Abstract
In eukaryotic cells, pre-mRNAs undergo extensive processing in the nucleus prior to export. Processing is subject to a quality-control mechanism that retains improperly processed transcripts at or near sites of transcription. A poly(A) tail added by the normal 3′-processing machinery is necessary but not sufficient for export. Retention depends on the exosome. In this study, we identify the poly(A)-binding protein, Pab1, and the poly(A) nuclease, PAN, as important factors that couple 3′ processing to export. Pab1 contains a nonessential leucine-rich nuclear export signal and shuttles between the nucleus and the cytoplasm. It can exit the nucleus either as cargo of exportin 1 or bound to mRNA. Pab1 is essential but several bypass suppressors have been identified. Deletion of PAB1 from these bypass suppressor strains results in exosome-dependent retention at sites of transcription. Retention is also seen in cells lacking PAN, which Pab1 is thought to recruit and which may be responsible for the final step of mRNA biogenesis, trimming of the poly(A) tail to the length found on newly exported mRNAs. The studies presented here suggest that proper loading of Pab1 onto mRNAs and final trimming of the tail allows release from transcription sites and couples pre-mRNA processing to export.
Keywords: mRNA transport, polyadenylation, exportin 1, nuclear export signal, gene expression, mRNA biogenesis
The gene expression pathway begins in the nucleus with modification of chromatin to make it accessible for transcription, and proceeds through several stages. These include initiation of transcription, elongation, pre-mRNA processing, transcription termination, export to the cytoplasm, translation, and eventual turnover of the mRNA. A large body of evidence indicates that these multiple steps are coordinated and mechanistically coupled rather than sequential (for review, see Manley 2002; Proudfoot et al. 2002), and that mRNAs are exported to the cytoplasm only after all nuclear processing events are completed successfully (for review, see Maniatis and Reed 2002; Stutz and Izaurralde 2003; McKee and Silver 2004). In yeast strains with defects affecting many stages of mRNA biogenesis, mRNA export is prevented and improperly processed mRNAs accumulate in the nucleus, where they are subsequently degraded (Birse et al. 1998; Brodsky and Silver 2000; Hilleren and Parker 2001; Jensen et al. 2001a; Strasser and Hurt 2001; Hammell et al. 2002; Jimeno et al. 2002; Strasser et al. 2002).
Almost all eukaryotic mRNAs are polyadenylated at their 3′ ends. The 3′ processing occurs in a coupled, two-step reaction that includes endonucleolytic cleavage of the nascent RNA, followed by the addition of a polyadenosine tract (for review, see Zhao et al. 1999; Jensen et al. 2003). These cleavage and polyadenylation reactions involve the coordinated action of at least 21 polypeptides, including poly(A) polymerase and multiple mRNA-binding proteins. Most of these proteins are also essential for termination, and some physically interact with pol II during transcription (Birse et al. 1998; Licatalosi et al. 2002; Calvo and Manley 2003).
In a screen for yeast mutants defective in mRNA export, we identified mutants affecting poly(A)-polymerase (Pap1) and other 3′-processing factors (Hammell et al. 2002). Surprisingly, in cells of many of these mutants, the mRNAs produced were cleaved at the proper site and polyadenylated, but were not exported. Cis-acting mutations of cleavage and polyadenylation signals also leads to accumulation of mRNAs (Custodio et al. 1999; Hammell et al. 2002). In both yeast and metazoan cells, inefficient export occurs when a cis-acting ribozyme is used to generate the 3′ end of an mRNA (Dower and Rosbash 2002), even when a template-encoded poly(A) tract is located very near the ribozyme cleavage site (Huang and Carmichael 1996). Together, these studies indicate that passage through the normal 3′-processing machinery facilitates mRNA export, but does not indicate how 3′ processing is coupled mechanistically to mRNA export.
RNAs not exported from the nucleus are turned over by components of an evolutionarily conserved multisubunit complex, the exosome (Bousquet-Antonelli et al. 2000; Burkard and Butler 2000; for review, see Mitchell and Tollervey 2000, 2001, 2003). Most of the 11 subunits of this complex are thought to have 3′–5′ exonuclease activity and have been implicated in processing a diverse set of substrates including snoRNAs, 5.8S rRNA, and improperly processed mRNAs (Briggs et al. 1998; Allmang et al. 1999; Bousquet-Antonelli et al. 2000; van Hoof et al. 2000; Libri et al. 2002; Torchet et al. 2002; Zenklusen et al. 2002; Phillips and Butler 2003). Recent studies indicate that a nuclear-specific subunit of the exosome, Rrp6, is mechanistically, physically, and/or genetically linked to several factors involved in mRNA transport and also to other factors required to process rRNAs and snoRNAs (Burkard and Butler 2000; Hilleren et al. 2001; Jensen et al. 2001a,b). Improperly processed ribonucleoprotein particles (mRNPs) accumulate at their sites of transcription (Hilleren and Parker 2001; Jensen et al. 2001b; Libri et al. 2002; Thomsen et al. 2003), and this accumulation does not occur in the absence of Rrp6 (Hilleren and Parker 2001; Jensen et al. 2001b). It has been proposed that the exosome actively retains and then degrades improperly processed and untransported mRNPs. However, it is not understood how the exosome determines that an mRNA has been incorrectly processed or improperly packaged, or how this prevents these mRNAs from being exported.
Metazoan genomes encode multiple poly(A)-binding proteins, but the Saccharomyces cerevisiae genome encodes a single related species, Pab1. Different poly(A)-binding proteins predominate in the nucleus and the cytoplasm of metazoan cells, whereas yeast Pab1 functions in both compartments (Sachs et al. 1986). It is an essential protein in S. cerevisiae, although no individual domain is absolutely required for viability (Sachs et al. 1987; Tarun and Sachs 1995, 1996; Amrani 1997; Minvielle-Sebastia et al. 1997; Coller et al. 1998; Kessler and Sachs 1998). At steady state, Pab1 is primarily cytoplasmic, but cell fractionation indicates that some Pab1 is nuclear (Sachs et al. 1986). Consistent with this location, some Pab1 copurifies with CFIA, a nuclear multisubunit complex that cleaves nascent mRNAs at their 3′ ends (Amrani et al. 1997; Minvielle-Sebastia et al. 1997). During 3′ processing, multiple Pab1s are loaded onto the poly(A) tail, and are exported as part of the mRNP. One nuclear function of Pab1 is modulation of the overall length of the poly(A) tail, which is initially synthesized at a length greater than is found on newly exported mRNA. The multisubunit poly(A) nuclease (PAN) is recruited by Pab1 and is thought to trim the poly(A) tail to the appropriate length. After export, Pab1 interacts with the translational machinery to stimulate translation initiation (Sachs 1990; Kessler and Sachs 1998).
Here, we report the results of several experiments conducted to investigate the roles that coupling between 3′ processing and mRNA export. Pab1 shuttles and is able to leave the nucleus by two routes, an exportin-1-dependent/mRNA-independent mechanism and by association with mRNA. It contains a nonessential, nuclear export signal (NES) near its N terminus, and information for nuclear import in its C-terminal region. Although PAB1 is essential, deletion of any one of several yeast genes bypasses the need for Pab1. The existence of these bypass suppressors permits studies of mRNA biogenesis in the absence of Pab1. We report that the absence of Pab1 leads to accumulation of polyadenylated mRNAs at sites of transcription in multiple bypass suppressor strains. Accumulation requires Rrp6; furthermore, deletion of RRP6 suppresses deletion of PAB1. Accumulation at sites of transcription also occurs in strains lacking subunits of the PAN nuclease. This indicates that the final steps in mRNA biogenesis, loading of Pab1 onto poly(A) tails, and trimming of tails to their proper length, overcome the retention function of the nuclear exosome and may actively contribute to release of mRNAs from sites of transcription.
Results
Pab1 shuttles in an Xpo1/Gsp1-dependent manner, and shuttling does not require ongoing mRNA production
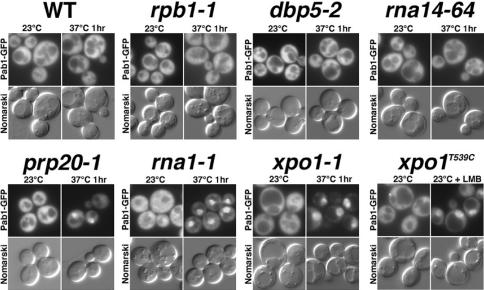
To investigate the nucleocytoplasmic trafficking of yeast Pab1, we prepared a functional Pab1–GFP fusion and examined the location of Pab1–GFP in wild-type cells and in several mutants defective for nucleocytoplasmic transport (Fig. 1). Pab1–GFP was detected primarily in the cytoplasm of wild-type cells. In cells carrying temperature-sensitive (ts) or leptomycin B-sensitive alleles of Xpo1, Pab1–GFP accumulated in nuclei under nonpermissive conditions. This indicates that Pab1 shuttles in an exportin-1-dependent manner. Consistent with the dependence of Pab1 shuttling on the protein export receptor Xpo1, Pab1–GFP accumulated rapidly in nuclei of cells carrying the prp20-1 and rna1-1 ts alleles. Prp20 and Rna1 are the guanine nucleotide exchange factor and GTPase activating protein, respectively, for Ran (Gsp1 in yeast), the small G protein that interacts directly with all karyopherins, including Xpo1, and is required for nucleocytoplasmic transport of most proteins and RNAs (for review, see Weis 2002).
Figure 1.
