Abstract
Translation initiation is a multistep process involving several canonical translation factors, which assemble at the 5′-end of the mRNA to promote the recruitment of the ribosome. Although the 3′ poly(A) tail of eukaryotic mRNAs and its major bound protein, the poly(A)-binding protein (PABP), have been studied extensively, their mechanism of action in translation is not well understood and is confounded by differences between in vivo and in vitro systems. Here, we provide direct evidence for the involvement of PABP in key steps of the translation initiation pathway. Using a new technique to deplete PABP from mammalian cell extracts, we show that extracts lacking PABP exhibit dramatically reduced rates of translation, reduced efficiency of 48S and 80S ribosome initiation complex formation, and impaired interaction of eIF4E with the mRNA cap structure. Supplementing PABP-depleted extracts with wild-type PABP completely rectified these deficiencies, whereas a mutant of PABP, M161A, which is incapable of interacting with eIF4G, failed to restore translation. In addition, a stronger inhibition (approximately twofold) of 80S as compared to 48S ribosome complex formation (∼65% vs. ∼35%, respectively) by PABP depletion suggests that PABP plays a direct role in 60S subunit joining. PABP can thus be considered a canonical translation initiation factor, integral to initiation complex formation at the 5′-end of mRNA.
Keywords: mRNA translation, poly(A)-binding protein (PABP), eukaryotic initiation factor 4G (eIF4G), ribosome binding, mRNA circularization
Initiation is the rate-limiting step in translation and is therefore a prime target for control. The initiation step consists of an ordered multistage process involving many translation factors and adapter proteins, which facilitate the recruitment of the ribosome to the mRNA (for review, see Gingras et al. 1999; Hershey and Merrick 2000). All eukaryotic mRNAs of nuclear origin possess a 5′-cap structure, and most contain a poly(A) tail (50–300 nucleotides [nt] in length). These structures are critical for efficient mRNA translation and are specifically recognized by proteins. The cap structure serves to recruit eIF4F, a multisubunit protein complex consisting of eIF4E, eIF4A, and eIF4G. eIF4E interacts directly with the cap structure. eIF4A is an ATP-dependent RNA helicase, which is thought to unwind secondary structure in the 5′-untranslated region of the mRNA. mRNA unwinding is markedly stimulated by an RNA-binding initiation factor, eIF4B. eIF4G is a large modular scaffolding protein, which contains binding sites for eIF4E, eIF4A, and eIF3, a protein that interacts directly with the small (40S) ribosomal subunit (for reviews, see Gingras et al. 1999; Hershey and Merrick 2000). eIF4G also interacts with the poly(A)-binding protein (PABP) in mammals, plants, and yeast (Tarun and Sachs 1996; Le et al. 1997; Imataka et al. 1998). PABP is a highly conserved protein, many copies of which cover the length of the poly(A) tail of the mRNA (for reviews, see Sachs 2000; Kahvejian et al. 2001; Mangus et al. 2003). It is a 636-amino acid protein that contains four RNA recognition motifs (RRMs), and a proline-rich C-terminal domain responsible for several protein–protein interactions. PABP is involved in multiple functions in the cell ranging from mRNA biogenesis to mRNA stabilization and translation (Gray et al. 2000; Grosset et al. 2000; Copeland and Wormington 2001).
The cap and poly(A) tail synergistically enhance translation of an mRNA in several systems derived from various species (Gallie 1991, 1998; Michel et al. 2000). The eIF4G/PABP interaction, which brings about the circularization of the mRNA, was suggested to stimulate translation (Sachs 2000; Wakiyama et al. 2000). Expression of an eIF4GI mutant defective in PABP binding in Xenopus oocytes reduces translation of polyadenylated mRNA and dramatically inhibits progesterone-induced maturation, underscoring the biological importance of the PABP/eIF4G interaction (Wakiyama et al. 2000). There are several possible mechanisms to explain how the PABP/eIF4G interaction stimulates translation: (1) promoting ribosome recycling; (2) stimulating 60S ribosome joining; and (3) increasing the affinity of eIF4F for the cap (i.e., 40S ribosome recruitment). Evidence consistent with the different mechanisms was derived chiefly from genetic and biochemical experiments in yeast (Sachs and Davis 1989; Tarun and Sachs 1995; Otero et al. 1999; Searfoss et al. 2001), and also from experiments in wheat germ extract (Le et al. 1997; Bi and Goss 2000).
Examples of mRNA circularization abound, attesting to its functional importance. The mRNAs of rotaviruses are capped but not polyadenylated, yet the virus has evolved a unique mechanism to achieve circularization. The viral Non-Structural Protein 3 (NSP3) circularizes the mRNA by simultaneously interacting with a specific sequence (UGACC) in the 3′-UTR and with eIF4G (Poncet et al. 1993; Piron et al. 1998). Recently, X-ray crystallographic studies solved the 3D structure of the NSP3/eIF4G interaction domain, and by extrapolation pointed to the amino acids responsible for the PABP/eIF4G interaction (Groft and Burley 2002). One particular surface residue in PABP, M161, is critical for the interaction between PABP and eIF4G. Mutation of M161 to an alanine abrogated the interaction of PABP with eIF4G without affecting its affinity for poly(A) RNA (Groft and Burley 2002).
Here, we sought to examine the mechanisms by which the mammalian PABP stimulates ribosome recruitment and translation. A novel assay to deplete PABP from nuclease-treated Krebs-2 cell-free translation extract was developed (Svitkin and Sonenberg 2004) and was used to study the function of PABP in translation assays, ribosome binding studies, and cross-linking experiments. We also investigated the ability of recombinant wild-type and mutant PABP M161A to restore translation. The results demonstrate that PABP enhances translation and stimulates ribosome recruitment to the mRNA both at the 40S ribosome subunit recruitment and 60S subunit joining steps, and that the interaction between PABP and eIF4G is essential for this enhancement. Cross-linking studies using PABP-depleted extracts demonstrated a role for PABP in the interaction of eIF4E with the cap. eIF4E cross-linking to the mRNA was reduced in PABP-depleted extracts and was restored by the addition of wild-type PABP, but not by PABP M161A. Cross-linking studies were also performed to examine the effects of the poly(A) tail on the affinity of eIF4E for the cap. The presence of a poly(A) tail enhanced eIF4E cross-linking to the cap. Our data provide compelling evidence that PABP is a bona fide translation initiation factor, and that the PABP/eIF4G interaction is crucial for the translational stimulatory effect that is conferred by the poly(A) tail in higher eukaryotes.
