG 26:2, MAG 20:0, MAG 16:0, and phosphatidylcholine (PC) 34:3 (most inclusive model), and those projected by RF were DAG 36:0, DAG 32:0, MAG 16:0, and PC 34:3 (a model in between), suggesting a high concordance in the final products of different classification methods.
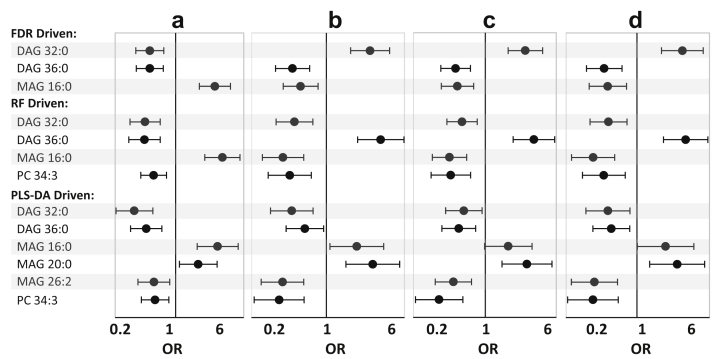
Figure 3 shows that the significance or direction of the risk associated with each 1 SD change in abundance was unchanged in unadjusted to fully adjusted models by age, sex, race, diabetes, hypertension, and congestive heart failure. According to the FDR-driven models and after full adjustment, each 1 SD increase in abundance of DAG 36:0 and DAG 32:0 was associated with the reduced risk of progression by 71% (95% CI: 43% to 85%, P < 0.001) and 66% (95% CI: 33% to 83%, P = 0.002), respectively. On the other hand, each 1 SD increase in abundance of MAG 16:0 was associated with increased risk of progression by 5.45-fold (95% CI: 2.51 to 11.86, P < 0.001), a result similar to the unadjusted model. Similar results were obtained from the RF- and PLS-DA-driven methods (Figure 3).
Figure 3.
Odds ratio and 95% confidence interval of progression to end-stage kidney disease according to the FDR proposed (top panel), RF proposed (middle panel), and PLS-DA proposed lipids (bottom panel) by change of each 1 SD in abundance of candidate lipids using (a) the logistic regression model in unadjusted model, (b) adjusted models by eGFR and other factors (age, sex, race, diabetes, hypertension, and congestive heart failure), (c) adjusted by the urine protein-to-creatinine ratio and other factors, and (d) adjusted by eGFR, urine protein-to-creatinine ratio, and other factors in the training set. DAG, diacylglycerol; eGFR, estimated glomerular filtration rate; FDR, false discovery rate; MAG, monoacylglycerol; PC, phosphatidylcholine; PLS-DA, partial least square-discriminant analysis; RF, Random Forest.
Classification Power and Internal Validation
Table 2 shows that in the training set, irrespective of the classification method, the addition of the multimarker panel to the base model (eGFR + UPCR) has significantly improved the c-statistic (P < 0.05). We also showed in Table 2 that such an improvement was reproducible in the test set by the application of the corresponding multivariable probabilistic model developed in the training set (P < 0.05). In addition, the net reclassification improvement and integrated discrimination improvement were highly significant in all multimarker lipid panels alone and when added to the base model in the training set (P ≤ 0.0005), an observation that was reproduced in the test set using the models that were developed in the training set. The statistically significant net reclassification improvement and integrated discrimination improvement imply that the sum of correctly classified progressors and correctly classified nonprogressors by the new models is significantly higher than what was obtained from the base model (eGFR + UPCR) alone.
Table 2.
Comparison of c-statistic, category-free continuous NRI, and IDI and their 95% confidence intervals in the “training set,” “test set,” and the entire cohort by base models, lipids, and their combinations to predict progression of chronic kidney disease to end-stage kidney disease
| Models | Training set (Npatient = 134) |
Test set (Npatient = 66) |
||||
|---|---|---|---|---|---|---|
| C (95% CI) | NRI | IDI (95% CI) | c (95% CI) | NRI | IDI (95% CI) | |
| Base (eGFR + UPCR) | 0.83 (0.76–0.90) | Sensitivity: 31/57 | – | 0.78 (0.67–0.89) | Sensitivity: 6/22 | – |
| Specificity: 41/77 | Specificity: 18/44 | |||||
| FDR-driven models | ||||||
| Lipids (n = 3) | 0.86 (0.79–0.92)a | Event NRI: 31/57 | 0.22 (0.15–0.29)b | 0.81 (0.70–0.93) | Event NRI: 10/22 | 0.28 (0.16–0.40)b |
| Nonevent NRI: 49/77 | Nonevent NRI: 26/44 | |||||
| Overall NRI: 1.18b | Overall NRI: 1.05b | |||||
| Lipids + base model | 0.92 (0.88–0.97)a | Event NRI: 53/57 | 0.23 (0.16–0.30)b | 0.91 (0.83–0.99)a | Event NRI: 12/22 | 0.38 (0.25–0.52)b |
| Nonevent NRI: 49/77 | Nonevent NRI: 30/44 | |||||
| Overall NRI: 1.34b | Overall NRI: 1.23b | |||||
| RF-driven models | ||||||
| Lipids (n = 4) | 0.89 (0.83–0.95)a | Event NRI: 35/57 | 0.28 (0.20–0.35)b | 0.85 (0.76–0.95)a | Event NRI: 10/22 | 0.28 (0.16–0.41)b |
| Nonevent NRI: 53/77 | Nonevent NRI: 26/44 | |||||
| Overall NRI: 1.30b | Overall NRI: 1.05b | |||||
| Lipids + base model | 0.94 (0.90–0.98)a | Event NRI: 41/57 | 0.30 (0.22–0.39)b | 0.93 (0.87–0.99)a | Event NRI: 12/22 | 0.40 (0.26–0.53)b |
| Nonevent NRI: 59/77 | Nonevent NRI: 30/44 | |||||
| Overall NRI: 1.49b | Overall NRI: 1.23b | |||||
| PLS-DA driven models | ||||||
| Lipids (n = 6) | 0.92 (0.87–0.97)a | Event NRI: 37/57 | 0.36 (0.25–0.42)b | 0.80 (0.69–0.91) | Event NRI: 8/22 | 0.20 (0.09–0.31)b |
| Nonevent NRI: 53/77 | Nonevent NRI:24/44 | |||||
| Overall NRI: 1.34b | Overall NRI: 0.91b | |||||
| Lipids + base model | 0.95 (0.92–0.99)a | Event NRI: 43/57 | 0.37 (0.28–0.45)b | 0.90 (0.82–0.97)a | Event NRI: 12/22 | 0.27 (0.15–0.39)b |
| Nonevent NRI: 65/77 | Nonevent NRI: 24/44 | |||||
| Overall NRI: 1.60b | Overall NRI: 1.09b | |||||
P values are comparisons with the corresponding base model. The components of FDR-driven model were DAG 36:0, DAG 32:0, MAG 16:0. The components of the RF-driven model were DAG 36:0, DAG 32:0, MAG 16:0, and PC 34:3. The components of the PLS-DA-driven model were DAG 36:0, DAG 32:0, MAG 26:2, MAG 20:0, MAG 16:0, and PC 34:3.