Pab1 shuttles in a Gsp1–GTP/Xpo1-dependent manner. Pab1–GFP localization was examined in several yeast strains with defects affecting RNA metabolism.
We also examined the distribution of Pab1 in additional strains carrying ts mutations affecting mRNA synthesis, processing, and export. We saw no change in the distribution of Pab1–GFP in rpb1-1 cells carrying a ts allele affecting the largest subunit of pol II, indicating that the bulk of Pab1–GFP does not depend on mRNA transport for export. Among mRNA export mutants, Pab1–GFP remained predominantly cytoplasmic in all of those examined, including mex67-5, mex67-6, mtr2-26, sub2-85, yra1-1, and rat8-2/dbp5-2 (encoding mutant mRNA export factors), in rat9-1/nup85-1, rat7-1/nup159-1, nup49-313, and rat2Δ/nup120Δ (encoding mutant nucleoporins important for mRNA export), or in rna14-63, rna15-58, or pcf11-1 (encoding mutant 3′-processing factors) (Fig. 1; our unpublished data).
Pab1 did not accumulate in 100% of cells when exportin 1-dependent protein export was blocked. Typically, only 40%–50% of cells showed complete mislocalization. In contrast, all of a model export cargo of Xpo1, NLS–NES–GFP2, or a shuttling mRNA export factor, Rat8–GFP, rapidly accumulates in the nuclei of all xpo1 mutant cells (Stade et al. 1997; Snay-Hodge et al. 1998). The absolute fraction of xpo1-1 cells accumulating Pab1–GFP at the nonpermissive temperature was greatly reduced as cells approached the late-log or stationary phase of growth (our unpublished data). These observations suggest that import of Pab1 may be regulated and sensitive to nutrient conditions.
Pab1's N terminus contains a leucine-rich nuclear export signal and its C-terminal region contains information for nuclear entry
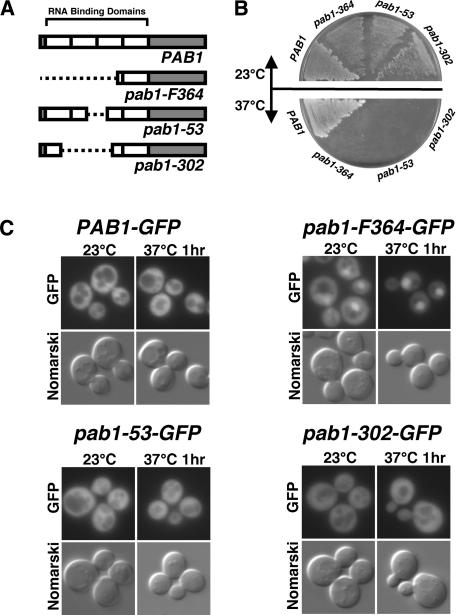
In addition to tagging wild-type Pab1 with GFP, we also tagged several PAB1 deletion mutants. Pab1 is a highly modular protein that contains a short N-terminal region, four evolutionarily conserved RNA recognition motifs (RRMs), and a C-terminal domain (Fig. 2A; Sachs et al. 1986; Sachs and Davis 1989). The mutants studied contain deletions of the N-terminal region or of different portions of the RNA-binding domains (RRMs) (Fig. 2A). Pab1-53 (lacking portions of RRM2 and RRM3), and Pab1-F364 (lacking the N-terminal domain, all of RRM1 and RRM2, and part of RRM3) are defective in several aspects of mRNA metabolism including poly(A) tail length control and mRNA stability (Sachs and Davis 1989; Morrissey et al. 1999). We constructed the pab1-302 allele, encoding a protein that lacks part of RRM 1 and RRM 3, and all of RRM2. Strains carrying the untagged alleles of pab1-53, pab1-364, and pab1-302 supported relatively robust growth at 23°C, but did not grow at 37°C (Fig. 2B).
Figure 2.
Localization of GFP fusions to several mutant forms of Pab1. (A) Schematic drawing of the domain organization of wild-type and several mutant forms of Pab1. (B) Growth properties of wild-type and mutant strains at 23°C and 37°C. (C) Distribution of wild-type Pab1–GFP (CMHy250) and GFP-tagged mutant Pab1 fusions (pab1-52–GFP [CMHy280], pab1-F364–GFP [CMHy281], and pab1-302–GFP [CMHy282]) at 23°C and following a 1-h shift to 37°C. Staining with DAPI confirmed that this accumulation is nuclear (data not shown).
Mutant Pab1–GFP constructs were transformed into wild-type yeast cells and the localization of each fusion protein was observed at 30°C and 37°C. Both Pab1-53–GFP and Pab1-302–GFP were predominantly cytoplasmic at both temperatures, similar to wild-type Pab1–GFP (Fig. 2C). In contrast, there was clear nuclear accumulation of Pab1-F364–GFP at both 23°C and 37°C (Fig. 2C). This indicates that the portion of Pab1 missing from Pab1-F364 either contains information for export, or alters the conformation of the remaining portion in a way that decreases its ability to be exported. The data also indicate that sequences within RRM4 and the C terminus have sufficient information for nuclear import of Pab1.
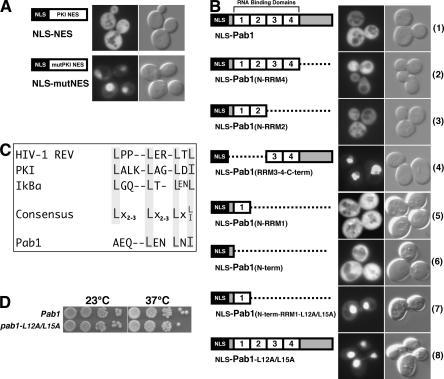
To determine whether Pab1 contained an active NES, we used an approach described by Stade et al. (1997) and examined the distribution of constructs containing GFP (some contain two GFPs to prevent diffusion through NPCs), the strong SV40 large T antigen nuclear localization signal (NLS), and either a wild-type NES or a mutant NES (nes). The construct with a wild-type NES (NLS–NES–GFP2) was found in both the nucleus and cytoplasm, reflecting active competition between the NLS and NES (Fig. 3A, top). In contrast, mutation of this NES resulted in constitutive nuclear localization (NLS–nes–GFP2) (Fig. 3A; Stade et al. 1997).
Figure 3.
Pab1 contains an active nuclear export signal (NES) near its N terminus. (A) Localization of NLS–NES–GFP2 and NLS–nes–GFP2 (mutant NES). (B) Schematic drawing of the domain organization of wild-type and several mutant forms of Pab1 and the distribution of GFP fusions to mutant Pab1s at 23°C. The mutant at the bottom encodes Pab1 carrying the L12A and L15A substitutions. (C) Alignment of previously characterized nuclear export signals and the putative NES from Pab1. (D) Growth behavior of a wild-type strain and one containing PAB1–L12A/L15A grown at 23°C or 37°C.
We replaced the NES and one of the GFPs with information encoding wild-type and mutant Pab1s. Insertion of the entire coding region into this reporter (NLS–Pab1–GFP) resulted in primarily cytoplasmic distribution identical to that of Pab1–GFP (Figs. 2C, 3B, line 1). This indicates that the export information contained within Pab1 functions well even when an NLS is fused to its N terminus. Deletion of either RRM3 + RRM4 + C terminus (Fig. 3B, line 3) or the C terminus alone (Fig. 3B, line 2) did not affect distribution, and indicates that a functional NES is located in the N-terminal portion of Pab1. Nuclear accumulation of a fusion of GFP and an NLS to RRM3–RRM4–C terminus (Fig. 3B, line 4) confirmed that sequences within the N-terminal half are required for Pab1 export.
To define the Pab1 NES more precisely, we fused either the short N-terminal domain, or this domain and RRM1 to the NLS and GFP. Both fusions were found throughout the cell (Fig. 3B, lines 5 and 6), indicating that information in the N-terminal 39 amino acids is sufficient for export. This region contains a segment similar to known leucine-rich NESs (Fig. 3C; Macara 2001). In the presence of wild-type Pab1, mutation of L12 and L15 to alanine (L12A/L15A) led to nuclear accumulation, in the context of both the N-terminal 112 amino acids (Fig. 3B, line 7) and full-length Pab1 (Pab1–L12A/L15A) (Fig. 3B, line 8). We conclude that Pab1 contains information for export at its N terminus. Interestingly, in a strain expressing only Pab1–L12A/L15A, the protein was found in both nuclear and cytoplasmic compartments (data not shown). Both cell growth (Fig. 3D) and mRNA export (our unpublished data) were indistinguishable from what was observed in wild-type cells. This indicates that the NES is not essential.
