Significance
Nucleotide sugars, the activated sugar donors essential for processes such as cell wall biosynthesis and protein and lipid glycosylation are predominantly made in the cytosol. However, a highly diverse range of glycosyltransferases that are located within the Golgi lumen, mediate the above-mentioned glycosylation reactions. Thus, transport of nucleotide sugars across the Golgi membrane into the lumen is crucial for growth and development of many species including microorganisms, plants, and humans. In this study, we identify and functionally characterize four UDP-arabinofuranose transporters from Arabidopsis that are responsible for the delivery of activated arabinose, a critical sugar of plant cell walls, glycoproteins, and signaling peptides.
Keywords: Golgi apparatus, nucleotide sugars, membrane transport, arabinose
Abstract
In plants, L-arabinose (Ara) is a key component of cell wall polymers, glycoproteins, as well as flavonoids, and signaling peptides. Whereas the majority of Ara found in plant glycans occurs as a furanose ring (Araf), the activated precursor has a pyranose ring configuration (UDP-Arap). The biosynthesis of UDP-Arap mainly occurs via the epimerization of UDP-xylose (UDP-Xyl) in the Golgi lumen. Given that the predominant Ara form found in plants is Araf, UDP-Arap must exit the Golgi to be interconverted into UDP-Araf by UDP-Ara mutases that are located outside on the cytosolic surface of the Golgi. Subsequently, UDP-Araf must be transported back into the lumen. This step is vital because glycosyltransferases, the enzymes mediating the glycosylation reactions, are located within the Golgi lumen, and UDP-Arap, synthesized within the Golgi, is not their preferred substrate. Thus, the transport of UDP-Araf into the Golgi is a prerequisite. Although this step is critical for cell wall biosynthesis and the glycosylation of proteins and signaling peptides, the identification of these transporters has remained elusive. In this study, we present data demonstrating the identification and characterization of a family of Golgi-localized UDP-Araf transporters in Arabidopsis. The application of a proteoliposome-based transport assay revealed that four members of the nucleotide sugar transporter (NST) family can efficiently transport UDP-Araf in vitro. Subsequent analysis of mutant lines affected in the function of these NSTs confirmed their role as UDP-Araf transporters in vivo.
The monosaccharide L-arabinose (Ara) is the second most abundant pentose found in plants and comprises between 10% and 20% of the noncellulosic polysaccharide fraction of Arabidopsis leaves (1). It is a key component of the pectic wall polymer rhamnogalacturonan-I (RG-I) as well as being found in hydroxyproline-rich glycoproteins (HRGPs) such as arabinogalactan proteins (AGPs) and extensins (2, 3). Both, RG-I and AGPs, share similar Ara-containing structures, which comprise galactan chains substituted with terminal Ara, and possess relatively large quantities of Ara in the form of α-1,5-linked and -branched arabinan (4, 5). Single Ara residues are also present in the complex pectic polymer rhamnogalacturonan-II (RG-II) (6). In grasses, the majority of Ara is found in arabinoxylan, where it occurs as a decoration of the xylan backbone (7). In dicots, such as Arabidopsis, xylan is less frequently arabinosylated. However, recently it was shown that an Arabidopsis AGP is covalently linked to pectin and arabinoxylan (8). Ara is also found in xyloglucans of some plant species (9). The XXGG-type xyloglucan found in Solanaceous species is characterized by branches extended with Ara instead of D-galactose (Gal) (10). Ara residues are also critical for signaling processes because mature signaling peptides such as CLAVATA3 require posttranslational arabinosylation in the Golgi apparatus to be biologically active and bind to their receptors (11). In addition to the occurrence of Ara in cell wall polysaccharides, glycoproteins, and secreted peptides, which are made in the Golgi lumen, it is also found in the cytosol-biosynthesized flavonoids such as quercetin glycoside (12) and myricetin glycoside (13).