CI, confidence interval; DAG, diacylglycerol; eFGR, estimated glomerular filtration rate; FDR, false discovery rate; IDI, integrated discrimination improvement; MAG, monoacylglycerol; NRI, net reclassification improvement; PC, phosphatidylcholine; PLS-DA, partial least square-discriminant analysis; RF, Random Forest; UPCR, urine protein-to-creatinine ratio.
P < 0.05, bP < 0.001.
Ten-Fold Cross-validation in the Entire Cohort
Other than internal validation by random splitting of the cohort to training and test sets, we additionally compared the c-statistic of the 10-fold cross-validated models with prevalidated models in the entire cohort. Accordingly, the c-statistic of the multimarker panel alone or after the addition to the base model was not significantly different in the original and validated models (Supplementary Figure S2).
Differentially Regulated Classes of Lipids in the Entire Cohort
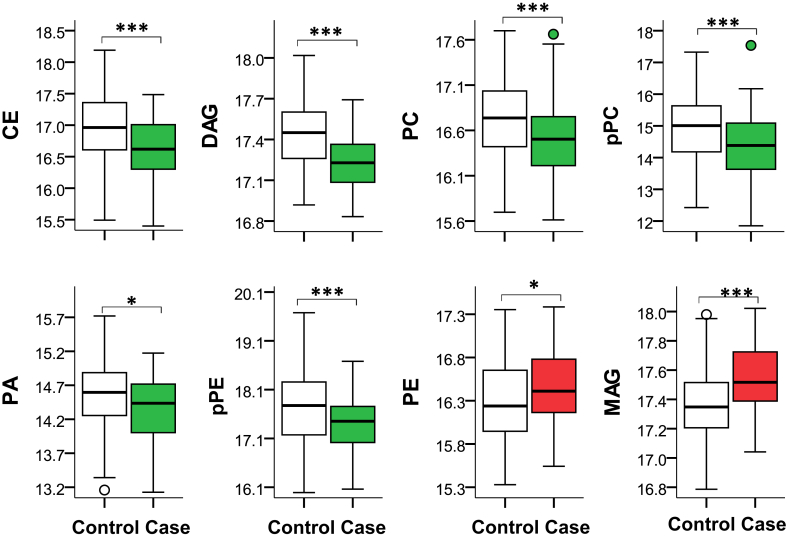
A comparison of intraclass mean peak intensities of each lipid class by progression to ESKD shows lower class level peak intensities in CE, plasmenyl-phosphatidylethanolamine (pPE), and ceramides in progressors irrespective of significance of individual lipids (Supplementary Table S5). However, as the lack of significance in other classes might be driven by a larger number of metabolites that have not reached statistical significance (with the net effect of leading the class level difference toward null), in the next step only the mean of the top differentially regulated lipids that passed the FDR threshold (P < 0.05) was compared (Figure 4). Accordingly, the intraclass mean of differentially regulated metabolites in MAG and PE was higher in progressors, but the mean of CE, DAG, PC, plasmenyl-PC (pPC), pPE, and phosphatidic acid was lower in progressors as compared with nonprogressors.
Figure 4.
Intraclass comparison of the distribution of the mean of log2 peak intensities of significant metabolites that passed the FDR threshold (q < 0.05) by case and control groups; CE: n = 10; DAG: n = 7; PC: n = 2; pPC: n = 2; PA: n = 4; pPE: n = 2; PE: n = 6; MAG: n = 2. The box represents median and interquartile ranges, and error bars present 1.5-fold × the interquartile range below the 25th and above the 75th percentile. Means were compared using the t-test, *P < 0.05; ***P < 0.001. CE, cholesterol esters; DAG, diacylglycerol; FDR, false discovery rate; MAG, monoacylglycerol; PA, phosphatidic acid; pPC, plasmenyl-phosphatidylcholine; pPE, plasmenyl-phosphatidylethanolamine.
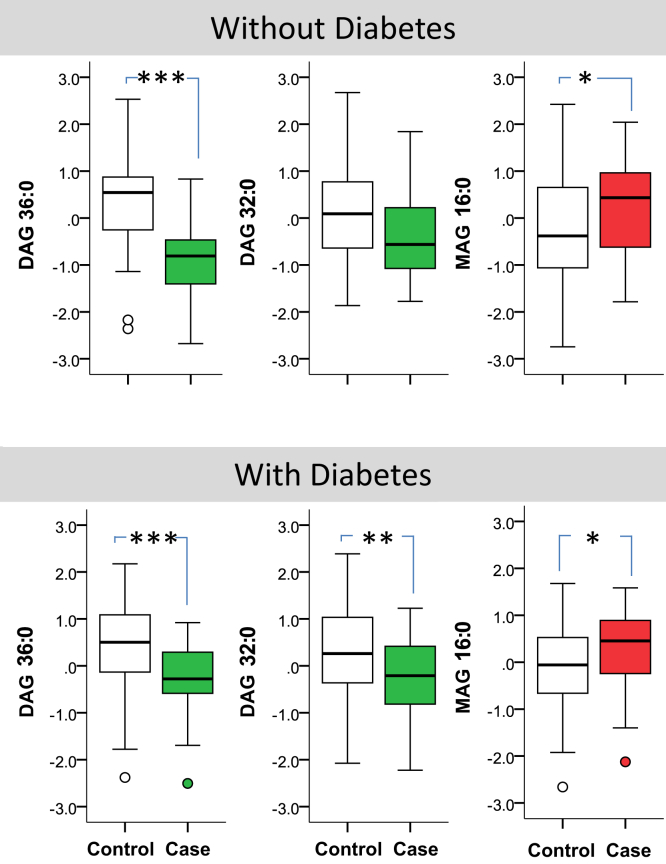
Subgroup Analysis by Diabetes
Figure 5 shows the distribution of the FDR-driven panel by diabetes in the entire cohort. Overall, the pattern of association of the proposed lipids with CKD progression in patients with and without diabetes was similar.
