Abstract
Electrostatic effects are ubiquitous in protein interactions and are found to be pervasive in the complement system as well. The interaction between complement fragment C3d and complement receptor 2 (CR2) has evolved to become a link between innate and adaptive immunity. Electrostatic interactions have been suggested to be the driving factor for the association of the C3d:CR2 complex. In this study, we investigate the effects of ionic strength and mutagenesis on the association of C3d:CR2 through Brownian dynamics simulations. We demonstrate that the formation of the C3d:CR2 complex is ionic strength-dependent, suggesting the presence of long-range electrostatic steering that accelerates the complex formation. Electrostatic steering occurs through the interaction of an acidic surface patch in C3d and the positively charged CR2 and is supported by the effects of mutations within the acidic patch of C3d that slow or diminish association. Our data are in agreement with previous experimental mutagenesis and binding studies and computational studies. Although the C3d acidic patch may be locally destabilizing because of unfavorable Coulombic interactions of like charges, it contributes to the acceleration of association. Therefore, acceleration of function through electrostatic steering takes precedence to stability. The site of interaction between C3d and CR2 has been the target for delivery of CR2-bound nanoparticle, antibody, and small molecule biomarkers, as well as potential therapeutics. A detailed knowledge of the physicochemical basis of C3d:CR2 association may be necessary to accelerate biomarker and drug discovery efforts.
Introduction
Electrostatics plays an important role in accelerating biomolecular reactions, such as diffusional encounters and catalytic processes.1−5 Both long-range and short-range electrostatic interactions have been shown to affect the protein–protein association rate6−8 and have demonstrated important contributions to improving the efficiency of enzymatic reactions, often by several orders of magnitude.9,10 An area of research that has received significant attention in recent computational studies is the role of electrostatics in the function and regulation of the complement system, as well as the electrostatic mechanisms that bacterial and viral proteins have evolved to infiltrate host cells or evade the immune system.11−20
The complement system is a vital component of innate immunity, acting as a rapid-response surveillance system that identifies and eliminates, or contributes to the elimination of foreign pathogens through the processes of inflammation, opsonization, phagocytosis, and direct cell lysis.21 In addition, the complement system contributes to clearance of apoptotic cells, damaged cells and cellular debris, and immune complexes.22−26 The complement system is tightly regulated to discriminate self-from nonself,27 and when such regulation fails, the complement system contributes to autoimmune and inflammatory diseases.28−30 Overall, the complement system senses and responds to danger signals, contributing to host homeostasis.23,25
A direct result of complement activation is the role it plays as a link between innate and adaptive immunity, through the interaction between complement fragment C3d and complement receptor 2 (CR2). This is a property of mammals and higher species, because invertebrates have complement immune response but lack adaptive immunity. The formation of the C3d:CR2 complex contributes to the formation of the B cell receptor–coreceptor complex and to the enhancement of B cell-mediated antibody production by up to 3–4 orders of magnitude.31−33 Due to the importance of the C3d:CR2 complex to the development of autoantibodies, the C3d:CR2 interaction is also implicated in the pathology of autoimmune and inflammatory diseases. Thus, a comprehensive understanding of the nature of the C3d:CR2 interaction not only will contribute to mechanistic knowledge of a fundamental immune response process, but also can serve as the basis for improvements in therapeutic development.
Complement activation occurs through three different pathways: the classical, alternative, and lectin pathways. All three pathways converge at complement component C3.34 Complement C3 undergoes a series of cleavage steps that activate, inactivate, and redirect its activation. The first cleavage step produces the opsonin fragment C3b that covalently (through a thioester bond) attaches to pathogens and other surfaces, tagging them for recognition and elimination by phagocytic cells, and the fragment C3a that contributes to inflammatory response and phagocytosis. The second cleavage step produces the so-called inactivated C3b, iC3b, which also contributes to phagocytosis. The final cleavage steps produce C3c and C3dg, which is immediately transformed to C3d. Although C3d is the final cleavage product that remains on cell surfaces for the life of the cell, it is not just a degradation product. Nature has evolved mechanisms that utilize antigen-bound C3d and B cell expressed CR2 as a site of interaction between innate and adaptive immunity. B cell expressed antibodies also opsonize pathogens by binding to antigens on pathogen surfaces. Thus, the combined function of B cell bound antibodies and the CR2-C3d complex cross-link B cells to pathogens, forming the so-called B cell receptor (antibody)–coreceptor (CR2) complex. This cross-linking initiates a cascade of intracellular signaling reactions, involving protein kinases.
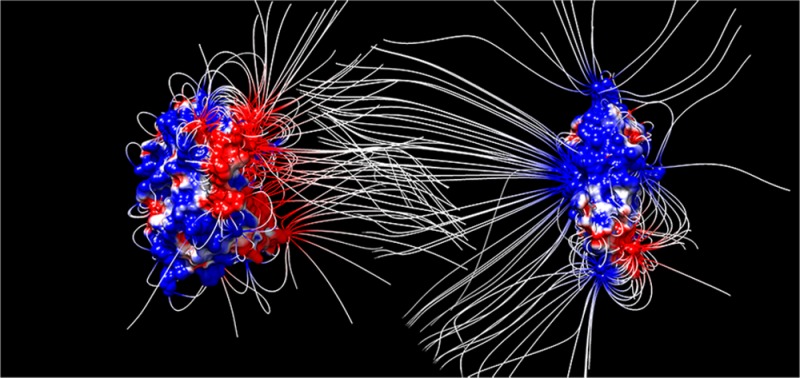
Several structural and computational studies have proposed that the interaction between C3d and CR2 is predominantly electrostatic in nature,35−39 occurring through a negatively charged patch on a concave surface of C3d and the first two modules of CR2 that are positively charged (Figure 1). Experimental studies involving mutagenesis, pH, and ionic strength effects are in agreement with the dominant role of electrostatics in the association between C3d and CR2.35,36,40−42 Although there was ambiguity and controversy for several years because of an older nonphysiological crystallographic structure of the C3d:CR2 complex, this controversy is now resolved with new crystallographic, mutagenesis and binding, and computational data.19,36,41,43 A recent study has evaluated the physicochemical origins and strength of the C3d:CR2 interaction, using the physiological and the controversial crystallographic structures, and has demonstrated the electrostatic mechanism of binding.19 This and earlier studies37−39 have proposed a two-step model for C3d:CR2 association, consisting of recognition and binding for highly and oppositely charged proteins. This model was based on earlier work on electrostatic steering in enzymatic reactions and protein interactions by McCammon and co-workers.1−10,44−46 During the recognition step, long-range electrostatic interactions between protein macrodipoles accelerate the formation of a transient encounter complex, followed by the binding step, which is marked by the stabilization of the bound complex through short-range, pairwise polar and nonpolar interactions and entropic effects.