Results
Krebs-2 cell extracts depleted of PABP are translationally impaired
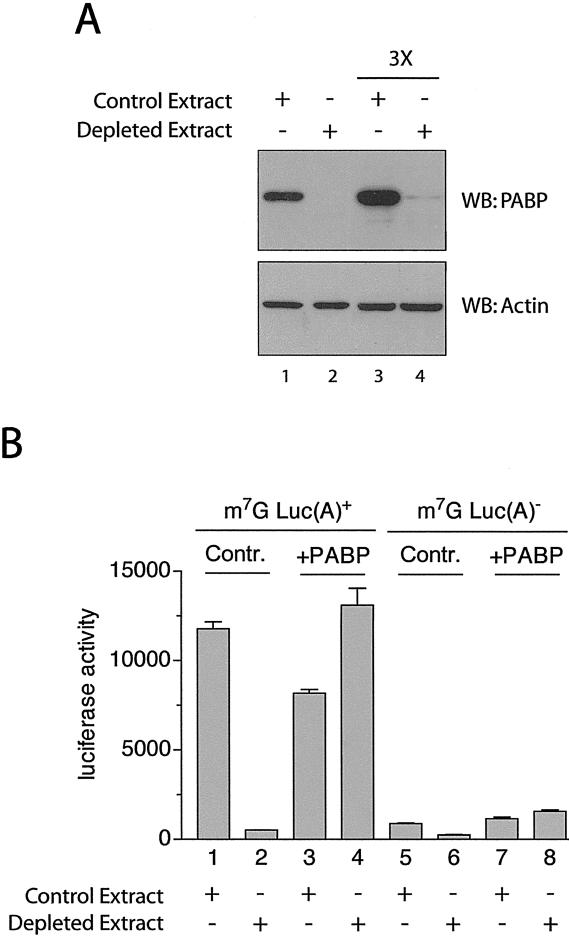
To directly analyze the effects of PABP on translation, PABP was depleted from a nuclease treated Krebs-2 extract by incubating it with GST-Paip2 coupled to glutathione-Sepharose beads (Svitkin and Sonenberg 2004). Paip2 is a protein that binds specifically and avidly to PABP, removes it from the poly(A) tail, and consequently inhibits translation (Khaleghpour et al. 2001a,b). The supernatant was analyzed for the presence of PABP by SDS-PAGE followed by Western blotting with an anti-PABP antibody (Afonina et al. 1997). Mock-depleted (control) extract treated with GST-coupled glutathione beads was analyzed as a control (Fig. 1A, lane 1). PABP was almost completely (>95%) eliminated from the extract treated with glutathione-Sepharose beads coupled to GST-Paip2 (Fig. 1A, cf. lanes 2 and 1). To demonstrate that the observed absence of PABP is not simply due to the antibody's detection limit, we also loaded three times more extract (Fig. 1A, lanes 3,4, 3×). Significantly, the levels of other canonical translation factors (i.e., eIF4E, eIF4A, and especially eIF4G) are not affected by the depletion of PABP by this method (Svitkin and Sonenberg 2004; data not shown). Furthermore, treatment with GST-coupled glutathione beads only marginally reduced the amount of PABP in the extract (data not shown). The ability of Krebs-2 extract to translate poly(A)+ mRNA decreased ∼16-fold as a consequence of PABP depletion (Fig. 1B). Importantly, addition of physiological amounts of recombinant PABP (10 μg/mL) rescued translation, confirming that the process of PABP depletion does not result in the removal or inactivation of other critical components of the translational machinery. PABP depletion also inhibited translation of poly(A)– mRNA three- to fourfold, and this inhibition was also reversed by PABP (Fig. 1B). PABP is therefore required for efficient translation, and it can stimulate translation both in cis and in trans [i.e., either via its interaction with the poly(A) tail or on its own, without the poly(A) tail].
Figure 1.
Depletion of PABP from Krebs-2 extracts results in diminished translation. (A, top panel) Western blot (SDS-10% PAGE) analysis of extracts treated with GST (Control Extract, lanes 1,3) or GST-Paip2 (Depleted Extract, lanes 2,4) probed using anti-PABP polyclonal antibody. (Bottom panel) Same membrane probed with an anti-β-actin antibody (Sigma) as a loading control. Three times more extract was loaded in lanes 3 and 4 (marked 3×) versus lanes 1 and 2, to reach the antibody's detection limit (lane 4). (B) Capped poly(A)+ (1–4) or capped poly(A)– (5–8) luciferase mRNA (2 μg/mL) was translated in Krebs-2 extracts that were either depleted of PABP (Depleted Extract) or mock-depleted (Control Extract). The reactions were supplemented with either recombinant human PABP (10 μg/mL) or control buffer, as indicated. Luciferase activity was measured as described in Materials and Methods and is expressed in relative light units (RLU) × 103.
Mutation in PABP that selectively inhibits its binding to eIF4G
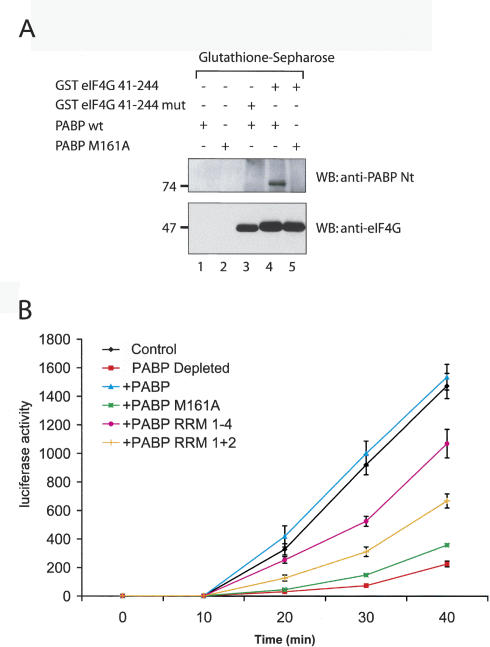
In vitro pull-down experiments were performed to assess the affinity of a PABP mutant, PABP M161A, for eIF4G. GST-eIF4G (41–244), containing the PABP-binding site in eIF4G, or a mutant that cannot bind PABP [these proteins were described as GST-eIF4G (1–204) and GST-eIF4G (1–204) mut, respectively (Imataka et al. 1998), prior to the discovery of an extra 40 amino acids at the N terminus of eIF4G (Byrd et al. 2002)] were immobilized on glutathione-Sepharose beads and incubated with PABP wild type and PABP M161A. Previously, RRM 1 + 2 of PABP with the M161A mutation was shown not to bind to eIF4G (Groft and Burley 2002). As expected, PABP M161A failed to coprecipitate with GST-eIF4G (41–244) (Fig. 2A, top panel, lane 5), whereas PABP wild type did (Fig. 2A, top panel, lane 4). A filter binding assay was carried out with the same proteins to compare their affinities for poly(A) RNA. PABP wild type and PABP M161A bound to poly(A) with similar affinities (data not shown), in agreement with the report of Groft and Burley (2002), indicating that the mutation did not cause protein misfolding or denaturation.
Figure 2.
Interaction with eIF4G is critical for PABP to stimulate translation. (A) PABP M161A cannot bind to eIF4G. PABP wild type and PABP M161A were incubated with glutathione-Sepharose beads alone (lanes 1,2), or with beads coupled to GST eIF4G 41–244 (lanes 4,5), washed with binding buffer, and eluted with Laemmli buffer. (Lane 3) GST eIF4G 41–244mut, which cannot bind to PABP, was used as a control. Eluted proteins were resolved on an SDS-10% PAGE. Western blot analysis was performed using anti-PABP antibody (top panel) or anti-eIF4G antibody (bottom panel). The positions of molecular weight markers are indicated on the left. (B) Translation of a capped luciferase poly(A)+ mRNA in PABP-depleted Krebs-2 extract supplemented with different forms of PABP. Luciferase activity was measured at several time points in mock-depleted (control, black line) or depleted extracts supplemented with no protein (PABP Depleted, red line), or supplemented with equimolar amounts of wild-type PABP (blue line), PABP M161A (green line), PABP RRM 1–4 (fuchsia line), or PABP RRM 1 + 2 (orange line). Luciferase activity was measured as described in Materials and Methods and is expressed in RLU × 103, as an average of four experiments.
The PABP–eIF4G interaction is essential for efficient translation
To determine the importance of eIF4G for PABP-mediated stimulation of translation, in vitro translation experiments were carried out using PABP-depleted Krebs-2 cell extracts with luciferase reporter mRNA (Svitkin and Sonenberg 2004), supplemented with PABP wild type or mutants. Translation in extracts lacking PABP was inhibited approximately sevenfold at 40 min of translation (Fig. 2B, cf. GST-Paip2 and GST). The addition of recombinant PABP (10 μg/mL) fully restored translation (Fig. 2B). In sharp contrast, the addition of PABP M161A (10 μg/mL) to the depleted extract failed to stimulate translation (Fig. 2B). The addition of equimolar amounts of PABP RRM 1–4 or PABP RRM 1 + 2 (5 and 2.5 μg/mL, respectively) stimulated translation five- and threefold, respectively, at 40 min (Fig. 2B). Although the latter fragments of PABP bind poly(A) RNA as well as PABP wild type and possess the eIF4G-binding site (Imataka et al. 1998), they do not rescue translation as well as PABP, presumably because they may be unable to recruit other stimulatory factors to the initiation complex. In conclusion, the PABP/eIF4G interaction is required for PABP to mediate its stimulatory effects on translation.