Figure 5.
Comparing the distribution of the false discovery rate proposed lipids in cases and controls in patients with and without diabetes. The box represents median and interquartile ranges, and error bars present 1.5-fold × the interquartile range below the 25th and above the 75th percentile. Means were compared using the t-test, *P < 0.05; **P < 0.01, ***P < 0.001. DAG, diacylglycerol; MAG, monoacylglycerol.
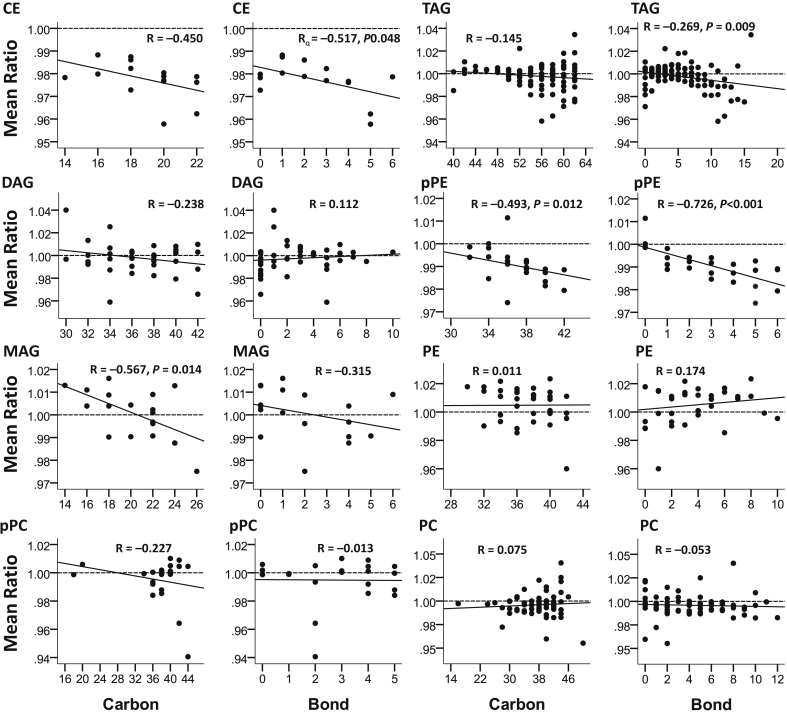
Relationship With Double Bond and Lipid Carbon Numbers
Figure 6 shows that overall there was a trend toward lower abundance of longer chain lipids within CE, DAG, MAG, pPC, and pPE in progressors as compared with nonprogressors, reaching statistical significance in MAG and pPE class (P ≤ 0.014). Such a trend was not observed in triacylglycerol, PE, and PC class. Similarly, there was a trend toward lower abundance of CE, MAG, triacylglycerol, and pPE lipids with higher number of double bonds in progressors as compared with nonprogressors, reaching statistical significance in CE, triacylglycerol, and pPE (P ≤ 0.048). Such a trend was not observed in DAG, pPC, PE, and PC class.
Figure 6.
ESKD:non-ESKD mean ratio of log2 peak intensities by the number of bonds and carbon number in different classes of lipids. Only statistically significant P values are shown. CE, cholesterol ester; DAG, diacylglycerol; ESKD, end-stage kidney disease; MAG, monoacylglycerol; pPC, plasmenyl-phosphatidylcholine; pPE, plasmenyl-phosphatidylethanolamine; TAG, triacylglycerol.
Class Enrichment Analysis, Lipid Correlation Network
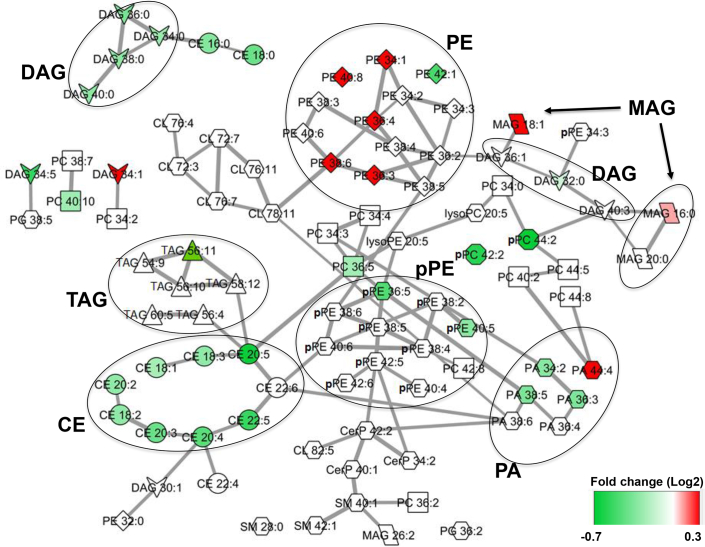
Table 3 demonstrates that the top 3 most significantly enriched classes of lipids using the entire cohort are CEs, DAGs, and PEs. A high-level overview correlation of each lipid class with other relevant metabolites in the metabolic network along with their corresponding involved genes, reactions, and enzymes according to the Kyoto Encyclopedia of Genes and Genomes reveals a number of genes in the network that have already been linked to kidney disease (Supplementary Figure S3). Figure 7 illustrates the map of the correlation network between and within members of the top differentially regulated lipids of various lipid classes driving the separation of progressors from nonprogressors in our study (Supplementary Table S6).
Table 3.