Overexpression of Rat8/Dbp5 corrects mislocalization of Pab1 in xpo1-1 cells
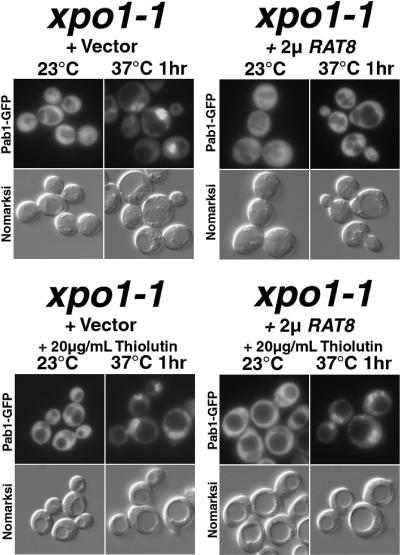
Mutations affecting Xpo1 prevent the efficient export of NES-containing proteins, large ribosomal subunits, and mRNA transcripts (Stade et al. 1997; Ho et al. 2000; Gadal et al. 2001), although the defect in mRNA export is thought to be indirect (Pasquinelli et al. 1997; Weis 1998; Neville and Rosbash 1999). Overexpression of the DEAD-box protein, Rat8/Dbp5, suppresses both the mRNA export and 3′-processing defects of xpo1-1 cells (Hodge et al. 1999; Hammell et al. 2002), but not the general block to protein export. When Rat8/Dbp5 was overexpressed in xpo1-1 cells, nuclear accumulation of Pab1 was suppressed (Fig. 4). When these cells were preincubated with thiolutin (20 μg/mL) to inhibit transcription, overexpression of Rat8/Dbp5 no longer corrected the nuclear accumulation phenotype of Pab1 (Fig. 4). This indicates that ongoing transcription is required for Rat8/Dbp5 overexpression to prevent nuclear accumulation of Pab1–GFP and suggests that overexpression of Rat8/Dbp5 allows export of Pab1 in an Xpo1-independent manner, but dependent upon ongoing mRNA production and export. We conclude that Pab1 can exit the nucleus by at least two routes, as a cargo (direct or indirect) for Xpo1 or with mRNA.
Figure 4.
Overexpression of Rat8/Dbp5 suppresses mislocalization of Pab1–GFP in xpo1-1 cells shifted to 37°C. (Top) xpo1-1 cells overexpressing Rat8. (Bottom) Thiolutin (20 μg/mL) was added to xpo1-1 cells; after 20 min, cells were shifted to 37°C.
PAB1 interacts genetically with RRP6, encoding a component of the nuclear exosome
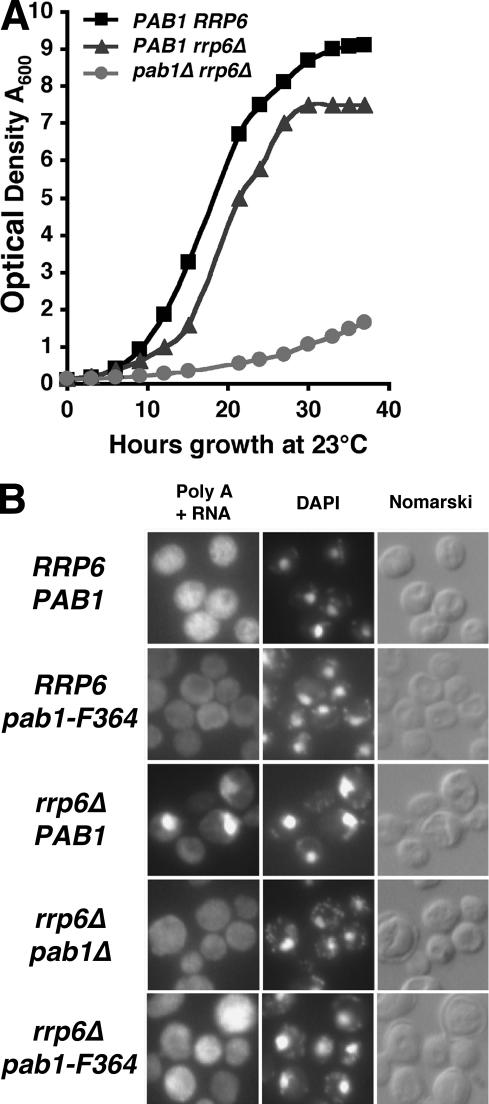
The 3′-processing machinery is thought to load Pab1 onto the poly(A) tail during processing. This loading may be aberrant in strains carrying mutations affecting 3′-processing factors. Similarly, loading of Pab1 may be defective in strains with hyperpolyadenylated tails, since one function of Pab1 is to control the trimming of the poly(A) tail to its normal length following initial polymerization of a slightly longer poly(A) tail (Sachs and Davis 1989; Caponigro and Parker 1995; Morrissey et al. 1999). mRNAs that are not exported are thought to be targets for the nuclear exosome. Therefore, we crossed strains carrying various mutant alleles of PAB1 with an rrp6Δ strain to see how the lack of Rrp6 would affect these mutants. Interestingly, we found that the pab1-55 allele (deletion of C terminus) (Morrissey et al. 1999) partially suppressed the growth defect of rrp6Δ cells at 37°C, although growth was very slow (our unpublished data). In no cases was there synthetic lethality. Therefore, we constructed a diploid carrying chromosomal disruptions of both RRP6 and PAB1. Haploids containing these two deletions (and a PAB1URA3CEN plasmid) were grown in nonselective medium and plated on medium containing 5-fluoroorotic acid (5-FOA) to select for cells that are able to loose the PAB1URA3 plasmid. pab1Δrrp6Δ colonies were isolated from 5-FOA plates, indicating that deletion of RRP6 bypassed the need for Pab1. Figure 5A shows the growth rates of pab1Δ, rrp6Δ, and pab1Δrrp6Δ cells. Although viable, pab1Δrrp6Δ cells grew slowly (doubling time of ∼10 h) as compared with either a wild-type or rrp6Δ strain (doubling times of 3 and 3.5 h, respectively). Interestingly, suppression can occur in both directions; deletion of rrp6 suppresses deletion of pab at 23°C and a C-terminally truncated Pab1 partially suppresses rrp6Δ at 37°C.
Figure 5.
(A) Growth curves for wild-type (FY86), rrp6Δ (CMHy250), and pab1Δrrp6Δ (CHMy251) cells in synthetic medium at 23°C. (B) Depletion of Rrp6 results in the nuclear accumulation of mRNA. Wild-type or mutant cells were grown to mid-log phase at 23°C, then fixed, permeabilized, and probed with an oligo(dT50) probe to localize poly(A) RNAs. The poly A [oligo(dT)], DAPI, and Normarski images of the same fields are shown. Shown are results using strains FY86 (PAB1 RRP6), EDy208 (pab1-F364), CMHy250 (rrp6Δ), CMHy251 (rrp6Δpab1Δ), and CMHy255 (rrp6Δpab1-F364).
We also examined mRNA export in rrp6Δ and pab1Δrrp6Δ cells. Approximately 60%–70% of cells lacking Rrp6 showed strong nuclear accumulation of polyadenylated RNA, and in cells showing this, the signal intensity was quite strong (Fig. 5B). Combining either a deletion of PAB1 or the pab1-F364 mutation (see Fig. 2A) with deletion of RRP6 largely overcame the poly(A) nuclear accumulation phenotype (Fig. 5B).
Deletion of PAB1 results in inefficient release of mRNAs from sites of transcription
If Pab1 were important for mRNA export, one might expect to see nuclear accumulation of mRNA when Pab1 is depleted or at 37°C in strains with ts alleles of PAB1. No nuclear accumulation of mRNA was detected following depletion of Pab1 (Kadowaki et al. 1994a) or at 37°C in strains carrying various ts pab1 alleles (D.C. Amberg, C.M. Hammell, and C.N. Cole, unpubl.). In these experiments, very little polyadenylated mRNA can be detected by in situ hybridization, and this presumably reflects rapid turnover of mRNA in cells lacking Pab1 (data not shown). One function of Pab1 is to protect mRNA from the endogenous turnover machinery. To see whether mRNA export defects resulted from absence of Pab1, we examined production and localization of the stress-inducible SSA4 transcript (encoding Hsp70) in several strains. Because pab1Δ cells are not viable, we examined pab1Δ cells carrying a disruption of SPB8, RPL39, or PAT1, known bypass suppressors of pab1Δ (Boeck et al. 1998; Bonnerot et al. 2000; Wyers et al. 2000). Spb8 and Pat1 participate in cytoplasmic mRNA turnover, and their absence most likely increases the overall level of mRNA enough to allow slow growth in cells lacking Pab1. Rpl39 is a component of the large ribosomal subunit. Cells were grown at 23°C, shifted to 42°C while in logarithmic growth, and analyzed by in situ hybridization to localize SSA4 mRNA.
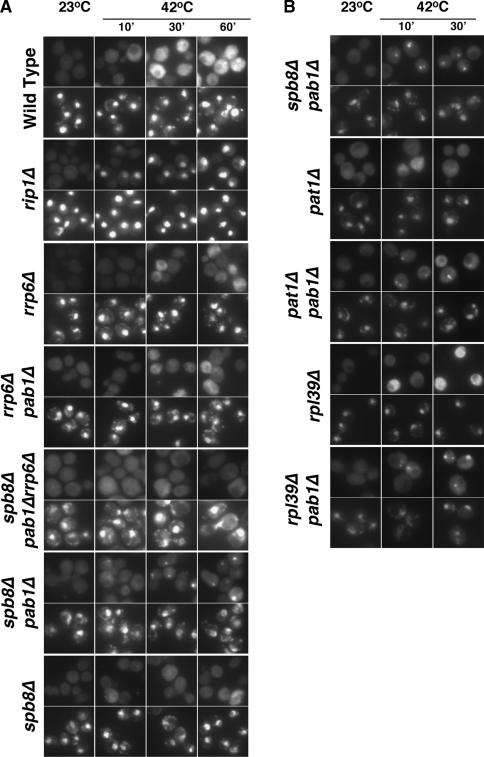
As expected, SSA4 mRNA was induced rapidly and exported efficiently in wild-type cells. Within 15 min, SSA4 mRNA can be seen throughout these cells (Fig. 6A). Cells lacking the nucleoporin, Rip1p/Nup42p, are defective for export of all RNA following heat shock (Saavedra et al. 1997). As previously reported by others (Hilleren and Parker 2001; Hilleren et al. 2001; Jensen et al. 2001a,b), SSA4 mRNA could be detected in bright intranuclear foci within 15 min of the temperature shift. Interestingly, in spb8Δpab1Δ, pat1Δpab1Δ, and rpl39Δpab1Δ cells (Fig. 6B), SSA4 mRNA was also detected in bright intranuclear foci, whereas SSA4 mRNA was localized throughout the cell in spb8Δ, pat1Δ, and rpl39Δ cells. The intranuclear foci seen in rip1Δ, spb8Δpab1Δ, pat1Δpab1Δ, and rpl39Δpab1Δ cells persisted for at least 1 h (data not shown). Intranuclear foci were seen in nearly 100% of rip1Δ and spb8Δpab1Δ cells, in ∼50%–60% of pat1Δpab1Δ cells, and in ∼25%–30% of rpl39Δpab1Δ cells.