PABP stimulates 80S ribosome initiation complex formation
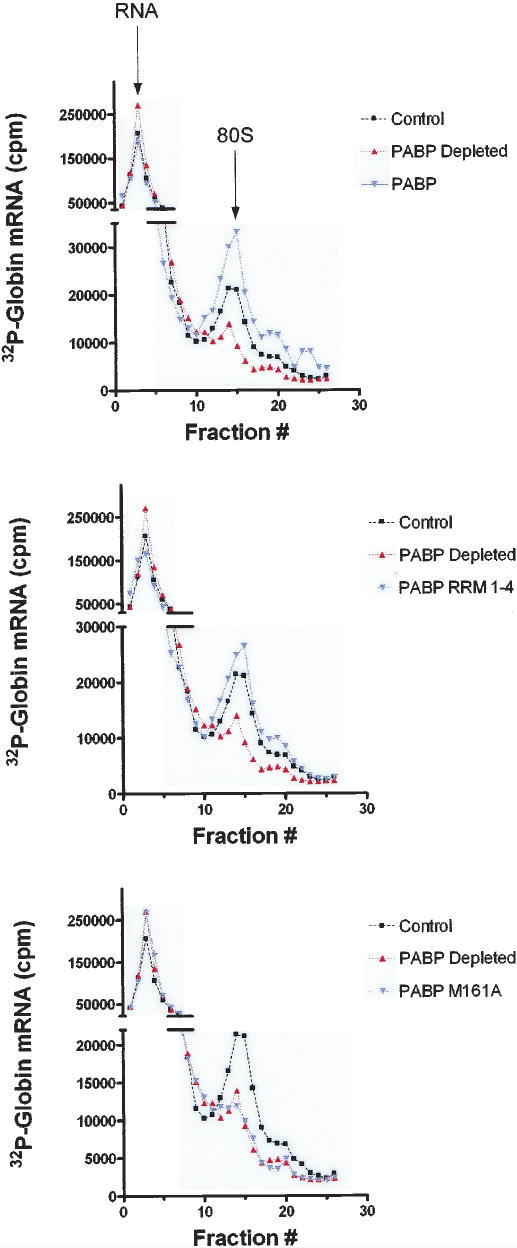
To investigate whether the translational deficiency resulting from the absence of PABP was due to a defect in initiation, 80S ribosome binding studies were carried out using PABP- and mock-depleted Krebs-2 cell extracts. Extracts were incubated with radiolabeled globin mRNA in the presence of cycloheximide, and analyzed on a 10%–15% sucrose density gradient in a high-salt buffer (HSB) (Lodish and Rose 1977) to reduce nonspecific interactions. PABP-depleted extracts exhibited a reduction in 80S ribosomal complex formation (∼65% relative to control, in a representative experiment, which was repeated four times, with variations of <10%) (Fig. 3, PABP Depleted) as compared to mock-depleted extracts (Fig. 3, Control). The addition of 120 ng of recombinant PABP (Fig. 3, top panel, PABP) or 72 ng of PABP RRM(1–4) (Fig. 3, middle panel, PABP RRM 1–4) to the depleted extracts not only restored, but even stimulated 80S initiation complex formation (by 174% and 140%, respectively, relative to control). Addition of 120 ng of PABP M161A failed to restore 80S initiation complex formation (Fig. 3, bottom panel, PABP M161A). PABP is therefore required for optimal initiation complex formation, for in its absence, the assembly of the 80S ribosomal initiation complex is severely impaired. Also, since PABP M161A failed to functionally substitute for wild-type PABP, PABP must interact with eIF4G to promote 80S initiation complex formation.
Figure 3.
80S ribosome initiation complex formation is deficient in PABP-depleted extracts (representative experiment of four independent experiments). Ribosome initiation complex profiles of control extract (black line, squares), PABP-depleted extract (red line, triangles), and supplemented extract (blue line, inverted triangles) are overlaid for comparison. Extracts were supplemented with equimolar amounts of either wild-type PABP (10 μg/mL; top panel), PABP RRM 1–4 (6 μg/mL; middle panel), or PABP M161A (10 μg/mL; lower panel). Peaks corresponding to unbound RNA and to RNA in complex with 80S ribosomes are indicated with arrows (top panel).
PABP stimulates 48S initiation complex formation
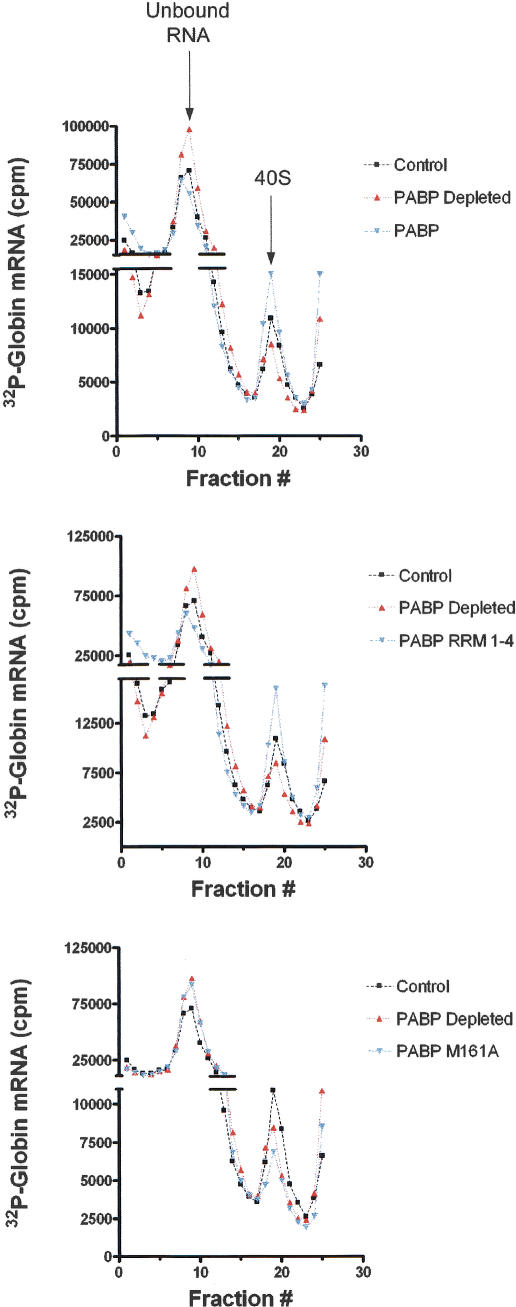
We wished to determine whether the reduction in 80S ribosome initiation complex formation was a result of reduced 40S ribosome recruitment. 40S ribosome binding studies were thus carried out in PABP-depleted Krebs-2 cell extracts. Extracts were incubated with radiolabeled globin mRNA in the presence of GMPPNP and analyzed on a 10%–15% sucrose density gradient prepared in a low-salt buffer (LSB) (Gray and Hentze 1994). PABP-depleted extracts exhibited a reduction in 48S initiation complex formation (34% reduction relative to control, in a representative experiment, which was repeated four times, with variations of <10%) (Fig. 4, PABP Depleted) as compared to mock-depleted extracts (Fig. 4, Control). The addition of 100 ng of recombinant PABP (Fig. 4, top panel, PABP) or 60 ng of PABP RRM(1–4) (Fig. 4, middle panel, PABP RRM 1–4) to the depleted extract restored 40S recruitment by 157% and 150%, respectively, relative to control, thus stimulating it to a higher level than in the mock-depleted extract. The addition of 120 ng of PABP M161A failed to restore 40S initiation complex formation (Fig. 4, bottom panel, PABP M161A), and even decreased complex formation by 39% relative to the depleted extract (decreased by 58% vs. control) (Fig. 4, bottom panel, PABP Depleted). Thus, PABP M161A appears to exert a dominant-negative effect on 40S ribosome recruitment. These results indicate that PABP is required for efficient 40S ribosome recruitment to the mRNA, and that the PABP/eIF4G interaction plays a role in 40S ribosome recruitment. An important conclusion from these experiments is that the reduction in 40S ribosome recruitment upon PABP depletion only partly accounts for the inhibition of 80S initiation complex formation. This indicates that PABP may affect 80S initiation complex formation both indirectly at an early step (via inhibition of 40S ribosome recruitment) and directly at later steps (e.g., at the step of 60S subunit joining). This will be further addressed in the Discussion.