Figure 1.
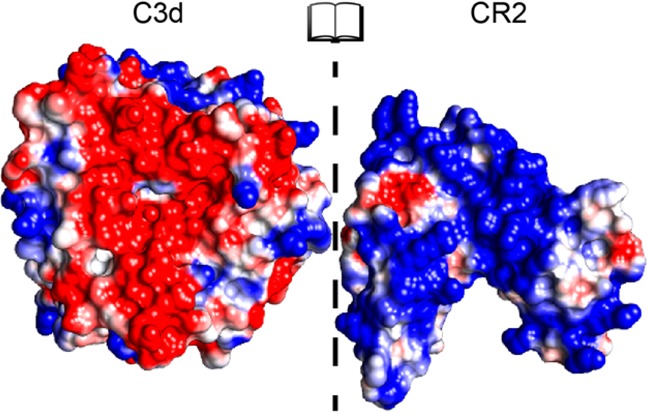
Electrostatic potentials mapped onto the protein surfaces of C3d and CR2 in open book representation. Electrostatic potentials were calculated at ionic strength corresponding to 150 mM monovalent counterion concentration. The color transitions from red to white to blue represent electrostatic potential values of −5 kT/e to 0 kT/e to 5 kT/e.
Another recent study used the concept of “electrostatic hotspots” to evaluate the origin of the C3d–CR2 interaction throughout evolution.47 An “electrostatic hotspot” was defined as a surface patch of like-charged residues that is resistant to perturbation. Such hotspots contribute to the formation of the encounter complex and rapid association. Because of the high concentration of like charges, a hotspot forms an unfavorable electrostatic environment, which is amenable to the formation of favorable interactions with proteins that have areas with complementary charges when such an encounter occurs. The study was performed using C3d sequences from 24 species, homology modeling based on the most recent (physiological) crystallographic structure, perturbations based on alanine scan of ionizable residues and molecular dynamics simulations, Poisson–Boltzmann electrostatic calculations, and electrostatic potential similarity clustering. The study proposed that C3d has two “electrostatic hotspots” located at opposite faces; one hotspot is predominantly positively charged and contains the thioester bond that opsonizes pathogen surfaces, and the other hotspot is predominantly negatively charged and forms the concave surface that is the site of interaction with CR2. The study concluded that the appearance of the negatively charged hotspot coincides with the onset of adaptive immunity at the level of jawless fish and beyond, and it is stronger in mammals, but it is not present in invertebrates. Therefore, C3d evolved its electrostatic properties to acquire the negatively charged hotspot to interact with the positively charged CR2, resulting to enhancement of adaptive immunity.
Given the large number of background studies on the electrostatic character of the C3d-CR2 interaction, and that the formation of the encounter complex is a diffusion-limited process,48 we initiated a Brownian dynamics (BD) simulation study to evaluate kon reaction rate constants for C3d:CR2 complexes. We perform our study for native C3d and CR2, as well as for a number of mutants with available experimental binding data. We evaluate the ionic strength dependence of the C3d:CR2 interaction to investigate the impact of salt concentration on electrostatic screening. We utilize computational mutagenesis to elucidate the contributions of specific mutants, with known experimental binding data, to the association of the C3d:CR2 complex. We demonstrate the inverse relationship between the association rate constant and ionic strength.We also find that mutations of residues shown to enhance or hinder binding in experimental binding data35,40,41 result in slower or higher association rate constants, respectively. The examined mutations involved ionizable residues at the binding interface and were introduced to disrupt association. Our results are in agreement with the experimental data, as well as with a previous computational study,19 and they indicate that the electrostatic steering accelerates the interaction between C3d and CR2.
Methods
Protein Structure Preparation
We utilized the more recent crystallographic structure of the C3d:CR2 complex (Protein Data Bank, PDB, code: 3OED)36 in our study. From the three-dimensional coordinates of this structure, we used chains A and C, corresponding to C3d and CR2, respectively, as they had better electron density and lower B-factor compared to chains B and D of another complex present in the structure. It should be noted that the structure of CR2 contains only the two modules that contact C3d, SCR1, and SCR2, out of a total of 15 or 16 SCR modules. C3d consists of 292 amino acids with a net charge of −1e while CR2 consists of 130 amino acids with a net charge of +8e. To alleviate crystal packing effects in the crystallographic structure, 25 000 steps of conjugate-gradient energy minimization were performed using NAMD.49
Subsequent to energy minimization, missing hydrogens, atomic radii, and partial charges were added to the coordinates of the structure, using PDB2PQR version 2.050 and the PARSE force field,51 thus converting the PDB file to a PQR file. No atypical protonation states were observed using PROPKA.52,53 Histidine residues were neutral with a hydrogen attached to Nδ1 atom. Computational mutagenesis was performed on the protein complex using the analysis of electrostatic similarities of proteins (AESOP) computational framework.15,39,54,55 Mutants were chosen from prior literature that had reported experimental binding data.35,40,41 Electrostatic potentials were calculated for the parent (wild-type) and mutated protein complexes using the Adaptive Poisson–Boltzmann Solver (APBS) version 1.4.56 The number of grid points was set to 129 × 161 × 161. Coarse and fine mesh dimensions were set to 1000 Å × 1000 Å × 1000 and 150 Å × 150 Å × 150 Å, respectively, as discussed previously.54 Protein and solvent dielectric values were set to 20 and 78.54, respectively.54 Ionic strengths corresponding to monovalent counterion concentrations of 50, 75, 100, 125, 150, 200, and 300 mM were used for the evaluation of ionic strength dependence of parent C3d:CR2 and the alanine scan mutants. An ionic concentration of 150 mM was used in the electrostatic analysis of the alanine scan mutants.