The number of metabolites within each lipid class that has passed the FDR threshold in the entire cohort
| Class | Pathways | N detected | N with P < 0.05 | N with q < 0.05 | Fisher exact P value |
|---|---|---|---|---|---|
| CE | Bile acid biosynthesis | 15 | 12 | 10 | 1.99 × 10–9 |
| DAG | Glycerophospholipid | 41 | 11 | 7 | 0.018 |
| PE | Glycerophospholipid, phosphatidylinositol phosphate | 35 | 14 | 6 | 0.029 |
| PA | Glycerophospholipid, phosphatidylinositol phosphate | 21 | 6 | 4 | 0.053 |
| PC | Glycerophospholipid, arachidonic acid metabolism, linoleate metabolism | 71 | 16 | 2 | 0.208 |
| SM | Glycosphingolipid metabolism | 30 | 3 | 0 | 0.257 |
| MAG | Glycerophospholipid | 18 | 5 | 2 | 0.368 |
| Lyso PC | – | 23 | 2 | 0 | 0.395 |
| PI | Phosphatidylinositol phosphate | 13 | 0 | 0 | 0.613 |
| Lyso PE | Glycerophospholipid | 16 | 1 | 0 | 0.618 |
| Plasmenyl PC | Glycerophospholipid | 22 | 2 | 2 | 0.663 |
| Plasmenyl PE | Glycerophospholipid | 25 | 11 | 2 | 0.694 |
| CerP | Glycosphingolipid | 10 | 4 | 0 | 1.000 |
| PS | Glycerophospholipid | 3 | 0 | 0 | 1.000 |
| PG | Glycerophospholipid | 12 | 2 | 0 | 1.000 |
| CL | Glycerophospholipid | 62 | 7 | 0 | 0.015 |
| TAG | Glycerophospholipid | 93 | 6 | 1 | 0.012 |
| Total detected | 510 | 102 | 36 | ||
The q value was obtained based on the Benjamini-Hochberg procedure for multiple testing of all identified lipids in the entire cohort. Fisher exact P value tests, if any particular class of lipids had differentially higher number of lipids that have passed the statistical threshold of the q value as compared with the rest of the identified lipids combined (class enrichment by the overrepresentation method). The statistically significant P values of overrepresented classes are shown in bold.
CE, cholesterol ester; Cer-P, ceramides; CL, cardiolipin; DAG, diacylglycerol; FDR, false discovery rate; MAG, monoacylglycerol; PA, phosphatidic acid; PE, phosphatidylethanolamine; PG, phosphoglycerol; PI, phosphatidylinositol; PS, phosphatidylserine; SM, sphingomyelin; TAG, triacylglycerol.
Figure 7.
The correlation network displaying metabolic differences between progressors and nonprogressors according to the top 102 differentially regulated lipids. Node color reflects fold changes. Lipids that have passed the FDR statistical significance with significantly lower and higher abundance in progressors are shown in green and red, respectively. Edge thickness represents the significance of adjusted partial correlation coefficients between the nodes (Supplementary Table S6). In most cases, the correlations within the same class of lipids are stronger than interclass correlations that are evident from the network structure. CE, cholesterol ester; DAG, diacylglycerol; FDR, false discovery rate; MAG, monoacylglycerol; PA, phosphatidic acid; pPE, plasmenyl-phosphatidylethanolamine; TAG, triacylglycerol.
Discussion
In this study, DAG and CE in the baseline samples were among the 2 most predictive classes of lipids for the separation of progressors from nonprogressors to ESKD, as they had the largest number of metabolites that passed the FDR threshold. At the class level, among metabolites that passed the FDR threshold, on average DAGs, CEs, PCs, pPCs, pPEs, and phosphatidic acids had lower abundance whereas PEs and MAGs had higher abundance in progressors. At the metabolite level, the top-hit independent predictors of progression uniformly picked up by all 3 classification methods were DAG36:0, DAG32:0, and MAG16:0. DAG36:0 and DAG32:0 were associated with lower and MAG16:0 associated with higher odds of progression to ESKD independent of eGFR and UPCR. The net reclassification improvement and integrated discrimination improvement indices of the lipids panels showed highly statistically significant improvement in classification, and the probabilistic models projected by the addition of a multimarker panel presented a significantly higher c-statistic above and beyond what was achieved from eGFR and UPCR and combined an observation that stood the internal validation.
These findings provide a basis for examining serum lipid levels as a minimally invasive tool for the identification of kidneys at risk of progression to ESKD. A systematic examination of serum levels of lipid species for their prognostic value is the first logical step toward the identification of novel pathways of damage amenable to intervention, particularly in the presence of conflicting results from association studies of dyslipidemia and progression of kidney disease in human studies.6, 7, 8, 9, 10 These studies are in part a reflection of the underappreciation of significant intraclass biological diversity and variations in lipid classes beyond traditional lipids panels such as lipoproteins. Although lipoproteins are complex particles that carry almost all the lipid classes, the determination of a level of LDL and high-density lipoprotein may not address the complexity of the lipidome. In a case-control observation, Reis et al.18 compared lipidomic profiles of LDL in 10 normocholestrolemic patients with CKD stages 4 and 5 with 10 healthy individuals. Triacylglycerols and N-acyl taurines were significantly higher in CKD, whereas PCs, pPEs, sulfatides, ceramides, and cholesterol sulfate were significantly lower in patients with CKD.18 These results are compatible with our findings, wherein from the list of differentially regulated species, MAGs were higher, and CEs, PCs, pPCs, pPEs, and phosphatidic acids were lower in progressors to ESKD. However, a limitation of the Reis study is that it was limited to LDL, but not other lipoprotein particles. Looking at serum gives a more comprehensive profile that might be the reason why we have identified additional lipids. What further highlights the significance of our findings is the predictive power of the proposed panel at earlier stage CKD (stage 2 or 3) to predict progression to ESKD. To our knowledge, this study is the first to show the predictive power of a distinct panel of lipids from the application of an unbiased lipidomics search strategy for the prediction of incident ESKD at CKD stages 2 and 3.
In this study, we also observed decreased abundance of longer acyl chains and polyunsaturated complex lipids in progressors. This is aligned with meta-analysis of clinical trials on beneficial effects of polyunsaturated fatty acid supplementation on urinary protein excretion and biomarkers of kidney injury.37, 38 We are unaware of any report on a protective effect of longer acyl chain carbons from kidney injury, and therefore the observation of lower abundance of longer acyl chain carbons in progressors is likely a novel observation. The mechanisms may involve differentially altered enzymatic pathways such as elongase, dietary effects, and alterations in liver metabolism besides other unknown mechanisms.