Figure 4.
40S ribosome initiation complex formation is deficient in PABP-depleted extracts (representative experiment of four independent experiments). 40S initiation complex profiles of control extract (black line, squares), PABP-depleted extract (red line, triangles), and PABP-depleted extract supplemented with PABP (blue line, inverted triangles) are overlaid for comparison. Extracts were supplemented with equimolar amounts of either wild-type PABP (10 μg/mL; top panel), PABP RRM 1–4 (6 μg/mL; middle panel), or PABP M161A (10 μg/mL; lower panel). Peaks corresponding to unbound RNA and to RNA in complex with 40S ribosomes are indicated with arrows (top panel).
PABP stimulates initiation factor recruitment to the mRNA
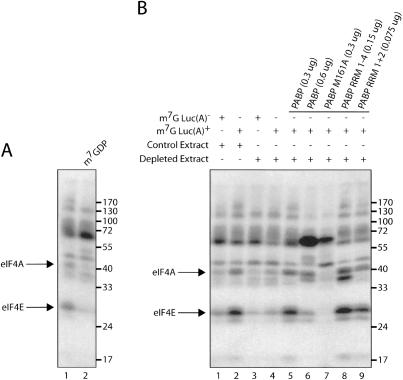
40S ribosome binding to the mRNA is effected by the eIF4 proteins. eIF4E binds directly to the cap structure and can be cross-linked to it in an ATP-independent manner (Sonenberg et al. 1978). However, several of the other eIF4 proteins, namely, eIF4A and eIF4B, become juxtaposed to the cap structure in the presence of eIF4E and ATP, and subsequently, can also be covalently cross-linked to the cap (Sonenberg 1981; Edery et al. 1983). Cross-linking experiments were carried out with luciferase mRNA 32P-labeled at the 5′-cap (m7Gp*ppG, where the α-phosphate is labeled) to determine the effects of PABP on the interaction of eIF4E, eIF4A, and eIF4B with the 5′-cap structure. In these experiments we used reticulocyte lysate, as the use of Krebs-2 extract resulted in the nonspecific cross-linking of many proteins (data not shown). Also, we chose a chemical cross-linking method that is superior to UV cross-linking for detecting the eIF4E–cap interaction, but results in feeble eIF4B cross-linking (Pelletier and Sonenberg 1985). eIF4E and eIF4A cross-linked to poly(A)– mRNA (Fig. 5A, lane 1). The identities of these proteins were confirmed by the fact that their binding was specifically inhibited by the cap analog, m7GDP (as demonstrated previously in many reports: Sonenberg 1981; Lee and Sonenberg 1982; Edery et al. 1983), whereas the nonspecific cross-linking of other proteins (e.g., molecular weight of ∼50 and ∼60 kDa) was not inhibited (Fig. 5A, cf. lanes 2 and 1). In the PABP-depleted lysate, eIF4E and eIF4A cross-linking was severely impaired (Fig. 5B, cf. lanes 1,2 and 3,4). Strikingly, the addition of a poly(A) tail does not stimulate eIF4E and eIF4A cross-linking in depleted extracts (Fig. 5B, cf. lanes 3 and 4), whereas in mock-depleted lysates it does. Therefore, the poly(A) tail can mediate its stimulatory effect only in the presence of PABP. Significantly, the addition of equimolar amounts of wild-type PABP, or truncation mutants of PABP (RRM 1–4 or RRM 1 + 2) to depleted extracts restored eIF4E and eIF4A cross-linking to the cap structure (Fig. 5B, lanes 5,8,9, respectively). Supplementing the extract with excess PABP (twofold), inhibits eIF4E and eIF4A cross-linking (Fig. 5B, cf. lanes 6 and 5). These data are consistent with our observation that adding increasing amounts of PABP to nondepleted extracts progressively inhibits translation (data not shown). Supplementing depleted extracts with PABP M161A (0.3 μg) did not restore eIF4E and eIF4A cross-linking, but rather dramatically inhibited factor binding to levels below those observed in PABP-depleted extracts (Fig. 5B, cf. lanes 7 and 4).
Figure 5.
PABP mediates poly(A)-tail-induced initiation factor binding to the m7G cap. (A) Cap-labeled Luc(A)– and Luc(A)+ mRNA were incubated with reticulocyte lysate. Proteins were cross-linked to mRNA by oxidation and subsequent reduction. Bands corresponding to eIF4E and eIF4A are indicated with arrows on the left. The addition of cap analog is indicated above. (B) Cap-labeled Luc(A)– and Luc(A)+ mRNA were incubated with Mock-Depleted (control extract) and PABP-Depleted (depleted extract) reticulocyte lysate. Extracts were supplemented with equimolar amounts of recombinant proteins (indicated above). The positions of molecular weight markers are indicated on the right.
Next, we wished to study the interaction of eIF4B with the cap. UV cross-linking experiments were thus carried out (as eIF4B cross-links better to the mRNA upon UV irradiation as compared to chemical cross-linking) (Pelletier and Sonenberg 1985) using polyadenylated and deadenylated luciferase mRNAs, which were 32P-labeled at the 5′-cap and incubated with a reticulocyte lysate. After UV irradiation, the extract was treated with a mixture of RNAse A/T1 and analyzed by SDS-PAGE. Using poly(A)– mRNA, only eIF4B was detected by autoradiography (eIF4B was shown before to be the prominent cross-linked protein in initiation factor preparations) (Fig. 6, lane 1; Pelletier and Sonenberg 1985). The cross-linking of this protein was inhibited by the cap analog, m7GDP (data not shown). When poly(A)300 RNA was used, several cross-linked proteins, which correspond to eIF4A, eIF4E, and possibly eIF4G (indicated by arrows and bullets) were detected, while the intensity of eIF4B was increased (Fig. 6, cf. lanes 1 and 3). Supplementing the extract with a dominant-negative form of eIF4A, which inhibits eIF4F function (Pause et al. 1994; Svitkin et al. 2001b), and consequently inhibits interactions of eIF4 proteins with the cap (Sonenberg 1981), caused a drastic reduction in the cross-linking of all initiation factors, without affecting the cross-linking of nonspecific bands (Fig. 6, lanes 2,4). Thus, a long poly(A) tail enhances eIF/cap binding by recruiting PABP to the mRNA.
Figure 6.