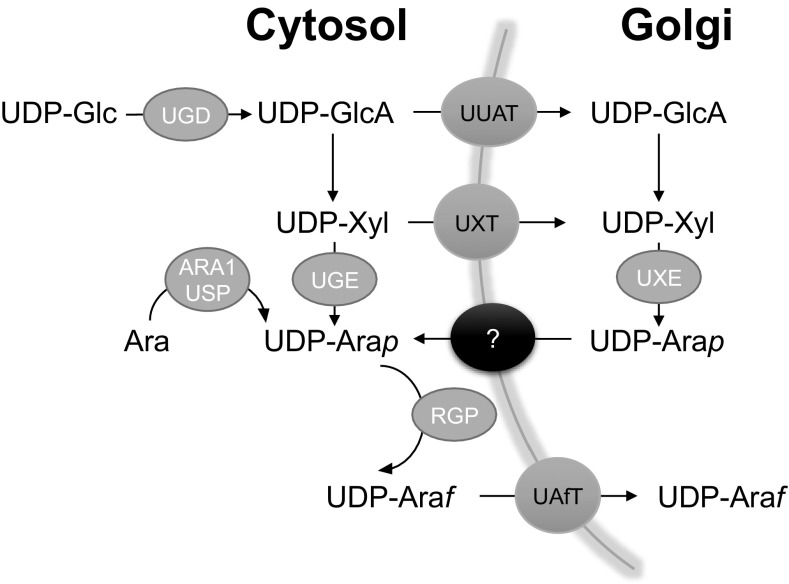
Ara occurs in two different ring forms, the furanose and the pyranose form. With few exceptions, for example, Ara found in RG-II sidechain B (6), the majority of Ara found in plants, occurs in the furanose form (Araf). This fact is somewhat surprising because in solution, Ara mainly exists in the more stable pyranose form (Arap). Likewise, the nucleotide sugar donor precursor for the biosynthesis of Ara-containing wall glycans is formed as a pyranose ring (UDP-Arap) via two different pathways. UDP-Arap can be formed either de novo by 4-epimerization of UDP-xylose (UDP-Xyl) in the Golgi lumen or via the Ara salvage pathway in the cytosol (14–16). Like most UDP sugars, most UDP-Arap is generated de novo in a sequential conversion from UDP-glucose (UDP-Glc), and subsequently via UDP-glucuronic acid (UDP-GlcA) and UDP-Xyl. The final interconversion step from UDP-Xyl to UDP-Arap is catalyzed by the UDP-Xyl 4-epimerase (UXE1). The Arabidopsis mutant murus4 (mur4), which was identified in a mutant screen, affects UXE1 activity (17) and has reduced amounts of cell wall Ara. UXE1 is localized within the Golgi and its catalytic domain is predicted to occur in the lumen (14). Thus, UDP-Arap synthesized via the luminal interconversion pathway provides the substrate for glycosylation reactions requiring UDP-Arap. Whereas it has been demonstrated that the cytosolic UDP-glucose 4-epimerases UGE1 and UGE3 can also catalyze the C-4 epimerization of UDP-Xyl to UDP-Arap in vitro (16), the uge1uge3 double mutant does not show any reduction in cell wall Ara content (18). Thus, a significant de novo interconversion pathway in the cytosol, generating UDP-Arap for Ara-containing polysaccharides assembled in the Golgi apparatus, is unlikely. Besides these de novo pathways, UDP-Arap can also be generated in the cytosol from free Ara released during degradation and metabolism of Ara-containing compounds and polymers. The Ara salvage pathway involves the sequential action of the substrate-specific arabinokinase I (19) and a UDP-sugar pyrophosphorylase with broad substrate specificity (15, 16). Given that the predominant form of Ara found in plants is Araf, UDP-Arap must be interconverted to UDP-Araf before polymer incorporation. The mutase responsible for this reaction was first identified in rice (20). In Arabidopsis, this reaction is catalyzed by at least three UDP-Ara mutases [reversibly glycosylated polypeptides (RGPs)], which have been shown to function in the cytosol, although they seem to be associated with the cytosolic side of the Golgi membrane (1). The UDP-Araf synthesized in the cytosol requires transport back into the lumen of the Golgi apparatus. This step is crucial, because the glycosyltransferases (GTs), the group of enzymes that mediate the biosynthesis of glycans for cell wall polymers, are located within this compartment (21) and the pyranose form, synthesized in the Golgi apparatus, is not the preferred donor substrate. Therefore, to enable the biosynthesis of Araf-containing wall polymers, transport of UDP-Araf from the cytosol into the lumen is essential (Fig. S1).
Fig. S1.