Brownian Dynamics Simulations
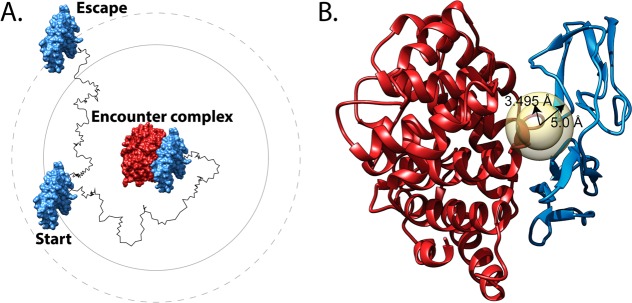
Brownian dynamics simulations and the corresponding rate calculations were performed using the BrownDye package,57 according to the Northrup–Allison–McCammon algorithm,44 which is based on the original Brownian dynamics algorithm by Ermak and McCammon.58 The two molecules start separated at a center-to-center radius (represented by the inner circle in Figure 2A), and the simulation progresses until termination due to formation of the encounter complex or due to the molecule reaching an escape radius (represented by the outer circle in Figure 2A). The center-to-center radius is calculated by BrownDye for each system, and through an improvement to the Northrup–Allison–McCammon algorithm, the escape radius is no longer a necessary input.59 Pairwise residue interactions with cutoff distances of 5.0 Å were calculated from the PQR files generated as described above. Criteria for a successful reaction required that at least two of the atom pairs from the calculated list of pairwise interactions approach within 3.495 Å of each other (Figure 2B).
Figure 2.
Schematic and molecular graphics illustrating the Brownian dynamics (BD) simulation of C3d:CR2 and the corresponding reaction criteria with C3d and CR2 in blue and red, respectively. (A) At the beginning of the BD simulation, CR2 starts at a center-to-center radius away from C3d, as represented by the inner circle. The simulation terminates when either the formation of the encounter complex occurs or if CR2 reaches an escape radius as represented by the outer circle. (B) The concentric circles represent the reaction criteria for the C3d:CR2 BD simulations where 5 Å is the cutoff for determining potential pairwise residue interactions between C3d and CR2. The circle with radius 3.495 Å represents the distance within which at least two atom pairs of the previously determined pairwise residue interactions must occur for a successful reaction.
Reaction criteria were selected to match known rate constants of C3d:CR2 association. Initially, the BrownDye program rates_of_distances was utilized to generate a list of reaction constants corresponding to minimum reaction distances. Then, the criteria were fine-tuned to the known association rate constant at 125 mM NaCl from experimental data.40 There are two experimental association rate constants, at 50 and 125 mM NaCl ionic strength, and we chose for calibration the value at 125 mM because of its proximity to the physiological ionic strength of 150 mM. Additional input files were generated using the program bd_top. BD simulations were carried out using the weighted-ensemble method to account for low probabilities of reactions.60 The association rate constant, kon, and corresponding reaction probabilities were calculated using weighted-ensemble simulations carried out for 2 000 000 steps with 200 copies of each system to guarantee convergence of the results. The acceleration of protein–protein association, as well as enzymatic reactions, by electrostatic steering has been explored previously, which suggested the feasibility of this study.61,62
Results and Discussion
Ionic Strength Dependence of Association
The weighted-ensemble BD simulations of C3d:CR2 demonstrate that under constraints of reaction criteria of 3.495 Å and at least two successful pairwise interactions, the association rate constant decreases with increasing ionic strength (Figure 3), which is expected when ionic screening of Coulombic interactions is present. The experimental data utilized for calibration of the reaction criteria is plotted in Figure 3 as well. Additionally, Figure 3 shows the ionic strength dependence of the Debye length, suggesting the importance of ionic screening for association. Both the ionic strength dependence and Debye length curves follow similar trends.
Figure 3.
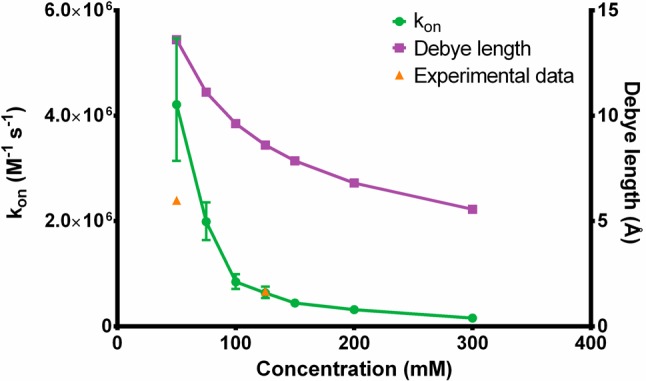
Association rate constant of CR2 binding to C3d and the calculated Debye length at varying ionic strengths. The mean association rate constant is represented by a green circle with 95% confidence intervals represented as error bars. The Debye length at each ionic strength is represented as a purple square. Experimentally known association rate constants at two ionic strengths, 50 mM and 125 mM NaCl, are plotted and represented as orange triangles.40 Note: the experimental data point at 125 mM was utilized for calibration of the BD simulation reaction criteria.
Because C3d and CR2 have surfaces that are both highly and oppositely charged (Figure 1), we expect that association follows the two-step model of recognition (formation of the intermediate encounter complex) and binding (formation of the final bound complex). This is demonstrated by the ionic strength dependence of the association, which is possible if electrostatics drives association. From our results, the acceleration of the interaction through electrostatic steering underscores the plausibility of the model. The kinetic rate constants acquired through these BD simulations reflect the recognition step, given the calculation limitation, such as the rigid body assumption of the protein structures and lack of modeling of short-range interactions such as salt bridges, van der Waals forces, and hydrogen bonding.63 These limitations may affect only the binding step of the aforementioned two-step model of protein–protein association and may not be necessary for the evaluation of electrostatic steering during the diffusion-limited recognition step.64
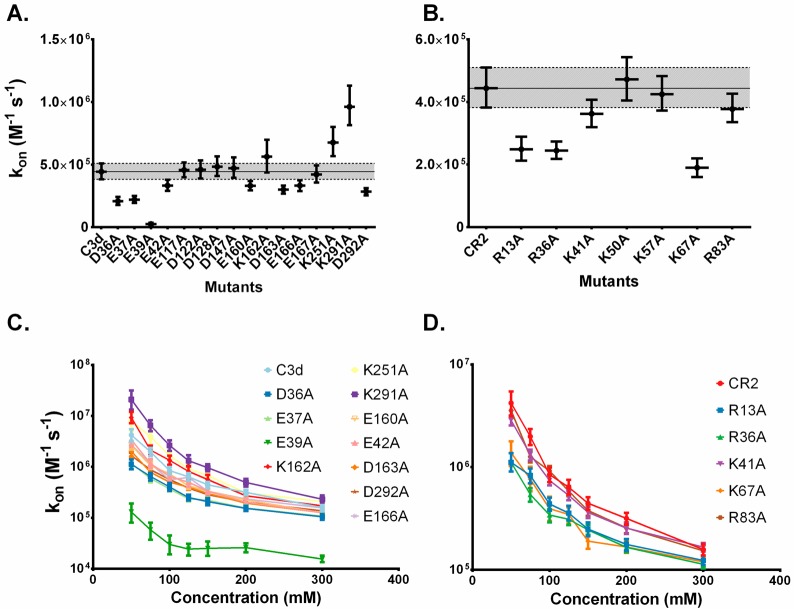
Impact of Specific Residues on Electrostatic Steering
We investigated the effects of electrostatic steering when various C3d:CR2 residues are mutated. Computational mutations were chosen from available experimental data and BD simulations were performed for each mutant system. We observe that mutations of certain residues result in a decrease in the association rate constant, or in other words, the residue is significant to the formation of the encounter complex of C3d:CR2 (Figure 4A,B). The reverse holds true as well where mutations of residues resulting in an increase in the association rate constant signify that the residue hinders the formation of the encounter complex. These results are in line with a previous computational alanine scan and electrostatic analysis study using AESOP19 with a few small discrepancies such as K251A on C3d and R83A on CR2, which are close to the threshold of the error. Thus, we establish that the acceleration of the formation of the encounter complex due to electrostatic steering is affected by individual charged residues contributions as well.