Figure 6.
Accumulation of SSA4 mRNA in wild-type and several mutant yeast strains. Cells in exponential growth (OD600 0.2–0.35) were shifted from room temperature to 42°C (heat shock) for the times shown, and the distribution of SSA4 mRNA determined by in situ hybridization. (A) (Wild-type) FY86; (rip1Δ) CHy120; (rrp6Δ) CMHy250; (rrp6Δpab1Δ) CHy251; (spb8Δpab1 Δrrp6Δ) EDy201; (spb8Δpab1Δ) yAS2315; (spb8Δ) yAS2324. (B) (spb8Δpab1Δ) yAS2315; (pat1Δ) EDy190; (pat1Δpab1Δ) EDy196; (rpl39Δ) EDy191; (rpl39Δpab1Δ) EDy200. For each strain, the top line of images shows SSA4 localization, and the bottom line of images shows the same cells stained with DAPI, which binds DNA and shows the location of the nucleus.
If these foci are sites of transcription, they would be expected to be absent if Rrp6 were also absent, because rrp6Δ cells lack the retention mechanism (Hilleren et al. 2001; Jensen et al. 2001a). To examine the dependence of nuclear foci in spb8Δpab1Δ cells on the exosome, we constructed a spb8Δpab1Δrrp6Δ strain. Consistent with these previous studies, SSA4 transcripts did not accumulate in foci in these cells (Fig. 6A). Similarly, we saw no accumulation in intranuclear foci in pab1Δrrp6Δ cells (Fig. 6A) or in pat1Δpab1Δrrp6Δ cells (data not shown).
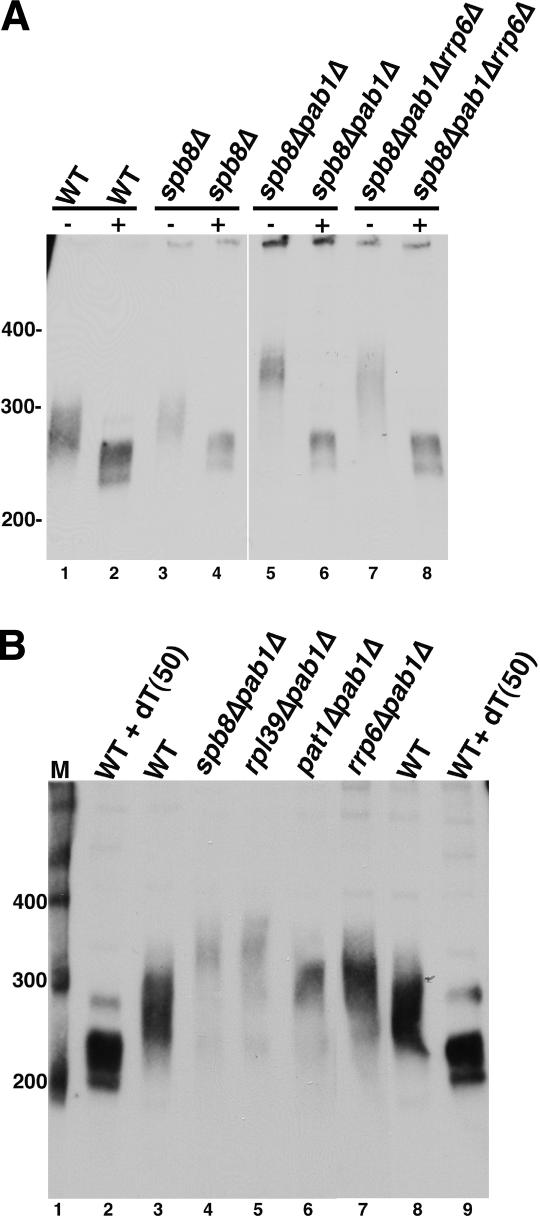
In many strains where bright intranuclear foci of SSA4 mRNA were detected, SSA4 mRNAs have unusually long poly(A) tails. We analyzed the poly(A) tail lengths of SSA4 mRNAs in wild-type and mutant strains using an RNase H cleavage assay. In spb8Δ cells, poly(A) tails were, on average, ∼20 nucleotides (nt) longer than in wild-type cells (Fig. 7A, cf. lanes 1 and 3). Even longer poly(A) tails (∼40 nt longer than in wild-type cells) were found in spb8Δpab1Δ cells (Fig. 7A, lane 5). These tails are about the same size as those found on SSA4 mRNAs in rip1Δ cells and in other strains where heat-shock mRNAs accumulated at sites of transcription. In spb8Δpab1Δrrp6Δ cells, tails averaged ∼10 nt shorter than in spb8Δpab1Δ cells (Fig. 7A, lane 7). This could reflect release of these transcripts from retention sites, followed by their partial degradation by nuclear deadenylation complexes (Tucker et al. 2001, 2002). The poly(A) tails of SSA4 mRNA in rpl39Δpab1Δ cells were approximately the same length as those in spb8Δpab1Δ cells (Fig. 7B, lanes 4,5), while poly(A) tails in pat1Δpab1Δ cells were approximately the same length as in wild-type cells. In all strains, the site of 3′ processing was the same (data not shown). This indicates that poly(A) tail length alone does not explain accumulation of SSA4 mRNAs at sites of transcription, since mRNAs found in intranuclear foci can have very long tails (rip1Δ, spb8Δpab1Δ in Fig. 7; nab2Δ cells, (Hector et al. 2002), tails of normal length (pat1Δpab1Δ), or no poly(A) tail (pap1-1) (Hilleren et al. 2001; Hammell et al. 2002).
Figure 7.
Hyperpolyadenylation of SSA4 mRNA in PAB1 null bypass strains. (A) Comparison of SSA4 mRNA poly(A) tail lengths in spb8Δ mutant strains. SSA4 mRNA RNase H assays were performed on 5 μg of total RNA from each strain either without (–) (lanes 1,3,5,7), or with (+) (lanes 2,4,6,8) 400 ng oligo (dT50). (Lanes 1,2) FY86 (WT). (Lanes 3,4) yAS2324 (spb8Δ). (Lanes 5,6) yAS2315 (spb8Δpab1Δ). (Lanes 7,8) yEDy201 (spb8Δpab1Δrrp6Δ). Lanes 1–4 represent a shorter exposure time as compared with lanes 5–8. (B) Comparison of SSA4 mRNA poly(A) tail lengths in pab1Δ bypass suppressor strains. SSA4 RNase H assays were performed on 5 μg of total RNA extracted from each strain after shifting to 42°C for 15 min. Strains used were yAS2315 (spb8Δpab1Δ, lane 3), EDy200 (rpl39Δpab1Δ, lane 5), EDy196 (pat1Δpab1Δ, lane 6), CMHy251 (rrp6Δpab1Δ, lane 7), FY86 (WT, lanes 3,8), and FY86 (WT) incubated with 400 ng oligo (dT50) prior to RNase digestion (lanes 2,9).
The PAN nuclease complex is required for efficient release of mRNAs from sites of transcription
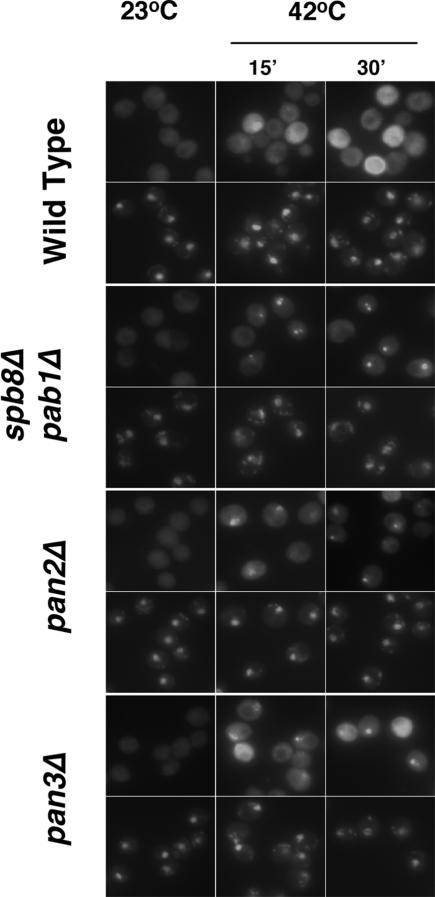
The final known nuclear step of mRNA biogenesis is trimming of poly(A) tails from their synthesized length to the slightly shorter length found on newly exported mRNAs. Pab1 recruits the multisubunit PAN nuclease complex, which is thought to perform this trimming (Brown et al. 1996; Brown and Sachs 1998). To determine whether the PAN nuclease plays a role in overcoming nuclear retention of mRNAs, we examined SSA4 mRNA distribution in strains lacking either of two nonessential subunits of PAN, Pan2 and Pan3. Absence of either resulted in accumulation of SSA4 mRNA in bright intranuclear foci (Fig. 8). Accumulation was detected in ∼50% of these cells. This indicates that quality control is monitored through this very late stage of nuclear mRNA biogenesis and that the PAN nuclease must perform its function before mRNAs can be released efficiently from transcription sites.