Diagram outlining the biosynthetic pathway of UDP-Araf and its transport across the Golgi membrane of plants. Characterized pathway members are identified by the gene names: UUAT, UDP-uronic acid transporter; UXT, UDP-Xyl transporter; RGP, UDP-Ara mutase (reversibly glycosylated polypeptide); ARA1, L-arabinokinase; USP, UDP-sugar pyrophosphorylase; UAfT, UDP-Araf transporter; UGE, UDP-glucose 4-epimerase; UXE, UDP-Xyl 4-epimerase; and UGD, UDP-glucose 6-dehydrogenase.
In plants, a family of NSTs has evolved to enable the transport of nucleotide sugars from the cytosol into the Golgi lumen. The Arabidopsis NST family contains 51 members falling into six subclades as part of the NST/triose phosphate translocator (TPT) superfamily (22). Some Arabidopsis NSTs localized in the Golgi and/or endoplasmic reticulum (ER) that transport GDP-mannose, UDP-galactose, UDP-glucose, UDP–N-acetylgalactosamine, and UDP–N-acetylglucosamine have been reported in the past decade (23). Recently, a robust assay to determine nucleotide sugar transport activities has been developed and enabled the characterization of the bifunctional UDP-rhamnose/UDP-galactose transporter family, the UDP-Xyl transporter family, the GDP-fucose transporter, and a UDP-uronic acid transporter (22, 24–26).
Whereas the compartmentalization of UDP-Ara interconverting mutases and GTs requiring UDP-Araf as activated donor substrate clearly argues for the existence of an UDP-Araf transporter, no such NST has yet been identified. Here, we report the characterization of the UDP-Araf transporter (UAfT) family from Arabidopsis. Using a refined transporter assay in conjunction with detection by mass spectrometry, we show that four members constituting the UAfT family are capable of efficiently transporting UDP-Araf. Analysis of the subcellular