Figure 4.
Effects of mutagenesis on association rate constant and ionic strength dependence. (A, B) The mean association rate at 150 mM ionic strength is plotted for each mutant for C3d and CR2 with 95% confidence intervals represented as error bars. The shaded region represents the upper and lower bound threshold of the association rate constant values of the parent (C3d or CR2). Computational mutations were performed based on previous experimentally generated mutants.35,40,41 (C, D) Ionic strength dependence of selected mutants demonstrating significant deviations from the wild type association rate constant. The mutant notation denotes the residue number surrounded by the replaced residue on the left and the replacing residue on the right.
Mutants displaying significant variance from the wild-type (outside the shaded area of Figure 4A,B) were selected for ionic strength dependence analysis (Figure 4C,D). We find that the mutants exhibit ionic strength dependence as well, and the trends are overall in line with what we would expect from the mutagenesis analysis of Figure 4A,B. As expected from the mutagenesis analysis, certain mutants demonstrate more drastic electrostatic steering, suggesting that they play a more significant role in the formation of the encounter complex.
Comparison with Prior Experimental and Computational Studies
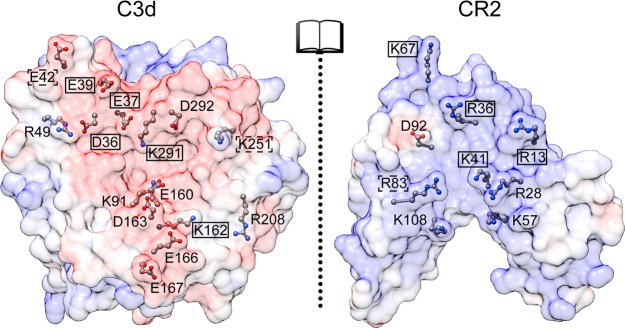
A recent computational study investigating the binding mode of C3d:CR2 using molecular dynamics (MD) simulations (both explicit-solvent and steered), including MM-GBSA analysis, and electrostatic calculations, including AESOP alanine scan analysis, quantifies why the acidic patch on C3d plays a key role in driving the C3d:CR2 interaction.19 In particular, two clusters of C3d residues (D36, E37, and E39; E160, K162, D163, E166, and E167) demonstrate significant contributions to electrostatic interactions, intermolecular interaction occupancies (hydrogen bonds, salt bridges, and nonpolar interactions) and steered MD (SMD) simulations. The computational study also emphasizes the importance of both the SCR1 and SCR2 domain to the stability and energetics of the complex. This is supported by the electrostatic and MD simulation analysis and also was strongly suggested by the slow unbinding of the SCR1 domain in SMD simulations, in contrast to the SCR2 domain.
Our results are generally in agreement with prior experimental and computational data. C3d residues at the acidic patch such as D36, E37, and E39 (Figure 5) were suggested to be important to binding by a 2000 rosette immunological assay study35 and a 2010 surface plasmon resonance study,41 in addition to the AESOP analysis.19 Our BD results support the importance of D36, E37, and E39 in binding as well. C3d residues E117, D122, D128, and D147, a cluster of residues not located at the acidic patch, demonstrated minimal contributions to electrostatic steering, in contrast to a mutagenesis study40 performed by authors of the older crystallographic C3d:CR2 structure.65 This is in line with the previous computational study19 and the 2000 and 2010 mutagenesis studies.35,41
Figure 5.
Molecular graphic of C3d:CR2 in open-book form and locations of residues with significant contributions to electrostatic steering. The surfaces of C3d and CR2 are rendered translucent and colored from red to white to blue according to electrostatic potential values (calculated at 150 mM ionic strength) from −5 kT/e to 0 kT/e to 5 kT/e. Residues are displayed in ball-and-stick form with atoms colored according to atom type (carbon in gray, oxygen in red, nitrogen in blue). Residues found to demonstrate significant contributions to electrostatic steering (falling outside the shaded area in Figure 4A,B) in this ionic strength dependence analysis and significant contributions to electrostatic interactions (greater or less than ±2.5 kJ/mol) in a previous AESOP computational alanine scan study19 are labeled with a solid black box. Residues found to demonstrate significant contributions to electrostatic steering in this ionic strength dependence analysis but less significant interactions (within ±2.5 kJ/mol) in the AESOP computational alanine scan study are labeled with a dashed black box. Additional residues found to have significant contributions to electrostatic interactions in the AESOP computational alanine scan study are labeled without a box.
C3d residues at the other acidic patch cluster (E160, D163, and E166) demonstrated significant contributions to electrostatic steering, in line with the previous computational study,19 but are at odds with the 2000 mutagenesis study. However, the influence of electrostatic steering of C3d residues K162 and E167 are in contrast to the previous computational study19 and 2000 mutagenesis study.35 Here, we found that removal of K162, which is located within the acidic patch of C3d, did not affect the electrostatic steering of the formation of the encounter complex as dramatically as it was suggested by the results of the 2000 rosette study35 and the previous AESOP study.19 It is likely that the role of K162 is to stabilize the negative surface patch, and its removal produces local structural rearrangements to optimize the remaining electrostatic interactions. Such structural effects are not taken into account by the rigid model BD and AESOP studies. We should keep in mind that the BD and AESOP analyses introduce theoretical perturbation to assess the importance of each mutated residue in binding of the parent proteins, and they do not aim to model the structures of the mutated proteins.