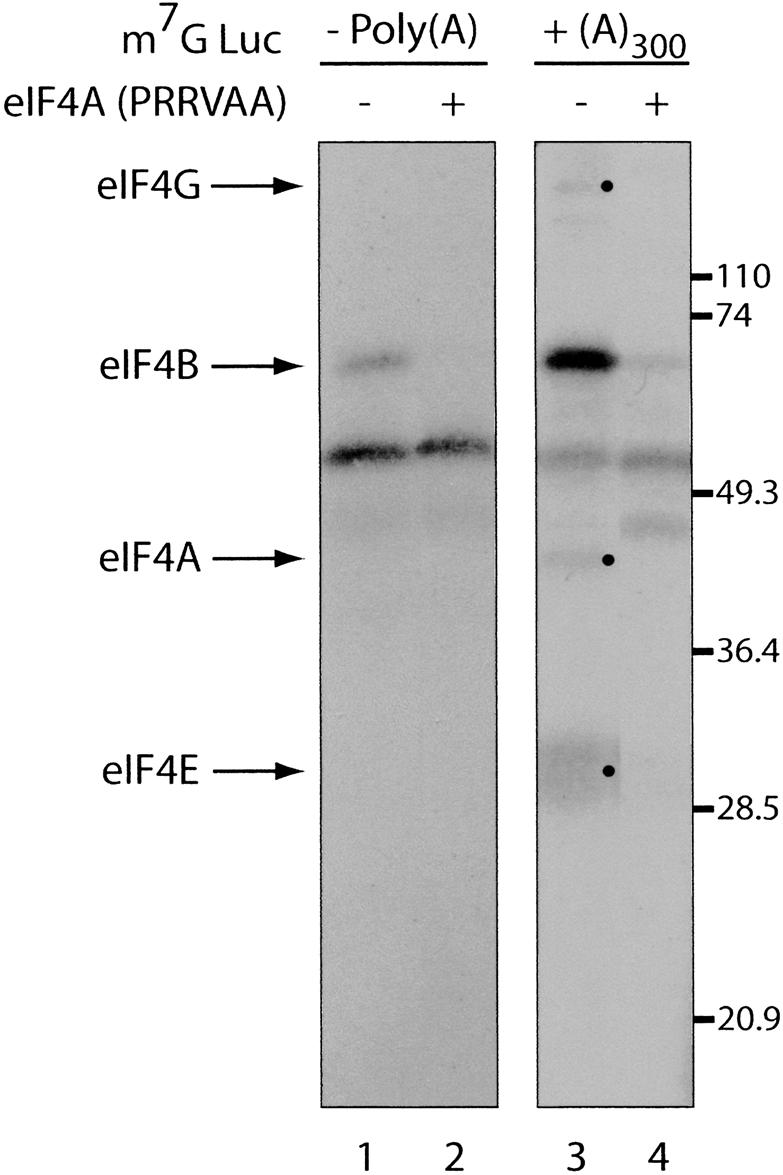
The poly(A) tail stimulates initiation factor binding to the m7G cap. Cap-labeled Luc(A)– (left panel) and Luc(A)+ (right panel) mRNA were incubated with rabbit reticulocyte lysate in the absence (lanes 1,3) or presence (lanes 2,4) of an eIF4A dominant-negative mutant (PRRVAA, 100 μg/mL) (Svitkin et al. 2001b), and subjected to UV cross-linking. Samples were analyzed by SDS-15% PAGE and bands were revealed by autoradiography. Bands corresponding to initiation factors are indicated with arrows on the left and bullets. The positions of molecular weight markers are indicated on the right.
Taken together, these data demonstrate that PABP mediates the stimulatory effects of an extended poly(A) tail on the interaction of the eIF4F group of initiation factors with the m7G cap structure. Furthermore, PABP stimulates the recruitment of eIF4 proteins to the cap by directly interacting with eIF4G, since PABP M161A is not capable of stimulating cross-linking of initiation factors to the cap structure.
Discussion
An ongoing debate exists regarding the mechanism by which PABP-induced circularization of the mRNA stimulates translation (Jacobson 1996; Sachs 2000; Kahvejian et al. 2001). Based on earlier data from experiments using yeast genetics, yeast translation extracts, and purified yeast proteins, several mechanisms have been postulated to explain the function of PABP in eukaryotic translation. One hypothesis posits that PABP promotes 60S ribosomal subunit joining at the start codon (Sachs and Davis 1989; Searfoss et al. 2001). Another model proposes that circularization stimulates ribosome recycling, by bringing the 5′- and 3′-ends of the mRNA together (Jacobson 1996). It has also been suggested that PABP promotes 40S initiation complex recruitment (Tarun and Sachs 1995), possibly by increasing the affinity of eIF4F for the cap structure or affecting other protein–protein or protein–RNA interactions (Le et al. 1997; Wei et al. 1998; Sachs 2000). Here, we address this question in the most direct manner by depleting PABP from Krebs-2 extract. The depleted extract does not support efficient translation of capped and polyadenylated mRNAs (Svitkin and Sonenberg 2004). Our method of depletion is highly efficient and specific, as >95% of PABP was depleted, and translation could be largely restored by addition of recombinant PABP.
Our data further bolster the idea that PABP stimulates translation by binding to eIF4G. Krebs-2 extracts depleted of PABP are severely impaired in translation of a reporter mRNA (Fig. 2B). Translation can be rescued by the addition of recombinant PABP protein, but not with a mutant form of the protein (PABP M161A), which cannot bind to eIF4G. Thus, the PABP/eIF4G, in mammalian cells (at least in vitro) and in Xenopus oocytes (Wakiyama et al. 2000) is clearly important. As stated above, stimulation of ribosome recycling is a possible explanation for the role of the PABP/eIF4G interaction. However, the time-course experiment using PABP M161A suggests that this cannot be the only mechanism. If solely ribosome recycling were affected by this mutation, the translation time course of the extract supplemented with PABP M161A would have resembled that of the extract with PABP wild type, in the early phase of translation. The curves would have only diverged at later time points, when new rounds of translation, which are presumably dependent on ribosome recycling, occur. Clearly, in our experiment, the two curves (PABP and PABP M161A) diverge at the onset of translation, indicating a deficiency in the initial rate of translation. Nevertheless, we cannot rule out the possibility that mRNA circularization, resulting from the interaction between PABP and eIF4G, also simulates translation by promoting ribosome recycling. The overall changes in translation in our time-course experiments can be attributed to PABP's effect on both initiation and on subsequent rounds of translation.
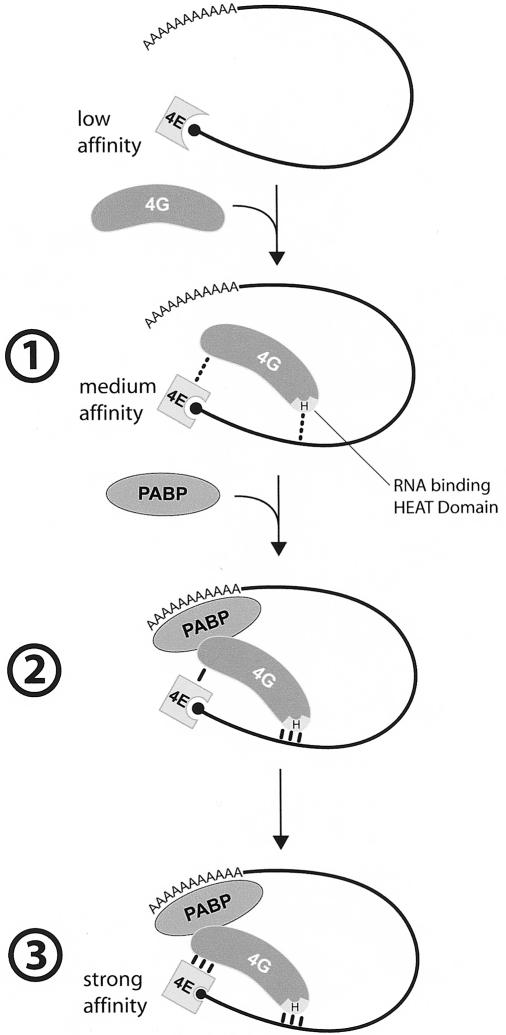
By performing protein cross-linking to the cap structure, we demonstrated PABP's direct effect on the initial recruitment of the translation machinery to the mRNA. Our results show that the PABP/poly(A) tail complex acts at a step prior to 43S preinitiation complex formation. As stated above, the interaction between PABP and eIF4G may stimulate other protein/protein or protein/RNA interactions. These interactions lie upstream of ribosome recruitment, and their stabilization will ultimately affect mRNA translatability. In the absence of a poly(A) tail, recruitment of initiation factors to the mRNA was markedly reduced when compared to the same mRNA with a long poly(A) tail (∼300 As). Similarly, depletion of PABP from mammalian extracts reduced initiation factor binding. Supplementing these extracts with recombinant PABP restored and even stimulated eIF4F cross-linking to the 5′-cap. The PABP/eIF4G interaction is necessary for this effect. Thus, PABP's binding to eIF4G may engender conformational changes that promote eIF4F activity. In plants, eIF4G appears to enhance PABP binding to poly(A) (Le et al. 1997), and in turn, PABP increases the affinity of eIF4F for the cap structure (Wei et al. 1998) and eIF4F helicase activity (Bi and Goss 2000). There are two possible mechanisms by which PABP can enhance the eIF4E affinity for the cap structure. One model may involve cooperative binding of PABP with eIF4G/4E to the mRNA. We have previously shown, using a cross-linking assay, that eIF4G increases the affinity of eIF4E for the cap (Haghighat and Sonenberg 1997). The RNA-binding domain in eIF4G must be present for this enhancement, inasmuch as an N-terminal fragment of eIF4G lacking this domain failed to enhance eIF4E interaction with the cap (A. Haghighat and N. Sonenberg, unpubl.). Thus, it is plausible that PABP enhances the eIF4G interaction with RNA, and consequently increases the affinity of eIF4E to the cap (see model in Fig. 7). This model is supported by the observation that PABP stimulates IRES function, and that this stimulation is abrogated upon eIF4G cleavage, which separates the PABP-binding site in eIF4G from the RNA-binding domain (Svitkin et al. 2001a). An alternative, but not mutually exclusive, model is based on experiments, which demonstrated that eIF4G stimulates eIF4E affinity for the cap through allosteric changes (von Der Haar et al. 2000; Gross et al. 2003). In a similar mechanism, binding of PABP to eIF4G might stimulate the eIF4E–cap interaction by producing conformational changes in eIF4G.