Figure 8.
Accumulation of SSA4 mRNA in pan2Δ and pan3Δ mutants at 42°C. pan2Δ (EDy268) and pan3Δ (EDy269) cells in exponential growth (OD600 0.2–0.35) were shifted from room temperature to 42°C for 15 or 30 min. The distribution of SSA4 mRNA was determined by in situ hybridization and compared with that observed in FY86 (WT) and yAS2315 (spb8Δpab1Δ) cells under identical conditions. For each strain, the top line of images shows SSA4 localization and the bottom line of images shows the same cells stained with DAPI, which binds DNA and shows the location of the nucleus.
Discussion
With few exceptions, mature mRNAs are exported from the nucleus only after they have completed successfully all processing steps (for review, see Jensen et al. 2003). How the cell determines which mRNAs are correctly processed and packaged, and therefore should be exported, is not known. Recent studies indicate that export-incompetent mRNAs are actively retained at or near their sites of transcription and subsequently degraded by the nuclear exosome (Hilleren et al. 2001; Jensen et al. 2001a,b). Presumably, export-competent mRNPs differ structurally or biochemically from those that are retained, and this allows them to escape from retention sites and avoid exosome-mediated degradation. Studies presented here indicate that poly(A)-binding protein, Pab1, and the poly(A) nuclease, PAN, play key roles during the final steps of mRNA biogenesis, allowing release of mRNAs from retention sites and their export to the cytoplasm.
Pab1 can leave the nucleus by both RNA-dependent and RNA-independent routes
Pab1 shuttles between the nucleus and the cytoplasm, and this shuttling is dependent on the exportin, Xpo1 (Fig. 1). Pabp, the Pab1 homolog from Schizosaccharomyces pombe, is known to shuttle and play a role in mRNA export (Thakurta et al. 2002); however, S. pombe Pabp is not essential, raising the possibility that S. pombe Pabp plays roles somewhat different from those of S. cerevisiae Pab1. Pab1 contains four RRMs. Deletion of a majority of these (e.g., pab1-F364) results in temperature sensitivity, partial mislocalization of Pab1-F364 to the nucleus at 30°C, and a severe mislocalization defect at 37°C (Fig. 2). Through further mutational analysis, we defined a leucine-rich NES near the N terminus (Fig. 3). Interestingly, this NES is not essential for viability, and cells where Pab1 without an NES is the only form of Pab1 present grow at near wild-type rates and without apparent defects in mRNA biogenesis.
We previously reported that Xpo1 interacts with Pab1 in the two-hybrid system (Hammell et al. 2002), but did not demonstrate a direct interaction. Karsten Weis's laboratory found that recombinant Pab1 and Xpo1 could be coprecipitated in a complex with RanGTP, indicate that Pab1 binds Xpo1 directly (K. Weis, pers. comm.). Several yeast mRNA-binding proteins (e.g., Npl3) can leave the nucleus solely when bound to mRNA and accumulate in nuclei when mRNA synthesis or export are prevented (Lee et al. 1996). Although Pab1 can leave the nucleus bound to poly(A) tails, it does not accumulate in nuclei when RNA export is blocked. We conclude that Pab1 can exit the nucleus by at least two mechanisms, associated with transcripts that have been 3′ processed correctly or as a cargo of Xpo1.
How can fully and accurately processed mRNAs be distinguished from those that should not be exported?
Since pre-mRNAs are capped at their 5′ ends very early during transcription (Jove and Manley 1984; Rasmussen and Lis 1993), a cap cannot allow discrimination between export-competent and export-incompetent RNAs. In yeast, most mRNAs lack introns. Therefore, splicing cannot provide an essential mark for export. Although most metazoan mRNAs are spliced, and splicing results in deposition of the exon junction complex (EJC) near splice junctions (LeHir et al. 2000), the EJC cannot be the sole key to export competence, since it will be deposited following each successful splicing event; yet, failure to complete even one splicing reaction generally results in retention. Most likely, mRNAs that do not complete splicing correctly are physically retained by association with spliceosomes and degraded.
Therefore, two other features of mRNAs, packaging into mRNPs and polyadenylation, have been the focus of studies to define the mechanisms by which export competence is achieved and signaled. Pre-mRNAs are packaged cotranscriptionally into mRNPs by associating with multiple mRNA-binding proteins and additional factors (McCracken et al. 1997; Komarnitsky et al. 2000; Lei et al. 2001; Lei and Silver 2002). Since many of these are added early during transcription, their mere presence on mRNA cannot be an essential signal for export competence. However, in strains carrying mutations affecting some mRNP proteins (e.g., Npl3, Nab2, Yra1, Sub2), polyadenylated RNA accumulates in nuclei (for review, see Jensen et al. 2003). Efficient recruitment of Npl3 to mRNAs requires that Npl3 be dephosphorylated by the nuclear protein phosphatase, Glc7. In cells where Npl3 cannot be dephosphorylated, there is reduced recruitment of Mex67 to the mRNA (Gilbert and Guthrie 2004). Mutations of YRA1 and SUB2 are also believed to reduce or prevent recruitment of Mex67 (Strasser and Hurt 2000, 2001). Thus, defects in one mRNA-binding protein appear to result in an overall change in the protein composition of the mRNP and probably also in its conformation. While export may require a particular mRNP configuration, we have little idea of what this means or how the cell examines mRNPs for having an exportable configuration.
We and others have shown that a poly(A) tail generated by the normal 3′-processing machinery is necessary, but not sufficient, for mRNA export (Brodsky and Silver 2000; Dower and Rosbash 2002; Hammell et al. 2002). Poly(A) tails are added through the action of macromolecular cleavage and polyadenylation complexes containing at least 21 different polypeptides in yeast (for review, see Zhao et al. 1999; Nedea et al. 2003). mRNAs with aberrant 3′ processing and 3′-end features accumulate in intranuclear foci (Hilleren et al. 2001; Jensen et al. 2001b; Dower and Rosbash 2002). Retention depends on the exosome (Hilleren et al. 2001), indicating that a mechanism exists either to release correctly processed mRNAs or to prevent them from associating with retention machinery.
mRNAs with hyperpolyadenylated tails (>100 nt) accumulate at transcription sites in several mutant yeast strains including mex67-5 and its heterodimerization partner, mtr2-1 (Kadowaki et al. 1994b; Hilleren and Parker 2001; Hilleren et al. 2001; Jensen et al. 2001b), suggesting that defects in mRNP organization or composition can affect 3′ processing. However, the poly(A) tails formed on SSA4 mRNA in yra1-1 and sub2-85 cells have approximately the same length as in wild-type cells (Hammell et al. 2002), yet the mRNP composition is also altered by these mutations. This suggests that 3′ processing is coordinated with mRNP formation, but the mechanism of coordination is uncertain. Another important question is what underlies production of hyperpolyadenylated mRNAs, since they are found in a diverse set of mutant strains including some affecting nucleoporins, the effectors of the Ran GTPase, and the DEAD-box protein, Dbp5.
Synthesis of the poly(A) tail is performed by poly(A) polymerase, which produces tails of ∼70–90 nt in yeast and 200–300 in metazoans. In all cases, the poly(A) tail is bound by multiple molecules of a poly(A)-binding protein (Pab1 in yeast and PABP2 in animals) (for review, see Mangus et al. 2003). In yeast, Pab1 recruits the PAN nuclease, which trims the tail to the length found on newly exported RNAs (55–72 nt) (Lowell et al. 1992; Brown and Sachs 1998; Mangus et al. 2004a,b).
While our studies support a model wherein PAN plays an important role in release of export-competent mRNPs from sites of transcription, we cannot distinguish between two possible mechanisms by which PAN might mediate release. The retention machinery might monitor the length of the poly(A) tail or the structure of the nucleoprotein complex present on the poly(A) tail. By one mechanism, Pab1 or the poly(A)/Pab1 ribonucleoprotein organization at the 3′ end of the mRNP could function as a release factor with the proper organization or structure requiring trimming of the poly(A) tail by PAN. Alternatively, PAN, a subunit of PAN, or a protein recruited by PAN could be a release factor that interacts with the retention machinery, thereby bringing about release of the mature mRNP. By this mechanism, an enzymatically inactive, but structurally normal PAN complex might be able to mediate release.
A second yeast protein that can bind poly(A) is Nab2 (Anderson et al. 1993). Since depletion or mutation of Nab2 also result in production of hyperpolyadenylated mRNAs and their accumulation at sites of transcription, Nab2 is also required for release. Overexpression of PAB1 (approximately twofold) suppressed the lethality of deleting NAB2 and allowed normal growth (Hector et al. 2002). Interestingly, the production of extended poly(A) tails was not suppressed, but mRNA export was restored, although nuclear accumulation of polyadenylated RNA still occurs. Hyperpolyadenylation was also observed in vitro using extracts lacking Pab1 (Amrani et al. 1997; Minvielle-Sebastia et al. 1997). Although hyperpolyadenylation was suppressed by adding recombinant Nab2 to the extract, the tails produced were longer than the 50–70-nt tails produced when Pab1 was present (Hector et al. 2002). This suggests that Nab2 helps to load Pab1 onto the poly(A) tail, allowing recruitment of PAN.