The importance of the SCR1 domain of CR2, which was overlooked by the original C3d:CR2 crystallographic structure, is further emphasized in our BD results, as evidenced by the effects of mutagenesis and electrostatic steering by CR2 residues R13, R36, and K41. This is in line with the previous computational study where it was established that the contributions of the SCR1 domain to the C3d:CR2 interface are significant.19
Pharmacological Significance
Understanding the physicochemical origins of the C3d:CR2 interaction is important for the design of biomarkers and therapeutic interventions. Recent studies have utilized information from the C3d–CR2 interaction to design C3d-binding biomarkers for imaging of complement activation, such as fluorescently labeled antibodies,66 small fluorescent molecules,67 and CR2-bound iron nanoparticles.68
The complement system has been a target of inhibition in several studies, but only two anticomplement drugs are currently in the clinic.69 Although, a lack of proper regulation of complement response has been implicated in several autoimmune and inflammatory diseases, inhibiting complement activation may reduce the efficacy of response to infection and injury. Recent studies have demonstrated in animal models effective targeted delivery of CR2-attached protein complement inhibitors, through the CR2:C3d interaction, to sites of local inflammation, which are abundant of C3d-opsonized tissues.70−75 Such inhibitors included fragments of the alternative pathway regulator Factor H, or the complement inhibitory protein Crry, but can also be low-affinity C3d-bound or CR2-bound peptidic or nonpeptidic molecules. Therefore, the C3d:CR2 interaction is an ideal target for therapeutic development because it allows for inhibition of complement response through the alternative pathway while retaining the ability to fight infection through the lectin and classical pathways. The C3d:CR2 interaction can also be a target for therapeutic intervention in cases of autoimmunity because it allows for inhibition of complement-enhanced adaptive immune response, through the inhibition of the formation of the B cell receptor–coreceptor complex.
Conclusion
We have investigated the role of electrostatic steering on the C3d:CR2 interaction, using BD simulations. We demonstrate that the predicted kon reaction rate constant depends on ionic strength, which is possible only if electrostatics contributes significantly to the C3d:CR2 interaction. We have also evaluated the contributions of specific ionizable residues to the electrostatic acceleration of the interaction through computational mutagenesis and ionic strength dependence analysis. We demonstrate that computational mutations of ionizable residues previously known from experimental studies to be significant to the C3d:CR2 interaction result in a reduced kon rate constant. Therefore, the replaced (original) residues contribute to the acceleration of the interaction in the native complex. These results are in agreement with a previous computational analysis, based on the calculation of electrostatic free energies of association for a family of experimentally known mutants and an alanine scan family of C3d:CR2 complexes.19 Interestingly, acidic residues within an evolutionarily significant “electrostatic hotspot” in C3d are the major contributors to the complex formation. As was suggested in a previous work, this acidic patch in the complement degradation product C3d may have evolved to establish a link between innate immunity (complement system) and adaptive immunity (B cell bound antibodies).47 Although this C3d “electrostatic hotspot” may be destabilizing local structure, it accelerates interaction with CR2 and function. As was suggested by Professor J. Andrew McCammon in his 2009 “Darwinian Biophysics” article, evolution favors speed and acceleration of function;76 and evolution favors function over stability.77
Acknowledgments
We are grateful to Professor Andrew McCammon for his seminal contributions to computational chemistry and biophysics and drug discovery. We are indebted to Professor McCammon for many years of mentorship in research (D.M. and G.A.H.) and during a stay at the McCammon lab in summer 2015 (R.R.M.) when this project was initiated. We thank the San Diego Supercomputing Center for financial support in the form of a summer fellowship (R.R.M.), which made this study possible. The work at the University of California, San Diego was supported in part by the National Institutes of Health and the National Biomedical Computation Resource.
Glossary
Abbreviations
- C3d
complement fragment 3d
- CR2
complement receptor 2
- SCR
short consensus repeat
- PDB
protein data bank
- BD
Brownian dynamics
- AESOP
analysis of electrostatic similarities of proteins computational framework
- APBS
adaptive Poisson–Boltzmann solver
The authors declare no competing financial interest.
References
- Sines J. J.; Allison S. A.; McCammon J. A. Point Charge Distributions and Electrostatic Steering in Enzyme/substrate Encounter: Brownian Dynamics of Modified Copper/zinc Superoxide Dismutases. Biochemistry 1990, 29, 9403–9412. 10.1021/bi00492a014. [DOI] [PubMed] [Google Scholar]
- Tan R. C.; Truong T. N.; McCammon J. A.; Sussman J. L. Acetylcholinesterase: Electrostatic Steering Increases the Rate of Ligand Binding. Biochemistry 1993, 32, 401–403. 10.1021/bi00053a003. [DOI] [PubMed] [Google Scholar]
- Gabdoulline R. R.; Wade R. C. Simulation of the Diffusional Association of Barnase and Barstar. Biophys. J. 1997, 72, 1917–1929. 10.1016/S0006-3495(97)78838-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elcock A. H.; McCammon J. A. Evidence for Electrostatic Channeling in a Fusion Protein of Malate Dehydrogenase and Citrate Synthase. Biochemistry 1996, 35, 12652–12658. 10.1021/bi9614747. [DOI] [PubMed] [Google Scholar]