Figure 7.
Model for PABP enhancement of eIF4E binding to the mRNA 5′-cap. (1) eIF4G enhances eIF4E affinity for the mRNA 5′-cap structure, through the interaction of the eIF4G HEAT (Marcotrigiano et al. 2001) domain (H) with mRNA. (2) PABP enhances eIF4G affinity for the mRNA. (3) eIF4G affinity for eIF4E is increased as a consequence of the increased eIF4G binding to the mRNA.
It is significant that the deficiency in 80S recruitment upon PABP depletion is more drastic than the reduction of 40S initiation complex formation observed under the same conditions. This may indicate that both 40S and 60S ribosomal subunit recruitment steps are separate targets of PABP. Searfoss et al. (2001) proposed that PABP derepresses 60S joining. They demonstrated that PABP inhibits two nonessential putative RNA helicases in yeast, Slh1p, and Ski2p, themselves inhibitors of eIF5B. Deletion of these two RNA helicases results in translational enhancement of poly(A)– mRNA to the level obtained with poly(A)+ mRNA. Importantly, Munroe and Jacobson (1990) first showed that the poly(A) tail stimulated 60S ribosomal subunit binding in a reticulocyte lysate. It is of interest that another example exists for an effect of the 3′ of mRNA on 60S subunit joining. Ostareck et al. (1997, 2001) have demonstrated that the 3′-UTR of Lox-15 mRNA stimulates 60S subunit joining via hnRNP K and E1 proteins. Sachs and Davis (1989) had shown that depletion of PABP by promoter inactivation in yeast resulted in an inhibition of translation at the level of 60S joining. Subsequently, Tarun and Sachs (1995) reported that PABP was involved in 40S recruitment in a poly(A)-dependent yeast assay. They proposed that under conditions of reduced 60S ribosome binding (e.g., deletion of eIF5 and eIF5B, or of enzymes involved in 60S ribosome biogenesis), 40S ribosome binding becomes the rate-limiting step and serves as a target of PABP-mediated translational stimulation (Tarun and Sachs 1995; Sachs 2000).
Taken together, our data indicate that PABP exerts its stimulatory effects at multiple stages of translation initiation in mammals. Why target several steps of initiation? Regulating both initiation factor binding to the mRNA (thus controlling 40S recruitment), and 60S joining permits a tighter control of initiation, by preventing nonspecific initiation events. Furthermore, the differences observed between yeast and mammalian systems (e.g., differences in the requirement for the PABP/eIF4G interaction, differential targets of PABP enhancement, the lack of Paip1 and Paip2 in yeast) represent the evolution of more complex poly(A)-tail-mediated translational control in higher eukaryotes. It is interesting that yeast eIF4G (TIF4631) does not interact with human PABP, despite the high degree of homology between human and yeast PABPs (Otero et al. 1999). Slight dissimilarities in amino acid sequence between the PABPs, and major differences between the PABP-binding sites in eIF4G homologs might account for this inability to interact. Thus, the manner by which eIF4G and PABP interact has changed, without disrupting the interaction per se, and potentially modifying its consequences and effects on the rest of the initiation apparatus.
Our experiments constitute the first direct and comprehensive study of PABP as a translation initiation factor. PABP clearly impinges on the earliest step of translation initiation and as a coactivator of translation, should be considered a canonical eukaryotic translation initiation factor.
Materials and methods
DNA vectors
The PABP used in these studies corresponds to GenBank accession number gi:4505575. This record was replaced by gi:BC023520.2 on April 14, 2004. The latter has three additional amino acids (212–214) located in RRM3. Consequently, we compared the activity of the two forms, and found them to behave similarly in our in vitro translation assays (data not shown). The pET-28b(+) PABP 1–190 (RRM 1 + 2) deletion construct containing a point mutation at position M161A was obtained from C. Groft and S. Burley (Laboratory of Molecular Biophysics and Howard Hughes Medical Institute, The Rockefeller University, New York). To generate the mutant in the context of full-length PABP (pET-28b PABPHis mutant), the construct was digested with EcoRI. A fragment resulting from the digestion of pET-3b PABPHis (previously described) (Khaleghpour et al. 2001a) with EcoRI was ligated into the digested pET-28b PABP (1–190) mutant vector. Resulting clones were screened by restriction enzyme analysis using NdeI or XhoI, to identify the clones containing the insert in the proper orientation.
Protein expression and purification
BL21 λDE3 bacteria were transformed with pET-28b PABPHis M161A. Protein expression and purification were carried out as previously described for PABPHis (Khaleghpour et al. 2001a).
Glutathione pull-down assay
Glutathione-Sepharose beads (500 μL; Amersham Biosciences) were washed in PBS, of which 20 μL (dry bed volume) was incubated with 5 μg of GST-eIF4G 41–244 (previously described as 1–204) (Imataka et al. 1998) or 5 μg of GST-eIF4G 41–244mut, which does not bind to PABP. Incubation was carried out for 30 min at 4°C in 300 μL of Binding Buffer (0.4% Triton X-100; 2.5 mM MgCl2; 20 mM Tris-HCl at pH 7.5; 10% glycerol; 0.1 mM EDTA; 1 mg/mL BSA) supplemented with Complete Protease Inhibitor cocktail tablets (Roche). After one wash with Binding Buffer, beads were incubated with 1 μg of PABP wild type or PABP M161A for 30 min at 4°C in 300 μL of Binding Buffer. The latter proteins were also incubated with 20 μL of glutathione-Sepharose beads, which were not coupled to GST-eIF4G, to control for nonspecific interactions. Beads were subsequently washed twice with Wash Buffer (same as Binding Buffer without Triton X-100 and protease tablets) and treated with 40 μL of Laemmli buffer. Proteins were analyzed by SDS-PAGE and revealed by Western blot analysis using anti-PABPNt and anti-eIF4GNt (kind gifts from H. Imataka, Protein Research Group, Genome Sciences Center, Riken (Yokohama) Tsurumi-Ku, Yokohama, Japan).