Antagonistic roles for Pab1 and Rrp6
Several bypass suppressors of pab1Δ have been identified (Sachs and Davis 1989; Caponigro and Parker 1995; Hatfield et al. 1996; Boeck et al. 1998; Mangus et al. 1998; Bonnerot et al. 2000; Winstall et al. 2000). Most are deletions or ts alleles of genes that encode proteins involved in cytoplasmic mRNA turnover (including Spb8, Dcp1, Mrt1, Mrt3), or which are part of 60S ribosomal subunits (Rpl39). Pab1 plays an important role in mRNA stability. Because there is much less polyadenylated mRNA in pab1 mutant strains, it has been difficult to determine how Pab1 affects mRNA export.
In these studies, we identified a disruption of RRP6 as a bypass suppressor of pab1Δ (Fig. 6). We suggest that Pab1 and Rrp6 play antagonistic roles in mRNA metabolism. Because Pab1 is essential, we used the pab1Δspb8Δ, pab1Δpat1Δ, and pab1Δrpl39Δ strains and detected induced SSA4 mRNA in intranuclear foci (Fig. 6). These foci were absent in spb8Δ, pat1Δ, and rpl39Δ cells, and were not detected in absence of Rrp6. This indicates that absence of Pab1 compromises release of transcripts from sites of transcription and, as shown in other studies (Hilleren and Parker 2001; Hilleren et al. 2001; Jensen et al. 2001b), Rrp6 is required for retention. Since we also saw retention at sites of transcription in strains that lack PAN nuclease, we propose that correctly loading Pab1 onto the poly(A) tail and recruitment of the PAN nuclease completes accurate 3′ processing and releases mature mRNAs for export. We hypothesize that mRNAs with hyperpolyadenylated tails, very short poly(A) tails, and no poly(A) tail have in common the absence of properly loaded Pab1, which leads to a failure to recruit PAN.
The nuclear retention of transcripts in pab1Δ bypass suppressor cells can explain previously noted alterations in mRNA metabolism in yeast strains carrying mutations or a deletion of PAB1. The absence of Pab1 in such double mutants apparently prevents transcripts from efficiently entering the normal mRNA decay pathway (Caponigro and Parker 1995; Morrissey et al. 1999). Pulse-chase analysis of mRNAs produced in wild-type cells indicates that mRNA degradation begins within 1–2 min after the initial pulse of transcription (Caponigro and Parker 1995). The brief time before an mRNA enters the degradation pathway implies that mRNA export normally occurs very rapidly after an mRNA is produced. When Pab1 was absent (e.g., in pab1Δxrn1Δ cells) there was a 15–20-min lag before degradation began, but mRNAs eventually entered the cytoplasmic decay pathway and were degraded with normal kinetics (Caponigro and Parker 1995).
rrp6Δ cells show very strong accumulation of poly(A) RNA in nuclei at 37°C. It was reported that polyadenylated snoRNAs were produced in exosome mutant cells; possibly, we are seeing both mRNAs and snoRNAs. This might indicate that Pab1 plays a role in snoRNA metabolism (van Hoof et al. 2000).
How might the exosome function to recognize defective mRNAs?
How the nuclear exosome retains and/or detects defective mRNAs is not known. The exosome is found in transcription domains (Andrulis et al. 2002; Libri et al. 2002; Zenklusen et al. 2002), and in yeast, it interacts genetically with poly(A) polymerase and mRNA-binding proteins (Burkard and Butler 2000). This positions it to degrade transcripts that remain in this vicinity. Any defect that prevents the movement of a transcript away from the transcription or processing regions would permit degradation by the exosome. Hilleren et al. (2001) and Libri et al. (2002) showed that defective mRNAs produced in specific mRNA export mutants are exported and translated if mutant cells lack Rrp6.
The emerging picture, on the basis of studies from many laboratories over many years, is that all nuclear events of mRNA biogenesis are linked to the mRNA export process. This coupling likely reflects the existence of an extremely large transcription–processing–surveillance–export complex. 3′-processing factors associate with transcribing pol II through the C-terminal domain (CTD) of the largest subunit of pol II in both yeast and metazoan cells (Barilla et al. 2001; Dichtl et al. 2002; Licatalosi et al. 2002). This positions them to recognize 3′-processing signals as soon as they are detected in the elongating mRNA. The dependence of transcription termination on proper 3′-processing reflects continued communication and physical association between pol II and the 3′-processing machinery. As long as contact is maintained among the 3′-processing machinery, mRNA, and pol II, the cleaved transcript will remain at the transcription/processing site.
Pab1 has been purified with CFIA, which mediates 3′ cleavage and polyadenylation (Amrani et al. 1997; Minvielle-Sebastia et al. 1997). Perhaps as one Pab1 is transferred to the poly(A) tail, additional Pab1 monomers are recruited by the 3′-processing machinery and loaded onto poly(A). This complex scenario may explain why so many proteins are required for proper 3′-end formation. These proteins need to bind pol II, recognize multiple signals in the RNA, direct cleavage to the proper site, recruit poly(A) polymerase, maintain contact and communication with the transcription complex, mediate the loading of multiple Pab1 molecules onto each poly(A) tail, recruit the poly(A)-dependent nuclease complex, and signal pol II that 3′ processing, polyadenylation, loading of Pab1, and trimming the poly(A) tail have all been completed correctly.
Materials and methods
Yeast strains and cell culture
The yeast strains and plasmids used in this study are listed in Supplementary Tables 1 and 2, respectively. Strains were cultured using standard methods (Sherman 1991), using rich (YPD) or defined (SC, synthetic complete) medium lacking the appropriate amino acids. Liquid and solid media included 2% dextrose, 2% galactose, or 2% raffinose as indicated. Transcriptional inhibition was achieved by incubating cells with thiolutin (final concentration of 20 μg/mL) for 20 min at room temperature prior to subsequent temperature shifts (Tipper 1973). Temperature sensitive (ts) mutants were grown at 23°C or 30°C (permissive temperatures) as indicated, or shifted to 37°C (nonpermissive temperature) for the indicated times. Genetic techniques were performed using standard methods (Sherman 1991).
Pab1 shuttling-assay plasmids
DNA fragments encoding the indicated amino acids of PAB1 were amplified by high fidelity PCR (Pfu-turbo, Stratagene) from pCMH035 with primers containing EcoRI (5′) and BamHI (3′) restriction sites. pKW430 (Stade et al. 1997) was digested with EcoRI and BamHI to remove the NES and one of the GFP open reading frames and replaced with EcoRI–BamHI fragments encoding different portions of PAB1. Mutagenesis of the Pab1 nuclear export signal (NES) was accomplished via the Quick-Change Site-Directed Mutagenesis Kit (Stratagene) with the following primers: NESmut1 and NESmut2 (see Supplementary Table 3). Changes in nucleotide sequences that result in L12>A and L15>A are indicated with lowercase letters.
Pab1 expression plasmids
The sequences of all primer pairs used in these studies are listed in Supplementary Table 3. Constructs expressed under the endogenous promoter pCMH144: (pab1-302 TRP1 CEN) allele was created by deletion of the HpaI–HpaI fragment of pCMH094. pCMH035 was constructed by PCR amplification of wild-type PAB1 (with primers containing PstI and BamHI ends) and GFP-(ADH1 3′UTR) (BamHI and PstI ends) from pFA6a–GFP-HIS3 (Puig et al. 2001). pCMH068, pCMH115, and pCMH145 were constructed in a similar manner by amplifying pab1-53 (Morrissey et al. 1999), pab1-F364 (Sachs and Davis 1989), and pab1302 open reading frames from their corresponding, untagged plasmids, and replacing the PstI–BamHI fragment of pCMH035, which contains the complete open reading frame of wild-type PAB1. Constructs expressed under the ADH1 promoter/3′-UTR were prepared as follows. The ADH1 5′ and 3′-untranslated regions (UTRs) were amplified from genomic DNA by PCR (Pfu-turbo; Stratagene) (Supplementary Table 3). The 5′-UTR was inserted into XhoI–ClaI sites of pRS313 (Sikorski and Hieter 1989), and the ADH1 3′-UTR (SpeI–SacI fragment), was subsequently subcloned into this vector to generate pED297. A full-length PAB1–GFP coding region was then amplified from pCMH035, digested with SmaI and XbaI, and inserted into between the Sma1 and SpeI sites of pED297 to generate pRS314ADH1–PAB1–GFP (pED325). The XhoI–SacI fragment from this plasmid was then subcloned between the XhoI and SacI sites of pRS315 (Sikorski and Hieter 1989) to produce pED327. Mutagenesis of the Pab1 NES was performed with primers NESmut1 and NESmut2, as described above, to create pED326 and pED328, respectively.
Construction of pab1Δrrp6Δ and other related yeast strains
A genomic disruption of RRP6 was created by transforming a PCR generated kanr::MX2 cassette into CMHy214 and selecting on G418 containing plates. Disruption was confirmed using PCR. The resulting strain (CMHy250) was grown in nonselective liquid medium and plated onto solid agar containing 5-fluoro-oritic acid (5-FOA) to select for cells lacking the covering URA3 PAB1–GFP plasmid, to produce CMHy251. To create strains CMHy253 through CMHy257, CMHy250 was transformed with the corresponding TRP1 CEN plasmid and plated on 5-FOA to obtain TRP+ ura3–cells.
In situ hybridization and microscopy
Detection of SSA4 mRNAs was performed as described previously (Saavedra et al. 1996). Fluorescence images were examined and photographed using an Axiophot microscope (Zeiss) and a CCD camera.