- Gabdoulline R. R.; Wade R. C. Biomolecular Diffusional Association. Curr. Opin. Struct. Biol. 2002, 12, 204–213. 10.1016/S0959-440X(02)00311-1. [DOI] [PubMed] [Google Scholar]
- Elcock A. H.; Sept D.; McCammon J. A. Computer Simulation of Protein–Protein Interactions. J. Phys. Chem. B 2001, 105, 1504–1518. 10.1021/jp003602d. [DOI] [Google Scholar]
- Chang C.-E.; Shen T.; Trylska J.; Tozzini V.; McCammon J. A. Gated Binding of Ligands to HIV-1 Protease: Brownian Dynamics Simulations in a Coarse-Grained Model. Biophys. J. 2006, 90, 3880–3885. 10.1529/biophysj.105.074575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaar A.; Dammer C.; Gabdoulline R. R.; Wade R. C.; Helms V. Diffusional Encounter of Barnase and Barstar. Biophys. J. 2006, 90, 1913–1924. 10.1529/biophysj.105.075507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade R. C.; Luty B. A.; Demchuk E.; Madura J. D.; Davis M. E.; Briggs J. M.; McCammon J. A. Simulation of Enzyme–substrate Encounter with Gated Active Sites. Nat. Struct. Biol. 1994, 1, 65–69. 10.1038/nsb0194-65. [DOI] [PubMed] [Google Scholar]
- Metzger V. T.; Eun C.; Kekenes-Huskey P. M.; Huber G.; McCammon J. A. Electrostatic Channeling in P. Falciparum DHFR-TS: Brownian Dynamics and Smoluchowski Modeling. Biophys. J. 2014, 107, 2394–2402. 10.1016/j.bpj.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; Morikis D. Immunophysical Properties and Prediction of Activities for Vaccinia Virus Complement Control Protein and Smallpox Inhibitor of Complement Enzymes Using Molecular Dynamics and Electrostatics. Biophys. J. 2006, 90, 3106–3119. 10.1529/biophysj.105.068130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyaram K.; Kieslich C. A.; Yadav V. N.; Morikis D.; Sahu A. Influence of Electrostatics on the Complement Regulatory Functions of Kaposica, the Complement Inhibitor of Kaposi’s Sarcoma-Associated Herpesvirus. J. Immunol. 2010, 184, 1956–1967. 10.4049/jimmunol.0903261. [DOI] [PubMed] [Google Scholar]
- El-Assaad A. M.; Kieslich C. A.; Gorham R. D. Jr.; Morikis D. Electrostatic Exploration of the C3d–FH4 Interaction Using a Computational Alanine Scan. Mol. Immunol. 2011, 48, 1844–1850. 10.1016/j.molimm.2011.05.007. [DOI] [PubMed] [Google Scholar]
- Kieslich C. A.; Vazquez H.; Goodman G. N.; de Victoria A. L.; Morikis D. The Effect of Electrostatics on Factor H Function and Related Pathologies. J. Mol. Graphics Modell. 2011, 29, 1047–1055. 10.1016/j.jmgm.2011.04.010. [DOI] [PubMed] [Google Scholar]
- Gorham R. D.; Kieslich C. A.; Morikis D. Electrostatic Clustering and Free Energy Calculations Provide a Foundation for Protein Design and Optimization. Ann. Biomed. Eng. 2011, 39, 1252–1263. 10.1007/s10439-010-0226-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieslich C. A.; Tamamis P.; Gorham R. D. Jr.; Lopez de Victoria A.; Sausman N. U.; Archontis G.; Morikis D. Exploring Protein-Protein and Protein-Ligand Interactions in the Immune System Using Molecular Dynamics and Continuum Electrostatics. Curr. Phys. Chem. 2012, 2, 324–343. 10.2174/1877946811202040324. [DOI] [Google Scholar]
- Gorham R. D.; Rodriguez W.; Morikis D. Molecular Analysis of the Interaction between Staphylococcal Virulence Factor Sbi-IV and Complement C3d. Biophys. J. 2014, 106, 1164–1173. 10.1016/j.bpj.2014.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojha H.; Panwar H. S.; Gorham R. D.; Morikis D.; Sahu A. Viral Regulators of Complement Activation: Structure, Function and Evolution. Mol. Immunol. 2014, 61, 89–99. 10.1016/j.molimm.2014.06.004. [DOI] [PubMed] [Google Scholar]
- Mohan R. R.; Gorham R. D. Jr.; Morikis D. A Theoretical View of the C3d:CR2 Binding Controversy. Mol. Immunol. 2015, 64, 112–122. 10.1016/j.molimm.2014.11.006. [DOI] [PubMed] [Google Scholar]
- Harrison R. E. S.; Gorham R. D.; Morikis D. Energetic Evaluation of Binding Modes in the C3d and Factor H (CCP 19–20) Complex. Protein Sci. 2015, 24, 789–802. 10.1002/pro.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roozendaal R.; Carroll M. C. Complement Receptors CD21 and CD35 in Humoral Immunity. Immunol. Rev. 2007, 219, 157–166. 10.1111/j.1600-065X.2007.00556.x. [DOI] [PubMed] [Google Scholar]
- Mackay I. R.; Rosen F. S.; Walport M. J. Complement. N. Engl. J. Med. 2001, 344, 1058–1066. 10.1056/NEJM200104053441406. [DOI] [PubMed] [Google Scholar]
- Merle N. S.; Church S. E.; Fremeaux-Bacchi V.; Roumenina L. T. Complement System Part I – Molecular Mechanisms of Activation and Regulation. Front. Immunol. 2015, 6, 262. 10.3389/fimmu.2015.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merle N. S.; Noe R.; Halbwachs-Mecarelli L.; Fremeaux-Bacchi V.; Roumenina L. T. Complement System Part II: Role in Immunity. Front. Immunol. 2015, 6, 257. 10.3389/fimmu.2015.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel P. F.; Skerka C. Complement Regulators and Inhibitory Proteins. Nat. Rev. Immunol. 2009, 9, 729–740. 10.1038/nri2620. [DOI] [PubMed] [Google Scholar]
- Kolev M.; Friec G. L.; Kemper C. Complement — Tapping into New Sites and Effector Systems. Nat. Rev. Immunol. 2014, 14, 811–820. 10.1038/nri3761. [DOI] [PubMed] [Google Scholar]
- Fearon D. T. The Complement System and Adaptive Immunity. Semin. Immunol. 1998, 10, 355–361. 10.1006/smim.1998.0137. [DOI] [PubMed] [Google Scholar]
- Ricklin D.; Lambris J. D. Complement in Immune and Inflammatory Disorders: Pathophysiological Mechanisms. J. Immunol. 2013, 190, 3831–3838. 10.4049/jimmunol.1203487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszewski M. K.; Atkinson J. P. Complement Regulators in Human Disease: Lessons from Modern Genetics. J. Intern. Med. 2015, 277, 294–305. 10.1111/joim.12338. [DOI] [PubMed] [Google Scholar]