The differentially regulated lipids in this study belong to the bile acid, glycerophospholipid, and phosphatidylinositol phosphate biosynthesis pathways. Although we have not explored the relationships between the lipids and the alterations in expression of the corresponding involved genes, reactions, and enzymes in the metabolic network, from the work of others, there is evidence to suggest that the involved genes and pathways might be differentially regulated in CKD (Supplementary Figure S3). Search in the Kyoto Encyclopedia of Genes and Genomes (Supplementary Figure S3) reveals a high-level view of such interrelationships. However, such a mapping strategy has important limitations. First, not every known lipid class is mapped in Kyoto Encyclopedia of Genes and Genomes, and second, the mapped lipids are limited to the class level of each lipid and therefore lack information on distinct lipids within each class, ignoring the intraclass diversity by carbon number or number of unsaturated carbons.
Unique aspects of our study include (a) identification of the top differentially regulated candidate predictors of progression, (b) demonstration of the intraclass diversity of lipids above and beyond the mapped lipids in Kyoto Encyclopedia of Genes and Genomes, and (c) elucidation of the lipidomic signature of progression in CKD for the first time in CRIC (Figure 7). Our network analysis highlights the links between DAGs, CEs, and PEs that are differentially expressed by cases and controls. The underlying mechanisms may involve the dysregulation of classic pathways of lipolysis including adenylate cyclase/Gs protein/protein kinase A/hormone-sensitive lipase cascade and/or phorbol myristate acetate/protein kinase-C (PKC)/mitogen-activated protein kinase signaling pathways.39 In particular, DAG is shown to be a cofactor of PKC stimulation40 and is linked with an inflammatory milieu41 and activation of the mitogen-activated protein kinase,42 nuclear factor κB,43 and vascular endothelial growth factor44, 45 signaling pathways. Similarly, cellular accumulation of CE is linked to the activation of PKC46 and mitogen-activated protein kinase.47 PKC promotes the synthesis of PE leading to differentially regulated degradation of membranous phospholipid and alteration of downstream membrane-protein physiological processes.48, 49, 50, 51, 52, 53, 54 Whether the lower serum level of differentially regulated DAGs and CEs in progressors reflects their increased utilization in cortical glomerular or tubular compartments and activating the above pathologic cascades of events and whether increased serum levels of PEs in progressors reflects renal tissue activation of PKC and inflammation55 could be subjects for further investigation. These mechanisms may also explain increased risk of cardiovascular diseases in CKD but requires confirmation in further studies.
This study has several strengths. It is powered with appropriate sample size for conduct of the proposed analyses. We followed a very strict quality control protocol and achieved excellent quality data evidenced by low coefficient of variation, low rate of missing values, and excellent reproducibility (Supplementary Figure S1). We took the advantage of outstanding infrastructure in CRIC with the prospectively ascertained outcome of ESKD allowing the identification of the proposed panel. There are limitations to the study as well. Although our results are consistent with the findings of Reis et al.,18 and the internal validation is suggestive of reproducibility, we are still in need of external validation of the findings in independent samples and/or cohorts. We have not studied the lipid alterations in kidney tissue as kidney biopsies were not available for analysis. As such the underpinning mechanisms of renal injury and the directionality with serum lipidomic signature require further investigation. In conclusion, the lipidomic signature of differentially regulated lipids separating progressors from nonprogressors to ESKD in a subset of the CRIC cohort included lower abundance of select number of DAGs, CEs, PCs, pPCs, pPEs, and phosphatidic acids, but higher abundance of select number of PEs and MAGs. From these differentially regulated lipids, a distinct multimarker panel was able to independently predict the progression of CKD to ESKD and improved the classification power of eGFR and UPCR when added to the base model. Further research is required for external validation, as well as serum-kidney tissue lipid alteration cross-talk.
Disclosure
EPR has the “Lipidomic Biomarkers of Diabetes” patent without receiving royalties. MRW gives consult to Akebia, Janssen, Astrazeneca, MSD, Relypsa, Lexicon, and Boehringer-Ingelheim. HF received lecture fees from Kyowa Kirin, Glaxo Smith Kline, and Boehringer-Ingelheim. All the other authors declared no competing interests.
Acknowledgments
Funding for the CRIC Study was obtained under a cooperative agreement from National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK): U01DK060990, U01DK060984, U01DK061022, U01DK061021, U01DK061028, U01DK060980, U01DK060963, and U01DK060902. In addition, this work is supported in part by grants from the National Institutes of Health: DK106523, DK094292, DK089503, DK082841, DK081943, DK097153, UL1TR000433, UL1TR000003, UL1TR000439, UL1 TR-000424, UL1RR029879, UL1 RR024131, M01 RR-16500, and GM103337. The authors also thank Dr. Matthias Kretzler for his remarks.
Footnotes
Supplementary Methods. Supplementary material includes details on the methods of sample preparation and tables of identified lipids.
Table S1. Comparison of baseline characteristics in case and control groups with the training and the test sets
Table S2. Identified lipids by type of adducts, experimental mass, and retention time in positive and negative modes. The mass accuracy was ±0.001 in positive mode and ±0.005 in negative mode, with an overall mass error of <2 parts per million
Table S3. List of processed lipids and their ranks according to the false discovery rate (FDR) corrected t-test, and unadjusted and adjusted logistic regression models in the training set. Adjustments are by eGFR, urine protein-to-creatinine ratio, age, sex, race, diabetes, hypertension, and congestive heart failure
Table S4. The top 10 metabolites loaded by various classification methods in the training set; partial least square-discriminant analysis (PLS-DA)
Table S5. Mean of log2 peak intensities of all detected lipids in each class by progression to end-stage kidney disease
Table S6. The partial correlation coefficients and adjusted P values of significant edges illustrated in Figure 7 after adjusting for the total number of bivariate correlations passing the Benjamini-Hochberg threshold of 0.1. The 108 edges out of 5151 possible bivariate correlations are shown among the top 102 nominally differentiated lipids in the entire cohort
Figure S1. (a) Batch variability of internal standards in test pools. (b) Batch variability of internal standards in pooled plasma.