RNA methods
RNase H assays were performed essentially as described by Boeck et al. (1998). Briefly, cells transformed with pCH16 (SSA4-URA3-2μ) were grown to mid-log phase (OD600 =0.2–0.35) and then shifted to 42°C for 15 min before extracting total RNA.
SSA4 RNase H cleavage analysis was performed on 5 μg of total RNA, which was incubated for 1 h at 30°C with 300 ng of oligonucleotide SSA4-21 (CCTGGGGCACCACCTGCAGCTC CG) complementary to sequences in the 3′ end of the SSA4 open reading frame, and 0.25 U of RNase H (GIBCO-BRL) in RNase H buffer (20 mM Tris-HCl at pH 7.4, 10 mM MgCl2, 0.5 mM EDTA, 50 mM NaCl, 1 mM dithiothreitol, 30 μg/mL of BSA). RNase H cleaves SSA4 mRNA ∼250 nt from the cleavage and polyadenylation site. Some samples also contained 400 ng of oligo(dT50). The differences in sizes of digestion products with and without oligo(dT50) indicates the length of the poly(A) tail. Reactions were stopped by the addition of 1 vol of RNA loading buffer (95% formamide, 0.05% bromophenol blue, 0.05% xylene cyanol). RNA samples were then electrophoresed through 5% polyacylamide gels before being electroblotted onto a GeneScreen Plus membrane (NEN Life Science Products). To visualize SSA4 mRNA bands, membranes were probed with a SSA4 DIG-DNA 3′-PCR fragment (Saavedra et al. 1996).
Acknowledgments
We thank members of the Cole lab for valuable discussions and Catherine Heath for critical review of the manuscript. We thank Karsten Weis for discussions and for communicating results of his laboratory's work prior to publication. We thank Ed Hurt, Pam Silver, Alan Sachs, Arlen Johnson, and Karsten Weis for strains and plasmids. This research was supported by grants from the National Institutes of Health (GM33998) and the National Science Foundation (MCB-9983378). Core facilities essential for this work were supported by a comprehensive cancer center core grant (CA16038) to the Norris Cotton Cancer Center. C.M.H. was supported by a training grant from the National Cancer Institute (CA-09658).
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1267005.
References
- Allmang C., Kufel, J., Chanfreau, G., Mitchell, P., Petfalski, E., and Tollervey, D. 1999. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 18: 5399–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrani N., Minet, M., Le Gouar, M., Lacroute, F., and Wyers, F. 1997. Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol. Cell. Biol. 17: 3694–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J.T., Wilson, S.M., Datar, K.V., and Swanson, M.S. 1993. NAB2: A yeast nuclear polyadenylated RNA-binding protein essential for cell viability. Mol. Cell. Biol. 13: 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis E.D., Werner, J., Nazarian, A., Erdjument-Bromage, H., Tempst, P., and Lis, J.T. 2002. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature 420: 837–841. [DOI] [PubMed] [Google Scholar]
- Barilla D., Lee, B.A., and Proudfoot, N.J. 2001. Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 98: 445–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse C.E., Minvielle-Sebastia, L., Lee, B.A., Keller, W., and Proudfoot, N.J. 1998. Coupling termination of transcription to messenger RNA maturation in yeast. Science 280: 298–301. [DOI] [PubMed] [Google Scholar]
- Boeck R., Lapeyre, B., Brown, C.E., and Sachs, A.B. 1998. Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol. Cell. Biol. 18: 5062–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnerot C., Boeck, R., and Lapeyre, B. 2000. The two proteins Pat1p (Mrt1p) and Spb8p interact in vivo, are required for mRNA decay, and are functionally linked to Pab1p. Mol. Cell. Biol. 20: 5939–5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet-Antonelli C., Presutti, C., and Tollervey, D. 2000. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102: 765–775. [DOI] [PubMed] [Google Scholar]
- Briggs M.W., Burkard, K.T., and Butler, J.S. 1998. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J. Biol. Chem. 273: 13255–13263. [DOI] [PubMed] [Google Scholar]
- Brodsky A.S. and Silver, P.A. 2000. Pre-mRNA processing factors are required for nuclear export. RNA 6: 1737–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.E. and Sachs, A.B. 1998. Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol. Cell. Biol. 18: 6548–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.E., Tarun Jr., S.Z., Boeck, R., and Sachs, A.B. 1996. PAN3 encodes a subunit of the Pab1p-dependent poly(A) nuclease in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 5744–5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard K.T. and Butler, J.S. 2000. A nuclear 3′-5′ exonuclease involved in mRNA degradation interacts with Poly(A) polymerase and the hnRNA protein Npl3p. Mol. Cell. Biol. 20: 604–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo O. and Manley, J. 2003. Strange bedfellows: Polyadenylation factors at the promoter. Genes & Dev. 17: 1321–1327. [DOI] [PubMed] [Google Scholar]
- Caponigro G. and Parker, R. 1995. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes & Dev. 9: 2421–2432. [DOI] [PubMed] [Google Scholar]
- Coller J.M., Gray, N.K., and Wickens, M.P. 1998. mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation. Genes & Dev. 12: 3226–3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custodio N., Carmo-Fonseca, M., Geraghty, F., Pereira, H., Grosveld, F., and Antoniou, M. 1999. Inefficient processing impairs release of RNA from the site of transcription. EMBO J. 18: 2855–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl B., Blank, D., Sadowski, M., Hubner, W., Weiser, S., and Keller, W. 2002. Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination. EMBO J. 21: 4125–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower K. and Rosbash, M. 2002. T7 RNA polymerase-directed transcripts are processed in yeast and link 3′ end formation to mRNA nuclear export. RNA 8: 686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal O., Strauss, D., Kessl, J., Trumpower, B., Tollervey, D., and Hurt, E. 2001. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 21: 3405–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W. and Guthrie, C. 2004. The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Mol. Cell 13: 201–212. [DOI] [PubMed] [Google Scholar]
- Hammell C.M., Gross, S., Zenklusen, D., Heath, C.V., Stutz, F., Moore, C., and Cole, C.N. 2002. Coupling of termination, 3′ processing, and mRNA export. Mol. Cell. Biol. 22: 6441–6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield L., Beelman, C.A., Stevens, A., and Parker, R. 1996. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 5830–5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hector R.E., Nykamp, K.R., Dheur, S., Anderson, J.T., Non, P.J., Urbinati, C.R., Wilson, S.M., Minvielle-Sebastia, L., and Swanson, M.S. 2002. Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. EMBO J. 21: 1800–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P. and Parker, R. 2001. Defects in the mRNA export factors Rat7p, Gle1p, Mex67p, and Rat8p cause hyperadenylation during 3′-end formation of nascent transcripts. RNA 7: 753–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P., McCarthy, T., Rosbash, M., Parker, R., and Jensen, T.H. 2001. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413: 538–542. [DOI] [PubMed] [Google Scholar]
- Ho J.H., Kallstrom, G., and Johnson, A.W. 2000. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell. Biol. 151: 1057–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge C.A., Colot, H.V., Stafford, P., and Cole, C.N. 1999. Rat8p/Dbp5p is a shuttling transport factor that interacts with Rat7p/Nup159p and Gle1p and suppresses the mRNA export defect of xpo1-1 cells. EMBO J. 18: 5778–5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. and Carmichael, G.G. 1996. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol. Cell. Biol. 16: 1534–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen T.H., Boulay, J., Rosbash, M., and Libri, D. 2001a. The DECD box putative ATPase Sub2p is an early mRNA export factor. Curr. Biol. 11: 1711–1715. [DOI] [PubMed] [Google Scholar]
- Jensen T.H., Patricio, K., McCarthy, T., and Rosbash, M. 2001b. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol. Cell 7: 887–898. [DOI] [PubMed] [Google Scholar]
- Jensen T.H., Dower, K., Libri, D., and Rosbash, M. 2003. Early formation of mRNP: License for export or quality control? Mol. Cell 11: 1129–1138. [DOI] [PubMed] [Google Scholar]
- Jimeno S., Rondon, A.G., Luna, R., and Aguilera, A. 2002. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J. 21: 3526–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jove R. and Manley, J.L. 1984. In vitro transcription from the adenovirus 2 major late promoter utilizing templates truncated at promoter-proximal sites. J. Biol. Chem. 259: 8513–8521. [PubMed] [Google Scholar]
- Kadowaki T., Chen, S., Hitomi, M., Jacobs, E., Kumagai, C., Liang, S., Schneiter, R., Singleton, D., Wisniewska, J., and Tartakoff, A.M. 1994a. Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants. J. Cell. Biol. 126: 649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T., Hitomi, M., Chen, S., and Tartakoff, A.M. 1994b. Nuclear mRNA accumulation causes nucleolar fragmentation in yeast mtr2 mutant. Mol. Biol. Cell 5: 1253–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler S.H. and Sachs, A.B. 1998. RNA recognition motif 2 of yeast Pab1p is required for its functional interaction with eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 18: 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky P., Cho, E.J., and Buratowski, S. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes & Dev. 14: 2452–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.S., Henry, M., and Silver, P.A. 1996. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes & Dev. 10: 1233–1246. [DOI] [PubMed] [Google Scholar]