Preparation and PABP depletion of Krebs-2 extracts
Krebs-2 extracts were prepared as previously described (Svitkin and Agol 1978; Svitkin and Sonenberg 2004). For the depletion of endogenous PABP protein, extracts were incubated with glutathione beads previously incubated with GST-Paip2 protein (Svitkin and Sonenberg 2004). The mixture was centrifuged, and the supernatant was analyzed by Western Blotting for the presence of PABP. Mock-depleted (control) extracts were treated with GST alone.
In vitro translation experiments
All translation experiments were performed as previously described (Khaleghpour et al. 2001b; Svitkin and Sonenberg 2004). Capped poly(A)+ and poly(A)– luciferase RNAs (Iizuka et al. 1994) were used to program translation reactions.
mRNA labeling for ribosome binding assays
Globin mRNA (Gibco Invitrogen Corporation, discontinued) was labeled using yeast poly(A) polymerase (USB Corporation) and [α-32P] Cordycepin-5′-Triphosphate (NEN) according to the 3′-end labeling of RNA protocol (P-74225B rev 03/07).
80S ribosome binding assays
80S ribosome binding studies were carried out based on the protocol from Lodish and Rose (1977). Briefly, Krebs-2 extracts were incubated with radiolabeled mRNA (106 cpm) in the presence of cycloheximide (0.6 mM) in a total volume of 37.5 μL for 15 min at 30°C. Reactions were stopped by diluting fourfold with ice-cold HSB (Lodish and Rose 1977). Total reactions were applied to cold 5-mL 15%–30% sucrose gradients prepared with HSB. Gradients were centrifuged in an SW55 rotor at 54,000 rpm at 4°C for 1 h 45 min. Fractions (0.2 mL) were collected and counted in CytoScint (ICN). Relative binding was assessed by weighing the paper under the 80S peaks using an analytical balance.
40S ribosome binding assays
40S ribosome binding studies were carried out as for 80S ribosomes except for the following modifications. GMPPNP (Sigma), a nonhydrolyzable analog of GTP, was used in conjunction with cycloheximide to prevent 60S subunit joining. Gradients (10%–30% sucrose) were prepared using a low-salt buffer (Gray and Hentze 1994) and centrifuged for 150 min. Binding was assessed by weighing the paper under the 40S peaks using an analytical balance.
mRNA preparation for cross-linking assays
Uncapped luciferase (Luc) RNA was obtained from Promega as part of the rabbit reticulocyte lysate translation kit. For the generation of Luc(A)+ mRNA, poly(A) tail extension was carried out using yeast poly(A) polymerase (USB Corporation) according to the Poly(A) tailing of RNA protocol (P-74225B rev 03/07). For the generation of Luc(A)– mRNA, Luc mRNA was hybridized with oligo(dT) and digested with RNAse H (MBI Fermentas) as suggested by the manufacturer.
mRNA labeling for cross-linking assays
Luc(A)+ and Luc(A)– mRNAs were radioactively labeled at the cap using vaccinia-virus guanylyltransferase (Ambion) with [α-32P]GTP (Perkin Elmer Life Sciences) and S-adenosyl methionine according to the manufacturer's instructions.
Chemical cross-linking assays
Chemical cross-linking was carried out as previously described (Sonenberg et al. 1978; Merrick and Sonenberg 1997). Briefly, cap-labeled mRNA was oxidized using 0.2 mM NaIO4 (freshly prepared) for 2 h in the dark. Oxidized mRNA (105 cpm) was subsequently incubated with RRL extract and incubated at 30°C for 20 min. Reduction of the Schiff base between the 5′ mRNA ribose and lysines in the proteins was carried out overnight at 4°C using 0.2 mM NaCNBH3. The mixture was treated with 0.1 mg/mL RNAse A to digest the RNA. Samples were analyzed by SDS-15% PAGE and bands were revealed by autoradiography.
UV cross-linking assays
Cap-labeled RNAs were incubated for 10 min at 33°C with rabbit reticulocyte lysate (RRL; Promega). Mixture was subjected to UV irradiation for 20 min at 4°C (Pelletier and Sonenberg 1985). The extract was incubated with a mixture of RNAse A and Micrococcal Nuclease for 30 min at 37°C. Samples were analyzed by SDS-15% PAGE and bands were revealed by autoradiography.
Acknowledgments
We thank C. Lister and I. Harvey for excellent technical assistance, J. Pelletier for advice and critical reading of the manuscript, and C. Groft and S. Burley for the mutant PABP. This research was supported by a Public Health Service Research Grant from the NIH to N.S. (Grant #5R01 GM66157-03). N.S. is a CIHR Distinguished Scientist and a Howard Hughes Medical Institute International Scholar. R.S. is a recipient of the McGill Chemical Biology Scholarship. A.K. is a recipient of the McGill Faculty of Medicine Internal Fellowship.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1262905.
References
- Afonina E., Neumann, M., and Pavlakis, G.N. 1997. Preferential binding of poly(A)-binding protein 1 to an inhibitory RNA element in the human immunodeficiency virus type 1 gag mRNA. J. Biol. Chem. 272: 2307–2311. [DOI] [PubMed] [Google Scholar]
- Bi X. and Goss, D.J. 2000. Wheat germ poly(A)-binding protein increases the ATPase and the RNA helicase activity of translation initiation factors eIF4A, eIF4B, and eIF-iso4F. J. Biol. Chem. 275: 17740–17746. [DOI] [PubMed] [Google Scholar]
- Byrd M.P., Zamora, M., and Lloyd, R.E. 2002. Generation of multiple isoforms of eukaryotic translation initiation factor 4GI by use of alternate translation initiation codons. Mol. Cell. Biol. 22: 4499–4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland P.R. and Wormington, M. 2001. The mechanism and regulation of deadenylation: Identification and characterization of Xenopus PARN. RNA 7: 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edery I., Humbelin, M., Darveau, A., Lee, K.A., Milburn, S., Hershey, J.W., Trachsel, H., and Sonenberg, N. 1983. Involvement of eukaryotic initiation factor 4A in the cap recognition process. J. Biol. Chem. 258: 11398–11403. [PubMed] [Google Scholar]
- Gallie D.R. 1991. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes & Dev. 5: 2108–2116. [DOI] [PubMed] [Google Scholar]
- ____. 1998. A tale of two termini: A functional interaction between the termini of an mRNA is a prerequisite for efficient translation initiation. Gene 216: 1–11. [DOI] [PubMed] [Google Scholar]
- Gingras A.-C., Raught, B., and Sonenberg, N. 1999. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68: 913–963. [DOI] [PubMed] [Google Scholar]
- Gray N.K. and Hentze, M.W. 1994. Iron regulatory protein prevents binding of the 43S translation pre-initiation complex to ferritin and eALAS mRNAs. EMBO J. 13: 3882–3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray N.K., Coller, J.M., Dickson, K.S., and Wickens, M. 2000. Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J. 19: 4723–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groft C.M. and Burley, S.K. 2002. Recognition of eIF4G by rotavirus NSP3 reveals a basis for mRNA circularization. Mol. Cell 9: 1273–1283. [DOI] [PubMed] [Google Scholar]
- Gross J.D., Moerke, N.J., von der Haar, T., Lugovskoy, A.A., Sachs, A.B., McCarthy, J.E., and Wagner, G. 2003. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell 115: 739–750. [DOI] [PubMed] [Google Scholar]
- Grosset C., Chen, C.Y., Xu, N., Sonenberg, N., Jacquemin-Sablon, H., and Shyu, A.B. 2000. A mechanism for translationally coupled mRNA turnover: Interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell 103: 29–40. [DOI] [PubMed] [Google Scholar]
- Haghighat A. and Sonenberg, N. 1997. eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J. Biol. Chem. 272: 21677–21680. [DOI] [PubMed] [Google Scholar]