Figure S2. The c-statistic of the original and 10-fold validated multimarker panel alone and after addition to the base model compared with the c-statistics of the base model (eGFR+UPCR).
Figure S3. Class level metabolic network represents 8 main classes of lipids that were detected in our samples. Limited representation of lipid metabolism in the most existing databases allows only a high level overview of relevant pathways; however, the Metscape network provides a broad list of reactions, enzymes, and genes involved in metabolism of different classes of lipids. A number of genes in this network including LCAT, LIPC, LPL, PLA2G1B, PLA2G2A, PLCD1, PLD2, SOAT1, PLA2G10, SOAT2, LIPG, PLCE1, and DGKH have been previously linked to kidney disease.
Supplementary material is linked to the online version of the paper at www.kireports.org.
Contributor Information
Farsad Afshinnia, Email: fafshin@med.umich.edu.
Subramaniam Pennathur, Email: spennath@umich.edu.
CRIC Study Investigators:
Lawrence J. Appel, Alan S. Go, John W. Kusek, James P. Lash, and Raymond R. Townsend
Supplementary Material
Supplementary material includes details on the methods of sample preparation and tables of identified lipids.
Comparison of baseline characteristics in case and control groups with the training and the test sets
Identified lipids by type of adducts, experimental mass, and retention time in positive and negative modes. The mass accuracy was ±0.001 in positive mode and ±0.005 in negative mode, with an overall mass error of <2 parts per million
List of processed lipids and their ranks according to the false discovery rate (FDR) corrected t-test, and unadjusted and adjusted logistic regression models in the training set. Adjustments are by eGFR, urine protein-to-creatinine ratio, age, sex, race, diabetes, hypertension, and congestive heart failure
The top 10 metabolites loaded by various classification methods in the training set; partial least square-discriminant analysis (PLS-DA)
Mean of log2 peak intensities of all detected lipids in each class by progression to end-stage kidney disease
The partial correlation coefficients and adjusted P values of significant edges illustrated in Figure 7 after adjusting for the total number of bivariate correlations passing the Benjamini-Hochberg threshold of 0.1. The 108 edges out of 5151 possible bivariate correlations are shown among the top 102 nominally differentiated lipids in the entire cohort
(a) Batch variability of internal standards in test pools. (b) Batch variability of internal standards in pooled plasma.
The c-statistic of the original and 10-fold validated multimarker panel alone and after addition to the base model compared with the c-statistics of the base model (eGFR+UPCR).
Class level metabolic network represents 8 main classes of lipids that were detected in our samples. Limited representation of lipid metabolism in the most existing databases allows only a high level overview of relevant pathways; however, the Metscape network provides a broad list of reactions, enzymes, and genes involved in metabolism of different classes of lipids. A number of genes in this network including LCAT, LIPC, LPL, PLA2G1B, PLA2G2A, PLCD1, PLD2, SOAT1, PLA2G10, SOAT2, LIPG, PLCE1, and DGKH have been previously linked to kidney disease.
References
- 1.Centers for Disease Control. National Chronic Kidney Disease Fact Sheet 2010. Available at: www.cdc.gov/diabetes/projects/pdfs/ckd_summary.pdf. Accessed September 8, 2016.
- 2.Levey A.S., Stevens L.A., Schmid C.H. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150:604–612. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rule A.D., Larson T.S., Bergstralh E.J. Using serum creatinine to estimate glomerular filtration rate: accuracy in good health and in chronic kidney disease. Ann Intern Med. 2004;141:929–937. doi: 10.7326/0003-4819-141-12-200412210-00009. [DOI] [PubMed] [Google Scholar]
- 4.Fahy E., Subramaniam S., Murphy R.C. Update of the LIPID MAPS comprehensive classification system for lipids. J Lipid Res. 2009;50(suppl):S9–S14. doi: 10.1194/jlr.R800095-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subramaniam S., Fahy E., Gupta S. Bioinformatics and systems biology of the lipidome. Chem Rev. 2011;111:6452–6490. doi: 10.1021/cr200295k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chawla V., Greene T., Beck G.J. Hyperlipidemia and long-term outcomes in nondiabetic chronic kidney disease. Clin J Am Soc Nephrol. 2010;5:1582–1587. doi: 10.2215/CJN.01450210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hadjadj S., Duly-Bouhanick B., Bekherraz A. Serum triglycerides are a predictive factor for the development and the progression of renal and retinal complications in patients with type 1 diabetes. Diabetes Metab. 2004;30:43–51. doi: 10.1016/s1262-3636(07)70088-5. [DOI] [PubMed] [Google Scholar]
- 8.Kaysen G.A. Lipid and lipoprotein metabolism in chronic kidney disease. J Ren Nutr. 2009;19:73–77. doi: 10.1053/j.jrn.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 9.Rahman M., Yang W., Akkina S. Relation of serum lipids and lipoproteins with progression of CKD: the CRIC study. Clin J Am Soc Nephrol. 2014;9:1190–1198. doi: 10.2215/CJN.09320913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samuelsson O., Mulec H., Knight-Gibson C. Lipoprotein abnormalities are associated with increased rate of progression of human chronic renal insufficiency. Nephrol Dial Transplant. 1997;12:1908–1915. doi: 10.1093/ndt/12.9.1908. [DOI] [PubMed] [Google Scholar]