- LeHir H., Izaurralde, E., Maquat, L.E., and Moore, M.J. 2000. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J. 19: 6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei E.P. and Silver, P.A. 2002. Intron status and 3′-end formation control cotranscriptional export of mRNA. Genes & Dev. 16: 2761–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei E.P., Krebber, H., and Silver, P.A. 2001. Messenger RNAs are recruited for nuclear export during transcription. Genes & Dev. 15: 1771–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D., Dower, K., Boulay, J., Thomsen, R., Rosbash, M., and Jensen, T.H. 2002. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol. Cell. Biol. 22: 8254–8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi D.D., Geiger, G., Minet, M., Schroeder, S., Cilli, K., McNeil, J.B., and Bentley, D.L. 2002. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol. Cell 9: 1101–1111. [DOI] [PubMed] [Google Scholar]
- Lowell J.E., Rudner, D.Z., and Sachs, A.B. 1992. 3′-UTR-dependent deadenylation by the yeast poly(A) nuclease. Genes & Dev. 6: 2088–2099. [DOI] [PubMed] [Google Scholar]
- Macara I.G. 2001. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65: 570–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus D.A., Amrani, N., and Jacobson, A. 1998. Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation. Mol. Cell. Biol. 18: 7383–7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus D.A., Evans, M.C., and Jacobson, A. 2003. Poly(A)-binding proteins: Multifunctional scaffolds for the posttranscriptional control of gene expression. Genome Biol. 4: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus D.A., Evans, M.C., Agrin, N.S., Smith, M., Gongidi, P., and Jacobson, A. 2004a. Positive and negative regulation of poly(A) nuclease. Mol. Cell. Biol. 24: 5521–5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus D.A., Smith, M.M., McSweeney, J.M., and Jacobson, A. 2004b. Identification of factors regulating poly(A) tail synthesis and maturation. Mol. Cell. Biol. 24: 4196–4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T. and Reed, R. 2002. An extensive network of coupling among gene expression machines. Nature 416: 499–506. [DOI] [PubMed] [Google Scholar]
- Manley J.L. 2002. Nuclear coupling: RNA processing reaches back to transcription. Nat. Struct. Biol. 9: 790–791. [DOI] [PubMed] [Google Scholar]
- McCracken S., Fong, N., Yankulov, K., Ballantyne, S., Pan, G., Greenblatt, J., Patterson, S.D., Wickens, M., and Bentley, D.L. 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385: 357–361. [DOI] [PubMed] [Google Scholar]
- McKee A.E. and Silver, P.A. 2004. REF-ereeing the cytoplasmic fate of mRNA via nuclear export. Dev. Cell 6: 740–742. [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia L., Preker, P.J., Wiederkehr, T., Strahm, Y., and Keller, W. 1997. The major yeast poly(A)-binding protein is associated with cleavage factor IA and functions in premessenger RNA 3′-end formation. Proc. Natl. Acad. Sci. 94: 7897–7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. and Tollervey, D. 2000. mRNA stability in eukaryotes. Curr. Opin. Genet. Dev. 10: 193–198. [DOI] [PubMed] [Google Scholar]
- ____. 2001. mRNA turnover. Curr. Opin. Cell. Biol. 13: 320–325. [DOI] [PubMed] [Google Scholar]
- ____. 2003. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′ → 5′ degradation. Mol. Cell 11: 1405–1413. [DOI] [PubMed] [Google Scholar]
- Morrissey J.P., Deardorff, J.A., Hebron, C., and Sachs, A.B. 1999. Decapping of stabilized, polyadenylated mRNA in yeast pab1 mutants. Yeast 15: 687–702. [DOI] [PubMed] [Google Scholar]
- Nedea E., He, X., Kim, M., Pootoolal, J., Zhong, G., Canadien, V., Hughes, T., Buratowski, S., Moore, C.L., and Greenblatt, J. 2003. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J. Biol. Chem. 278: 33000–33010. [DOI] [PubMed] [Google Scholar]
- Neville M. and Rosbash, M. 1999. The NES–Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J. 18: 3746–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli A.E., Powers, M.A., Lund, E., Forbes, D., and Dahlberg, J.E. 1997. Inhibition of mRNA export in vertebrate cells by nuclear export signal conjugates. Proc. Natl. Acad. Sci. 94: 14394–14399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips S. and Butler, J.S. 2003. Contribution of domain structure to the RNA 3′ end processing and degradation functions of the nuclear exosome subunit Rrp6p. RNA 9: 1098–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N.J., Furger, A., and Dye, M.J. 2002. Integrating mRNA processing with transcription. Cell 108: 501–512. [DOI] [PubMed] [Google Scholar]
- Puig O., Caspary, F., Rigaut, G., Rutz, B., Bouveret, E., Bragado-Nilsson, E., Wilm, M., and Seraphin, B. 2001. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods 24: 218–229. [DOI] [PubMed] [Google Scholar]
- Rasmussen E.B. and Lis, J.T. 1993. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc. Natl. Acad. Sci. 90: 7923–7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra C., Tung, K.S., Amberg, D.C., Hopper, A.K., and Cole, C.N. 1996. Regulation of mRNA export in response to stress in Saccharomyces cerevisiae. Genes & Dev. 10: 1608–1620. [DOI] [PubMed] [Google Scholar]
- Saavedra C.A., Hammell, C.M., Heath, C.V., and Cole, C.N. 1997. Yeast heat shock mRNAs are exported through a distinct pathway defined by Rip1p. Genes & Dev. 11: 2845–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A. 1990. The role of poly(A) in the translation and stability of mRNA. Curr. Opin. Cell. Biol. 2: 1092–1098. [DOI] [PubMed] [Google Scholar]
- Sachs A.B. and Davis, R.W. 1989. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell 58: 857–867. [DOI] [PubMed] [Google Scholar]
- Sachs A.B., Bond, M.W., and Kornberg, R.D. 1986. A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: Domain structure and expression. Cell 45: 827–835. [DOI] [PubMed] [Google Scholar]
- Sachs A.B., Davis, R.W., and Kornberg, R.D. 1987. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol. Cell. Biol. 7: 3268–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. 1991. Getting started with yeast. Meth. Enzymol. 194: 3–21. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter, P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snay-Hodge C.A., Colot, H.V., Goldstein, A.L., and Cole, C.N. 1998. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 17: 2663–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade K., Ford, C.S., Guthrie, C., and Weis, K. 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90: 1041–1050. [DOI] [PubMed] [Google Scholar]
- Strasser K. and Hurt, E. 2000. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 19: 410–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2001. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature 413: 648–652. [DOI] [PubMed] [Google Scholar]
- Strasser K., Masuda, S., Mason, P., Pfannstiel, J., Oppizzi, M., Rodriguez-Navarro, S., Rondon, A.G., Aguilera, A., Struhl, K., Reed, R., et al. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304–308. [DOI] [PubMed] [Google Scholar]
- Stutz F. and Izaurralde, E. 2003. The interplay of nuclear mRNP assembly, mRNA surveillance and export. Trends Cell. Biol. 13: 319–327. [DOI] [PubMed] [Google Scholar]
- Tarun S.Z. and Sachs, A.B. 1995. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes & Dev. 9: 2997–3007. [DOI] [PubMed] [Google Scholar]
- ____. 1996. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 15: 7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Thakurta A.G., Ho Yoon, J., and Dhar, R. 2002. Schizosaccharomyces pombe spPABP, a homologue of Saccharomyces cerevisiae Pab1p, is a non-essential, shuttling protein that facilitates mRNA export. Yeast 19: 803–810. [DOI] [PubMed] [Google Scholar]
- Thomsen R., Libri, D., Boulay, J., Rosbash, M., and Jensen, T.H. 2003. Localization of nuclear retained mRNAs in Saccharomyces cerevisiae. RNA 9: 1049–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipper D.J. 1973. Inhibition of yeast ribonucleic acid polymerases by thiolutin. J. Bacteriol. 116: 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchet C., Bousquet-Antonelli, C., Milligan, L., Thompson, E., Kufel, J., and Tollervey, D. 2002. Processing of 3′-extended read-through transcripts by the exosome can generate functional mRNAs. Mol. Cell 9: 1285–1296. [DOI] [PubMed] [Google Scholar]
- Tucker R., Valencia-Sanchez, M.A., Staples, R.R., Chen, J., Denis, C.L., and Parker, R. 2001. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic deadenylase in Saccharomyces cerevisiae. Cell 104: 377–386. [DOI] [PubMed] [Google Scholar]
- Tucker R., Staples, R.R., Valencia-Sanchez, M.A., Muhlrad, D., and Parker, R. 2002. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Not1p mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 21: 1427–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A., Lennertz, P., and Parker, R. 2000. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell Biol. 20: 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis K. 1998. Importins and exportins: How to get in and out of the nucleus. Trends Biochem. Sci. 23: 185–189. [DOI] [PubMed] [Google Scholar]
- ____. 2002. Nucleocytoplasmic transport: Cargo trafficking across the border. Curr. Opin. Cell. Biol. 14: 328–335. [DOI] [PubMed] [Google Scholar]
- Winstall E., Sadowski, M., Kuhn, U., Wahle, E., and Sachs, A.B. 2000. The Saccharomyces cerevisiae RNA-binding protein Rbp29 functions in cytoplasmic mRNA metabolism. J. Biol. Chem. 275: 21817–21826. [DOI] [PubMed] [Google Scholar]
- Wyers F., Minet, M., Dufour, M.E., Vo, L.T., and Lacroute, F. 2000. Deletion of the PAT1 gene affects translation initiation and suppresses a PAB1 gene deletion in yeast. Mol. Cell. Biol. 20: 3538–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D., Vinciguerra, P., Wyss, J.C., and Stutz, F. 2002. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 22: 8241–8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Hyman, L., and Moore, C. 1999. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63: 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]