- Morgan B. P.; Harris C. L. Complement, a Target for Therapy in Inflammatory and Degenerative Diseases. Nat. Rev. Drug Discovery 2015, 14, 857–877. 10.1038/nrd4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll M. C. The Complement System in Regulation of Adaptive Immunity. Nat. Immunol. 2004, 5, 981–986. 10.1038/ni1113. [DOI] [PubMed] [Google Scholar]
- Carroll M. C.; Isenman D. E. Regulation of Humoral Immunity by Complement. Immunity 2012, 37, 199–207. 10.1016/j.immuni.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holers V. M. Complement and Its Receptors: New Insights into Human Disease. Annu. Rev. Immunol. 2014, 32, 433–459. 10.1146/annurev-immunol-032713-120154. [DOI] [PubMed] [Google Scholar]
- Zipfel P. F.Complement: The Alternative Pathway. In eLS; John Wiley & Sons, Ltd: Chichester, UK, 2015; pp 1–10. [Google Scholar]
- Clemenza L.; Isenman D. E. Structure-Guided Identification of C3d Residues Essential for Its Binding to Complement Receptor 2 (CD21). J. Immunol. 2000, 165, 3839–3848. 10.4049/jimmunol.165.7.3839. [DOI] [PubMed] [Google Scholar]
- van den Elsen J. M. H.; Isenman D. E. A Crystal Structure of the Complex Between Human Complement Receptor 2 and Its Ligand C3d. Science 2011, 332, 608–611. 10.1126/science.1201954. [DOI] [PubMed] [Google Scholar]
- Morikis D.; Lambris J. D. The Electrostatic Nature of C3d-Complement Receptor 2 Association. J. Immunol. 2004, 172, 7537–7547. 10.4049/jimmunol.172.12.7537. [DOI] [PubMed] [Google Scholar]
- Zhang L.; Mallik B.; Morikis D. Immunophysical Exploration of C3d–CR2(CCP1–2) Interaction Using Molecular Dynamics and Electrostatics. J. Mol. Biol. 2007, 369, 567–583. 10.1016/j.jmb.2007.02.101. [DOI] [PubMed] [Google Scholar]
- Kieslich C. A.; Morikis D.; Yang J.; Gunopulos D. Automated Computational Framework for the Analysis of Electrostatic Similarities of Proteins. Biotechnol. Prog. 2011, 27, 316–325. 10.1002/btpr.541. [DOI] [PubMed] [Google Scholar]
- Hannan J. P.; Young K. A.; Guthridge J. M.; Asokan R.; Szakonyi G.; Chen X. S.; Holers V. M. Mutational Analysis of the Complement Receptor Type 2 (CR2/CD21)–C3d Interaction Reveals a Putative Charged SCR1 Binding Site for C3d. J. Mol. Biol. 2005, 346, 845–858. 10.1016/j.jmb.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Isenman D. E.; Leung E.; Mackay J. D.; Bagby S.; van den Elsen J. M. H. Mutational Analyses Reveal That the Staphylococcal Immune Evasion Molecule Sbi and Complement Receptor 2 (CR2) Share Overlapping Contact Residues on C3d: Implications for the Controversy Regarding the CR2/C3d Cocrystal Structure. J. Immunol. 2010, 184, 1946–1955. 10.4049/jimmunol.0902919. [DOI] [PubMed] [Google Scholar]
- Toapanta F. R.; DeAlmeida D. R.; Dunn M. D.; Ross T. M. C3d Adjuvant Activity Is Reduced by Altering Residues Involved in the Electronegative Binding of C3d to CR2. Immunol. Lett. 2010, 129, 32–38. 10.1016/j.imlet.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikis D. F1000Prime Recommendation of [van den Elsen JM and Isenman DE, Science 2011, 332(6029):608–11]. F1000Prime.com/10371956#eval14018054 (accessed Sep 24, 2014). [DOI] [PubMed]
- Northrup S. H.; Allison S. A.; McCammon J. A. Brownian Dynamics Simulation of Diffusion-influenced Bimolecular Reactions. J. Chem. Phys. 1984, 80, 1517–1524. 10.1063/1.446900. [DOI] [Google Scholar]
- Gabdoulline R. R.; Wade R. C. On the Protein-Protein Diffusional Encounter Complex. J. Mol. Recognit. 1999, 12, 226–234. . [DOI] [PubMed] [Google Scholar]
- Spaar A.; Helms V. Ionic Strength Effects on the Association Funnel of Barnase and Barstar Investigated by Brownian Dynamics Simulations. J. Non-Cryst. Solids 2006, 352, 4437–4444. 10.1016/j.jnoncrysol.2006.03.117. [DOI] [Google Scholar]
- Kieslich C. A.; Morikis D. The Two Sides of Complement C3d: Evolution of Electrostatics in a Link between Innate and Adaptive Immunity. PLoS Comput. Biol. 2012, 8, e1002840. 10.1371/journal.pcbi.1002840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elcock A. H. Molecular Simulations of Diffusion and Association in Multimacromolecular Systems. Methods Enzymol. 2004, 383, 166–198. 10.1016/S0076-6879(04)83008-8. [DOI] [PubMed] [Google Scholar]
- Phillips J. C.; Braun R.; Wang W.; Gumbart J.; Tajkhorshid E.; Villa E.; Chipot C.; Skeel R. D.; Kalé L.; Schulten K. Scalable Molecular Dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky T. J.; Nielsen J. E.; McCammon J. A.; Baker N. A. PDB2PQR: An Automated Pipeline for the Setup of Poisson-Boltzmann Electrostatics Calculations. Nucleic Acids Res. 2004, 32, W665–W667. 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitkoff D.; Sharp K. A.; Honig B. Accurate Calculation of Hydration Free Energies Using Macroscopic Solvent Models. J. Phys. Chem. 1994, 98, 1978–1988. 10.1021/j100058a043. [DOI] [Google Scholar]
- Olsson M. H. M.; Søndergaard C. R.; Rostkowski M.; Jensen J. H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. J. Chem. Theory Comput. 2011, 7, 525–537. 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- Søndergaard C. R.; Olsson M. H. M.; Rostkowski M.; Jensen J. H. Improved Treatment of Ligands and Coupling Effects in Empirical Calculation and Rationalization of pKa Values. J. Chem. Theory Comput. 2011, 7, 2284–2295. 10.1021/ct200133y. [DOI] [PubMed] [Google Scholar]
- Gorham R. D.; Kieslich C. A.; Nichols A.; Sausman N. U.; Foronda M.; Morikis D. An Evaluation of Poisson-Boltzmann Electrostatic Free Energy Calculations through Comparison with Experimental Mutagenesis Data. Biopolymers 2011, 95, 746–754. 10.1002/bip.21644. [DOI] [PubMed] [Google Scholar]
- Kieslich C. A.; Gorham R. D. Jr.; Morikis D. Is the Rigid-Body Assumption Reasonable?: Insights into the Effects of Dynamics on the Electrostatic Analysis of Barnase–barstar. J. Non-Cryst. Solids 2011, 357, 707–716. 10.1016/j.jnoncrysol.2010.05.087. [DOI] [Google Scholar]