localization indicates that all four members are present in the Golgi apparatus supporting their function as Golgi UDP-Araf transporters. An examination of plant lines overexpressing UDP-Araf transporter candidates confirmed an in vivo function as UDP-Araf transporters.
Results
Members of Clade VI of the NST Family Transport UDP-Araf.
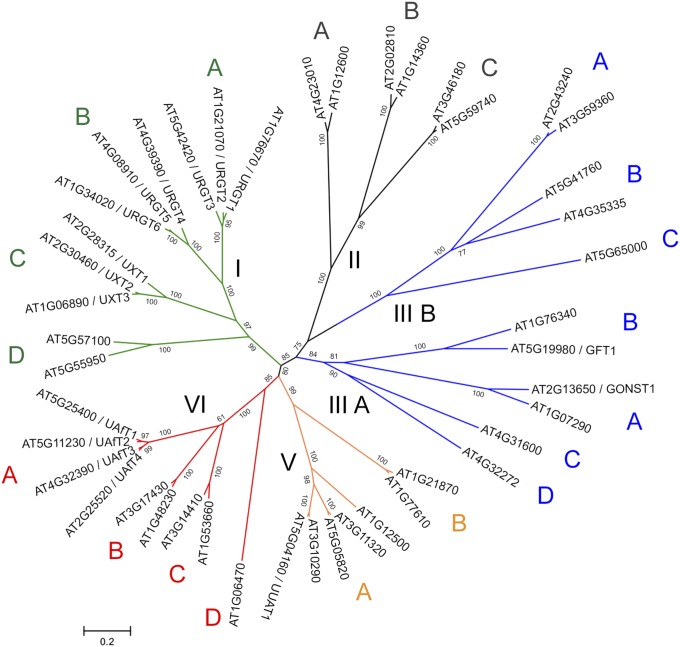
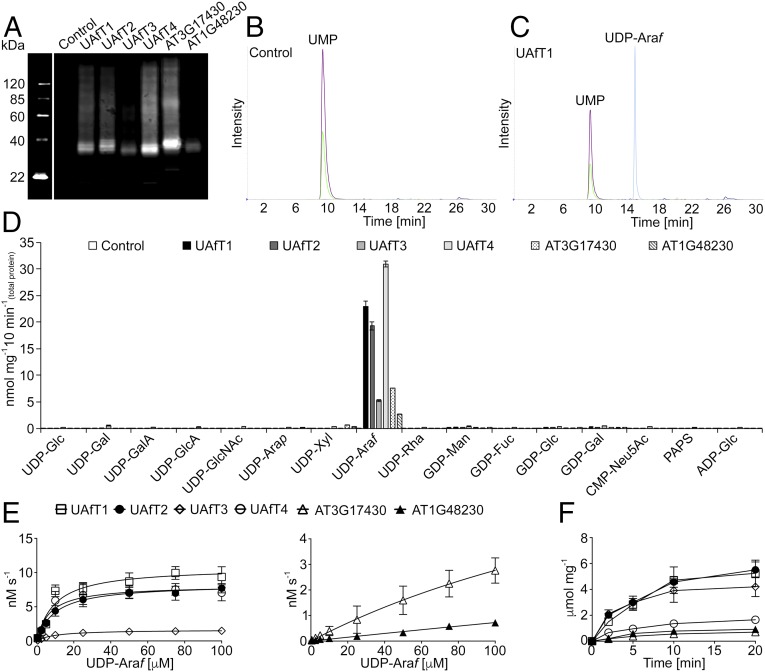
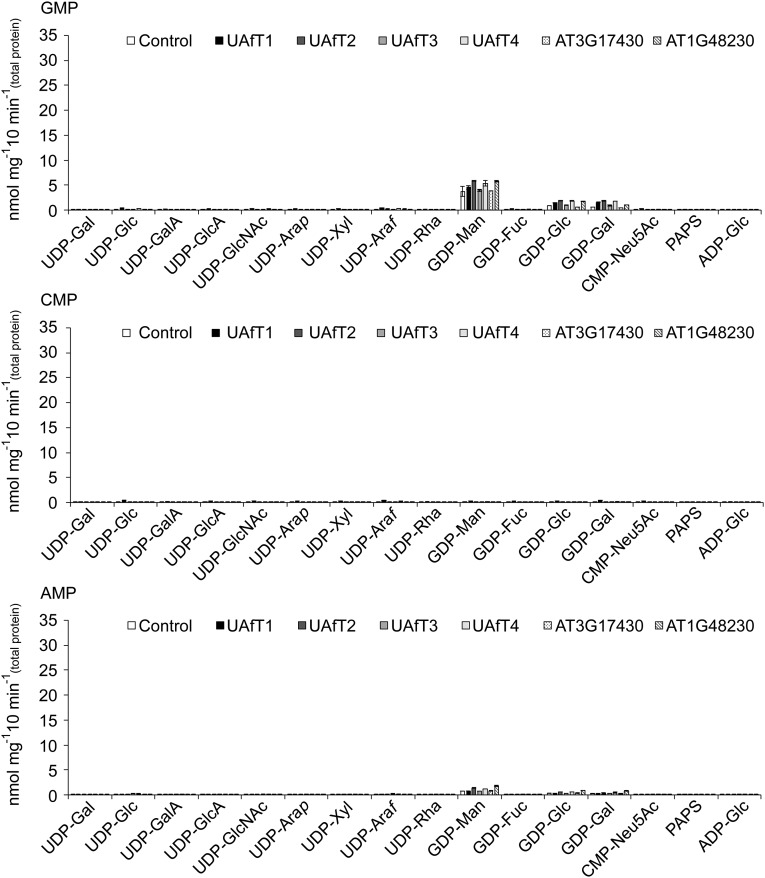
A substrate screen conducted on the entire Arabidopsis NST family identified members of clade VI subclade A (AT5G25400, AT5G11230, AT4G32390, and AT2G25520) and subclade B (AT3G17430 and AT1G48230) (Fig. S2) with specificities for UDP-Araf when proteoliposomes were preloaded with UMP. Each of these NSTs was heterologously expressed in yeast and its presence in microsomal preparations was assessed by immunoblotting (Fig. 1A). The ability of each NST to transport nucleotide sugar substrates was assessed by LC-MS/MS after conducting the transport assay with 16 nucleotide/nucleotide sugar substrates (listed in Table S1) against each of the putative exchange substrates UMP, GMP, CMP, or AMP. An empty vector control was used to assess background transport after removal of unincorporated substrates by gel filtration (Fig. 1B). A typical output after a single transport assay using proteoliposomes containing AT5G25400 preloaded with UMP is outlined in Fig. 1C. All six NST candidates demonstrated significant transport of UDP-Araf when proteoliposomes were preloaded with UMP (Fig. 1D). No apparent transport was observed when proteoliposomes were preloaded with GMP, CMP, or AMP. The observed GDP-sugar transport when proteoliposomes were preloaded with GMP can be accounted for by activity of the endogenous yeast GDP-Man transporter (Fig. S3).