- Hershey J.W.B. and Merrick, W.C. 2000. The pathway and mechanism of initiation of protein synthesis. In Translational control of gene expression (ed. M.B. Mathews), pp. 33–88. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Iizuka N., Najita, L., Franzusoff, A., and Sarnow, P. 1994. Capdependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol. Cell. Biol. 14: 7322–7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imataka H., Gradi, A., and Sonenberg, N. 1998. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 17: 7480–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson A. 1996. Poly(A) metabolism and translation: The closed-loop model. In Translational control (ed. N. Sonenberg), pp. 451–480. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Kahvejian A., Roy, G., and Sonenberg, N. 2001. The mRNA closed-loop model: The function of PABP and PABP-interacting proteins in mRNA translation. Cold Spring Harb. Symp. Quant. Biol. 66: 293–300. [DOI] [PubMed] [Google Scholar]
- Khaleghpour K., Kahvejian, A., De Crescenzo, G., Roy, G., Svitkin, Y.V., Imataka, H., O'Connor-McCourt, M., and Sonenberg, N. 2001a. Dual interactions of the translational repressor Paip2 with poly(A) binding protein. Mol. Cell. Biol. 21: 5200–5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaleghpour K., Svitkin, Y.V., Craig, A.W., DeMaria, C.T., Deo, R.C., Burley, S.K., and Sonenberg, N. 2001b. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol. Cell 7: 205–216. [DOI] [PubMed] [Google Scholar]
- Le H., Tanguay, R.L., Balasta, M.L., Wei, C.C., Browning, K.S., Metz, A.M., Goss, D.J., and Gallie, D.R. 1997. Translation initiation factors eIF-iso4G and eIF-4B interact with the poly(A)-binding protein and increase its RNA binding activity. J. Biol. Chem. 272: 16247–16255. [DOI] [PubMed] [Google Scholar]
- Lee K.A. and Sonenberg, N. 1982. Inactivation of cap-binding proteins accompanies the shut-off of host protein synthesis by poliovirus. Proc. Natl. Acad. Sci. 79: 3447–3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish H.F. and Rose, J.K. 1977. Relative importance of 7-methylguanosine in ribosome binding and translation of vesicular stomatitis virus mRNA in wheat germ and reticulocyte cell-free systems. J. Biol. Chem. 252: 1181–1188. [PubMed] [Google Scholar]
- Mangus D.A., Evans, M.C., and Jacobson, A. 2003. Poly(A)-binding proteins: Multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 4: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano J., Lomakin, I.B., Sonenberg, N., Pestova, T.V., Hellen, C.U., and Burley, S.K. 2001. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol. Cell 7: 193–203. [DOI] [PubMed] [Google Scholar]
- Merrick W.C. and Sonenberg, N. 1997. Assays for eukaryotic translation factors that bind mRNA. Methods 11: 333–342. [DOI] [PubMed] [Google Scholar]
- Michel Y.M., Poncet, D., Piron, M., Kean, K.M., and Borman, A.M. 2000. Cap-poly(A) synergy in mammalian cell-free extracts. Investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J. Biol. Chem. 275: 32268–32276. [DOI] [PubMed] [Google Scholar]
- Munroe D. and Jacobson, A. 1990. mRNA poly(A) tail, a 3′ enhancer of translational initiation. Mol. Cell. Biol. 10: 3441–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostareck D.H., Ostareck-Lederer, A., Wilm, M., Thiele, B.J., Mann, M., and Hentze, M.W. 1997. mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell 89: 597–606. [DOI] [PubMed] [Google Scholar]
- Ostareck D.H., Ostareck-Lederer, A., Shatsky, I.N., and Hentze, M.W. 2001. Lipoxygenase mRNA silencing in erythroid differentiation: The 3′UTR regulatory complex controls 60S ribosomal subunit joining. Cell 104: 281–290. [DOI] [PubMed] [Google Scholar]
- Otero L.J., Ashe, M.P., and Sachs, A.B. 1999. The yeast poly(A)-binding protein Pab1p stimulates in vitro poly(A)-dependent and cap-dependent translation by distinct mechanisms. EMBO J. 18: 3153–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause A., Methot, N., Svitkin, Y., Merrick, W.C., and Sonenberg, N. 1994. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 13: 1205–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J. and Sonenberg, N. 1985. Photochemical cross-linking of cap binding proteins to eucaryotic mRNAs: Effect of mRNA 5′ secondary structure. Mol. Cell. Biol. 5: 3222–3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piron M., Vende, P., Cohen, J., and Poncet, D. 1998. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J. 17: 5811–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncet D., Aponte, C., and Cohen, J. 1993. Rotavirus protein NSP3 (NS34) is bound to the 3′ end consensus sequence of viral mRNAs in infected cells. J. Virol. 67: 3159–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A. 2000. Physical and functional interactions between the mRNA cap structure and the poly(A) tail. in Translational control of gene expression (ed. M.B. Mathews), pp. 447–466. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sachs A.B. and Davis, R.W. 1989. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell 58: 857–867. [DOI] [PubMed] [Google Scholar]
- Searfoss A., Dever, T.E., and Wickner, R. 2001. Linking the 3′ poly(A) tail to the subunit joining step of translation initiation: Relations of pab1p, eukaryotic translation initiation factor 5b (Fun12p), and Ski2p–Slh1p. Mol. Cell. Biol. 21: 4900–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N. 1981. ATP/Mg++-dependent cross-linking of cap binding proteins to the 5′ end of eukaryotic mRNA. Nucleic Acids Res. 9: 1643–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N., Morgan, M.A., Merrick, W.C., and Shatkin, A.J. 1978. A polypeptide in eukaryotic initiation factors that crosslinks specifically to the 5′-terminal cap in mRNA. Proc. Natl. Acad. Sci. 75: 4843–4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin Y.V. and Agol, V.I. 1978. Complete translation of encephalomyocarditis virus RNA and faithful cleavage of virus-specific proteins in a cell-free system from Krebs-2 cells. FEBS Lett. 87: 7–11. [DOI] [PubMed] [Google Scholar]
- Svitkin Y.V. and Sonenberg, N. 2004. An efficient system for cap- and poly(A)-dependent translation in vitro. In mRNA processing and metabolism (ed. D.R. Schoenberg), pp. 155–170. Humana Press Inc., Totowa, NJ. [DOI] [PubMed]
- Svitkin Y.V., Imataka, H., Khaleghpour, K., Kahvejian, A., Liebig, H.D., and Sonenberg, N. 2001a. Poly(A)-binding protein interaction with elF4G stimulates picornavirus IRES-dependent translation. RNA 7: 1743–1752. [PMC free article] [PubMed] [Google Scholar]
- Svitkin Y.V., Pause, A., Haghighat, A., Pyronnet, S., Witherell, G., Belsham, G.J., and Sonenberg, N. 2001b. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA 7: 382–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun S.Z. and Sachs, A.B. 1995. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes & Dev. 9: 2997–3007. [DOI] [PubMed] [Google Scholar]
- ____. 1996. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 15: 7168–7177. [PMC free article] [PubMed] [Google Scholar]
- von Der Haar T., Ball, P.D., and McCarthy, J.E. 2000. Stabilization of eukaryotic initiation factor 4E binding to the mRNA 5′-cap by domains of eIF4G. J. Biol. Chem. 275: 30551–30555. [DOI] [PubMed] [Google Scholar]
- Wakiyama M., Imataka, H., and Sonenberg, N. 2000. Interaction of eIF4G with poly(A)-binding protein stimulates translation and is critical for Xenopus oocyte maturation. Curr. Biol. 10: 1147–1150. [DOI] [PubMed] [Google Scholar]
- Wei C.C., Balasta, M.L., Ren, J., and Goss, D.J. 1998. Wheat germ poly(A) binding protein enhances the binding affinity of eukaryotic initiation factor 4F and (iso)4F for cap analogues. Biochemistry 37: 1910–1916. [DOI] [PubMed] [Google Scholar]