- Baker N. A.; Sept D.; Joseph S.; Holst M. J.; McCammon J. A. Electrostatics of Nanosystems: Application to Microtubules and the Ribosome. Proc. Natl. Acad. Sci. U. S. A. 2001, 98, 10037–10041. 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber G. A.; McCammon J. A. Browndye: A Software Package for Brownian Dynamics. Comput. Phys. Commun. 2010, 181, 1896–1905. 10.1016/j.cpc.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermak D. L.; McCammon J. A. Brownian Dynamics with Hydrodynamic Interactions. J. Chem. Phys. 1978, 69, 1352–1360. 10.1063/1.436761. [DOI] [Google Scholar]
- Luty B. A.; McCammon J. A.; Zhou H.-X. Diffusive Reaction Rates from Brownian Dynamics Simulations: Replacing the Outer Cutoff Surface by an Analytical Treatment. J. Chem. Phys. 1992, 97, 5682–5686. 10.1063/1.463777. [DOI] [Google Scholar]
- Huber G. A.; Kim S. Weighted-Ensemble Brownian Dynamics Simulations for Protein Association Reactions. Biophys. J. 1996, 70, 97–110. 10.1016/S0006-3495(96)79552-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabdoulline R. R.; Wade R. C. Brownian Dynamics Simulation of Protein–Protein Diffusional Encounter. Methods 1998, 14, 329–341. 10.1006/meth.1998.0588. [DOI] [PubMed] [Google Scholar]
- Huang Y. M.; Huber G.; Andrew McCammon J. Electrostatic Steering Enhances the Rate of cAMP Binding to Phosphodiesterase: Brownian Dynamics Modeling: Brownian Dynamics Modeling. Protein Sci. 2015, 24, 1884–1889. 10.1002/pro.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Votapka L. W.; Amaro R. E. Multiscale Estimation of Binding Kinetics Using Brownian Dynamics, Molecular Dynamics and Milestoning. PLoS Comput. Biol. 2015, 11, e1004381. 10.1371/journal.pcbi.1004381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaar A.; Helms V. Free Energy Landscape of Protein–Protein Encounter Resulting from Brownian Dynamics Simulations of Barnase:Barstar. J. Chem. Theory Comput. 2005, 1, 723–736. 10.1021/ct050036n. [DOI] [PubMed] [Google Scholar]
- Szakonyi G.; Guthridge J. M.; Li D.; Young K.; Holers V. M.; Chen X. S. Structure of Complement Receptor 2 in Complex with Its C3d Ligand. Science 2001, 292, 1725–1728. 10.1126/science.1059118. [DOI] [PubMed] [Google Scholar]
- Thurman J. M.; Kulik L.; Orth H.; Wong M.; Renner B.; Sargsyan S. A.; Mitchell L. M.; Hourcade D. E.; Hannan J. P.; Kovacs J. M.; et al. Detection of Complement Activation Using Monoclonal Antibodies against C3d. J. Clin. Invest. 2013, 123, 2218–2230. 10.1172/JCI65861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorham R. D.; Nuñez V.; Lin J.; Rooijakkers S. H. M.; Vullev V. I.; Morikis D. Discovery of Small Molecules for Fluorescent Detection of Complement Activation Product C3d. J. Med. Chem. 2015, 58, 9535–9545. 10.1021/acs.jmedchem.5b01062. [DOI] [PubMed] [Google Scholar]
- Serkova N. J.; Renner B.; Larsen B. A.; Stoldt C. R.; Hasebroock K. M.; Bradshaw-Pierce E. L.; Holers V. M.; Thurman J. M. Renal Inflammation: Targeted Iron Oxide Nanoparticles for Molecular MR Imaging in Mice. Radiology 2010, 255, 517–526. 10.1148/radiol.09091134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklin D.; Lambris J. D. Complement in Immune and Inflammatory Disorders: Therapeutic Interventions. J. Immunol. 2013, 190, 3839–3847. 10.4049/jimmunol.1203200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.; He C.; Knaak C.; Guthridge J. M.; Holers V. M.; Tomlinson S. Complement Receptor 2–mediated Targeting of Complement Inhibitors to Sites of Complement Activation. J. Clin. Invest. 2003, 111, 1875–1885. 10.1172/JCI17348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson C.; Song H.; Lu B.; Qiao F.; Burns T. A.; Holers V. M.; Tsokos G. C.; Tomlinson S. Targeted Complement Inhibition by C3d Recognition Ameliorates Tissue Injury without Apparent Increase in Susceptibility to Infection. J. Clin. Invest. 2005, 115, 2444–2453. 10.1172/JCI25208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer B.; Long Q.; Coughlin B.; Wilson R. B.; Huang Y.; Qiao F.; Tang P. H.; Kunchithapautham K.; Gilkeson G. S.; Tomlinson S. A Targeted Inhibitor of the Alternative Complement Pathway Reduces Angiogenesis in a Mouse Model of Age-Related Macular Degeneration. Invest. Ophthalmol. Visual Sci. 2009, 50, 3056–3064. 10.1167/iovs.08-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C. Q.; Bai H.; Lin Z.; Risitano A. M.; Barlow P. N.; Ricklin D.; Lambris J. D. Rational Engineering of a Minimized Immune Inhibitor with Unique Triple-Targeting Properties. J. Immunol. 2013, 190, 5712–5721. 10.4049/jimmunol.1203548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridkis-Hareli M.; Storek M.; Mazsaroff I.; Risitano A. M.; Lundberg A. S.; Horvath C. J.; Holers V. M. Design and Development of TT30, a Novel C3d-Targeted C3/C5 Convertase Inhibitor for Treatment of Human Complement Alternative Pathway–mediated Diseases. Blood 2011, 118, 4705–4713. 10.1182/blood-2011-06-359646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holers V. M.; Rohrer B.; Tomlinson S.. CR2-Mediated Targeting of Complement Inhibitors: Bench-to-Bedside Using a Novel Strategy for Site-Specific Complement Modulation. In Complement Therapeutics; Lambris J. D., Holers V. M., Ricklin D., Eds.; Springer US: Boston, MA, 2013; Vol. 735, pp 137–154. [DOI] [PubMed] [Google Scholar]
- McCammon J. A. Darwinian Biophysics: Electrostatics and Evolution in the Kinetics of Molecular Binding. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 7683–7684. 10.1073/pnas.0902767106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber G.; Buckle A. M.; Fersht A. R. Stability and Function: Two Constraints in the Evolution of Barstar and Other Proteins. Structure 1994, 2, 945–951. 10.1016/S0969-2126(94)00096-4. [DOI] [PubMed] [Google Scholar]