- 11.Miller M., Stone N.J., Ballantyne C. Triglycerides and cardiovascular disease: a scientific statement from the American Heart Association. Circulation. 2011;123:2292–2333. doi: 10.1161/CIR.0b013e3182160726. [DOI] [PubMed] [Google Scholar]
- 12.de Boer I.H., Astor B.C., Kramer H. Lipoprotein abnormalities associated with mild impairment of kidney function in the multi-ethnic study of atherosclerosis. Clin J Am Soc Nephrol. 2008;3:125–132. doi: 10.2215/CJN.03390807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhee E.P., Cheng S., Larson M.G. Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. J Clin Invest. 2011;121:1402–1411. doi: 10.1172/JCI44442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dutta T., Chai H.S., Ward L.E. Concordance of changes in metabolic pathways based on plasma metabolomics and skeletal muscle transcriptomics in type 1 diabetes. Diabetes. 2012;61:1004–1016. doi: 10.2337/db11-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinterwirth H., Stegemann C., Mayr M. Lipidomics: quest for molecular lipid biomarkers in cardiovascular disease. Circ Cardiovasc Genet. 2014;7:941–954. doi: 10.1161/CIRCGENETICS.114.000550. [DOI] [PubMed] [Google Scholar]
- 16.Zhou X., Mao J., Ai J. Identification of plasma lipid biomarkers for prostate cancer by lipidomics and bioinformatics. PLoS One. 2012;7:e48889. doi: 10.1371/journal.pone.0048889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sas K.M., Karnovsky A., Michailidis G., Pennathur S. Metabolomics and diabetes: analytical and computational approaches. Diabetes. 2015;64:718–732. doi: 10.2337/db14-0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reis A., Rudnitskaya A., Chariyavilaskul P. Top-down lipidomics of low density lipoprotein reveal altered lipid profiles in advanced chronic kidney disease. J Lipid Res. 2015;56:413–422. doi: 10.1194/jlr.M055624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jassim F.A. Image denoising using interquartile range filter with local averaging. Int J Soft Comput Eng. 2013;2:2231–2307. [Google Scholar]
- 20.Feldman H.I., Appel L.J., Chertow G.M. The Chronic Renal Insufficiency Cohort (CRIC) Study: design and methods. J Am Soc Nephrol. 2003;14:S148–S153. doi: 10.1097/01.asn.0000070149.78399.ce. [DOI] [PubMed] [Google Scholar]
- 21.Lash J.P., Go A.S., Appel L.J. Chronic Renal Insufficiency Cohort (CRIC) Study: baseline characteristics and associations with kidney function. Clin J Am Soc Nephrol. 2009;4:1302–1311. doi: 10.2215/CJN.00070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altman N.S. An introduction to kernel and nearest-neighbor nonparametric regression. Am Stat. 1992;46:175–185. [Google Scholar]
- 23.Redestig H., Fukushima A., Stenlund H. Compensation for systematic cross-contribution improves normalization of mass spectrometry based metabolomics data. Anal Chem. 2009;81:7974–7980. doi: 10.1021/ac901143w. [DOI] [PubMed] [Google Scholar]
- 24.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benjamini Y., Drai D., Elmer G. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 26.Eriksson L., Antti H., Gottfries J. Using chemometrics for navigating in the large data sets of genomics, proteomics, and metabonomics (gpm) Anal Bioanal Chem. 2004;380:419–429. doi: 10.1007/s00216-004-2783-y. [DOI] [PubMed] [Google Scholar]
- 27.Eriksson L., Johansson E., Lindgren F. Megavariate analysis of hierarchical QSAR data. J Comput Aided Mol Des. 2002;16:711–726. doi: 10.1023/a:1022450725545. [DOI] [PubMed] [Google Scholar]
- 28.Svetnik V., Liaw A., Tong C. Random forest: a classification and regression tool for compound classification and QSAR modeling. J Chem Inf Comput Sci. 2003;43:1947–1958. doi: 10.1021/ci034160g. [DOI] [PubMed] [Google Scholar]
- 29.DeLong E.R., DeLong D.M., Clarke-Pearson D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 30.Karnovsky A., Weymouth T., Hull T. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics. 2012;28:373–380. doi: 10.1093/bioinformatics/btr661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jankova J, van de Geer S. Honest confidence regions and optimality in high-dimensional precision matrix estimation. Available at: https://arxiv.org/pdf/1507.02061v2.pdf. Accessed September 8, 2016.
- 32.Hanley J.A., McNeil B.J. A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology. 1983;148:839–843. doi: 10.1148/radiology.148.3.6878708. [DOI] [PubMed] [Google Scholar]
- 33.Obuchowski N.A., McClish D.K. Sample size determination for diagnostic accuracy studies involving binormal ROC curve indices. Stat Med. 1997;16:1529–1542. doi: 10.1002/(sici)1097-0258(19970715)16:13<1529::aid-sim565>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 34.Xia J., Mandal R., Sinelnikov I.V. MetaboAnalyst 2.0—a comprehensive server for metabolomic data analysis. Nucleic Acids Res. 2012;40:W127–W133. doi: 10.1093/nar/gks374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xia J., Psychogios N., Young N., Wishart D.S. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009;37:W652–W660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Livera AM, Bowne JB. Package ‘Metabolomics’. Available at: https://cran.r-project.org/web/packages/metabolomics/metabolomics.pdf. Accessed September 8, 2016.
- 37.Miller E.R., III, Juraschek S.P., Anderson C.A. The effects of n-3 long-chain polyunsaturated fatty acid supplementation on biomarkers of kidney injury in adults with diabetes: results of the GO-FISH trial. Diabetes Care. 2013;36:1462–1469. doi: 10.2337/dc12-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller E.R., 3rd, Juraschek S.P., Appel L.J. The effect of n-3 long-chain polyunsaturated fatty acid supplementation on urine protein excretion and kidney function: meta-analysis of clinical trials. Am J Clin Nutr. 2009;89:1937–1945. doi: 10.3945/ajcn.2008.26867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carmen G.Y., Victor S.M. Signalling mechanisms regulating lipolysis. Cell Signal. 2006;18:401–408. doi: 10.1016/j.cellsig.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 40.Mehta K.D., Radominska-Pandya A., Kapoor G.S. Critical role of diacylglycerol- and phospholipid-regulated protein kinase C epsilon in induction of low-density lipoprotein receptor transcription in response to depletion of cholesterol. Mol Cell Biol. 2002;22:3783–3793. doi: 10.1128/MCB.22.11.3783-3793.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reibman J., Korchak H.M., Vosshall L.B. Changes in diacylglycerol labeling, cell shape, and protein phosphorylation distinguish “triggering” from “activation” of human neutrophils. J Biol Chem. 1988;263:6322–6328. [PubMed] [Google Scholar]
- 42.van Dijk M., Muriana F.J., van Der Hoeven P.C. Diacylglycerol generated by exogenous phospholipase C activates the mitogen-activated protein kinase pathway independent of Ras- and phorbol ester-sensitive protein kinase C: dependence on protein kinase C-zeta. Biochem J. 1997;323(Pt 3):693–699. doi: 10.1042/bj3230693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamamoto H., Hanada K., Nishijima M. Involvement of diacylglycerol production in activation of nuclear factor kappaB by a CD14-mediated lipopolysaccharide stimulus. Biochem J. 1997;325(Pt 1):223–228. doi: 10.1042/bj3250223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baldanzi G., Mitola S., Cutrupi S. Activation of diacylglycerol kinase alpha is required for VEGF-induced angiogenic signaling in vitro. Oncogene. 2004;23:4828–4838. doi: 10.1038/sj.onc.1207633. [DOI] [PubMed] [Google Scholar]
- 45.Yang L., Xu Y., Li W. Diacylglycerol kinase (DGK) inhibitor II (R59949) could suppress retinal neovascularization and protect retinal astrocytes in an oxygen-induced retinopathy model. J Mol Neurosci. 2015;56:78–88. doi: 10.1007/s12031-014-0469-2. [DOI] [PubMed] [Google Scholar]
- 46.Maehira F., Harada K., Shimoji J. Age-related changes in the activation of aortic cholesteryl ester hydrolases by protein kinases in rats. Biochim Biophys Acta. 1998;1389:197–205. doi: 10.1016/s0005-2760(97)00154-9. [DOI] [PubMed] [Google Scholar]
- 47.Mei S., Gu H., Ward A. p38 mitogen-activated protein kinase (MAPK) promotes cholesterol ester accumulation in macrophages through inhibition of macroautophagy. J Biol Chem. 2012;287:11761–11768. doi: 10.1074/jbc.M111.333575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bazzi M.D., Youakim M.A., Nelsestuen G.L. Importance of phosphatidylethanolamine for association of protein kinase C and other cytoplasmic proteins with membranes. Biochemistry. 1992;31:1125–1134. doi: 10.1021/bi00119a022. [DOI] [PubMed] [Google Scholar]
- 49.Cook H.W., Ridgway N.D., Byers D.M. Involvement of phospholipase D and protein kinase C in phorbol ester and fatty acid stimulated turnover of phosphatidylcholine and phosphatidylethanolamine in neural cells. Biochim Biophys Acta. 1998;1390:103–117. doi: 10.1016/s0005-2760(97)00162-8. [DOI] [PubMed] [Google Scholar]
- 50.Kiss Z. Cooperative effects of ethanol and protein kinase C activators on phospholipase-D-mediated hydrolysis of phosphatidylethanolamine in NIH 3T3 fibroblasts. Biochim Biophys Acta. 1992;1175:88–94. doi: 10.1016/0167-4889(92)90013-2. [DOI] [PubMed] [Google Scholar]
- 51.Kiss Z. The long-term combined stimulatory effects of ethanol and phorbol ester on phosphatidylethanolamine hydrolysis are mediated by a phospholipase C and prevented by overexpressed alpha-protein kinase C in fibroblasts. Eur J Biochem. 1992;209:467–473. doi: 10.1111/j.1432-1033.1992.tb17311.x. [DOI] [PubMed] [Google Scholar]
- 52.Kiss Z. Expression of protein kinase C-beta promotes the stimulatory effect of phorbol ester on phosphatidylethanolamine synthesis. Arch Biochem Biophys. 1997;347:37–44. doi: 10.1006/abbi.1997.0308. [DOI] [PubMed] [Google Scholar]
- 53.Sesca E., Perletti G.P., Binasco V. Phosphatidylethanolamine N-methyltransferase 2 and CTP-phosphocholine cytidylyltransferase expressions are related with protein kinase C isozymes in developmental liver growth. Biochem Biophys Res Commun. 1996;229:158–162. doi: 10.1006/bbrc.1996.1773. [DOI] [PubMed] [Google Scholar]
- 54.Tomono M., Kiss Z. Vitamin K3 preferentially inhibits stimulation of phospholipase D-mediated hydrolysis of phosphatidylethanolamine by protein kinase C activators in NIH 3T3 fibroblasts. Arch Biochem Biophys. 1994;314:217–223. doi: 10.1006/abbi.1994.1432. [DOI] [PubMed] [Google Scholar]
- 55.Eros G., Varga G., Varadi R. Anti-inflammatory action of a phosphatidylcholine, phosphatidylethanolamine and N-acylphosphatidylethanolamine-enriched diet in carrageenan-induced pleurisy. Eur Surg Res. 2009;42:40–48. doi: 10.1159/000167856. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material includes details on the methods of sample preparation and tables of identified lipids.
Comparison of baseline characteristics in case and control groups with the training and the test sets
Identified lipids by type of adducts, experimental mass, and retention time in positive and negative modes. The mass accuracy was ±0.001 in positive mode and ±0.005 in negative mode, with an overall mass error of <2 parts per million
List of processed lipids and their ranks according to the false discovery rate (FDR) corrected t-test, and unadjusted and adjusted logistic regression models in the training set. Adjustments are by eGFR, urine protein-to-creatinine ratio, age, sex, race, diabetes, hypertension, and congestive heart failure
The top 10 metabolites loaded by various classification methods in the training set; partial least square-discriminant analysis (PLS-DA)
Mean of log2 peak intensities of all detected lipids in each class by progression to end-stage kidney disease
The partial correlation coefficients and adjusted P values of significant edges illustrated in Figure 7 after adjusting for the total number of bivariate correlations passing the Benjamini-Hochberg threshold of 0.1. The 108 edges out of 5151 possible bivariate correlations are shown among the top 102 nominally differentiated lipids in the entire cohort
(a) Batch variability of internal standards in test pools. (b) Batch variability of internal standards in pooled plasma.
The c-statistic of the original and 10-fold validated multimarker panel alone and after addition to the base model compared with the c-statistics of the base model (eGFR+UPCR).
Class level metabolic network represents 8 main classes of lipids that were detected in our samples. Limited representation of lipid metabolism in the most existing databases allows only a high level overview of relevant pathways; however, the Metscape network provides a broad list of reactions, enzymes, and genes involved in metabolism of different classes of lipids. A number of genes in this network including LCAT, LIPC, LPL, PLA2G1B, PLA2G2A, PLCD1, PLD2, SOAT1, PLA2G10, SOAT2, LIPG, PLCE1, and DGKH have been previously linked to kidney disease.