Fig. S2.
Phylogenetic tree of the Arabidopsis NST family. Sequences of the previously defined NST clades I, II, IIIA, IIIB, V, and VI were aligned and a phylogenetic tree was created using MEGA6. The subclades were assigned arbitrarily based on the clustering of sequences.
Fig. 1.
Determining the substrate specificity and kinetics of UAfT candidates. (A) Immunoblot analysis of microsomes isolated from yeast transformed with the empty vector (control), UAfT1–4, AT3G17430, and AT1G48230 (2.5 µg protein per lane). (B) Representative MRM analysis of proteoliposomes preloaded with 10 mM UMP generated from a control after conducting the transport assay with 16 nucleotide sugar substrates. (C) Representative MRM analysis of UAfT1-containing proteoliposomes preloaded with 10 mM UMP after conducting the transport assay with 16 nucleotide sugar substrates. (D) Quantification of nucleotide sugar transport into proteoliposomes preloaded with 10 mM UMP after incubation with 16 nucleotide sugar substrates. Data are the mean and SD of n = 4 assays. (E) Proteoliposomes containing each candidate preloaded with 10 mM UMP were incubated with UDP-Araf at varying concentration (0.5–100 μM) for 2 min at 25 °C. Data are the mean and SEM of n = 8 assays. (F) Proteoliposomes containing each candidate preloaded with 10 mM UMP were incubated with UDP-Araf at a concentration of 50 μM for the indicated time points at 25 °C. Values are normalized to the calculated NST content present in proteoliposome preparations. Data are the mean and SD of n = 8 assays. Legend as shown in E applies.
Fig. S3.
Exchange substrate specificities. Quantification of nucleotide sugar transport into proteoliposomes containing the indicated NST preloaded with 10 mM GMP, CMP, or AMP after incubation with 16 nucleotide sugar substrates at a concentration of 50 μM. Data represent the mean and SD of n = 4 assays.
Kinetic Parameters of UDP-Araf Transport.
The transport response of each NST candidate to UDP-Araf was then independently determined. Subclade A members appeared to be saturable in a concentration-dependent manner, as would be expected for protein-mediated transport across a membrane (Fig. 1E). In contrast, the two subclade B members did not show saturation with substrate concentrations up to 100 μM (Fig. 1E). All six NST candidates were saturable in a time-dependent manner (Fig. 1F). Analysis of these data resulted in apparent KMs for UDP-Araf for members of subclade A, ranging from 7 to 10 μM with resultant turnover rates (kcat) of 1–4 s−1 (Table 1). We were unable to obtain corresponding kinetic parameters for members of subclade B. Consequently, the four members of the Arabidopsis NST clade VI, subclade A were designated the functional identifier UDP-Arabinofuranose Transporters (UAfT), and comprise the Arabidopsis loci AT5G25400 (UAfT1), AT5G11230 (UAfT2), AT4G32390 (UAfT3), and AT2G25520 (UAfT4). Previously, we have compared the apparent KM with the physiological concentrations of the substrate in plant organs to support the in vitro kinetic values (22, 24). The concentration of UDP-Araf in various plant organs was determined to be in the range of 5–15 pmol mg−1 dry weight (22). Because UDP-Araf is synthesized within the cytosol (1), we have previously reasoned that given that the vacuole generally comprises the bulk of a plant cells volume, dry weight measurements would be somewhat analogous to the volume of the cell excluding the vacuole. Consequently, an estimate for the cellular concentrations of UDP-Araf within the plant cell would be between 5 and 15 μM. This physiological concentration range aligns well with the determined KM values outlined above and supports the notion that we have identified biologically relevant transporters.
Table 1.
Kinetic parameters for UDP-Araf transport into proteoliposomes
| Parameter | UAfT1 | UAfT2 | UAfT3 | UAfT4 | AT3G17430 | AT1G48230 |
| KM (µM) | 10 (1) | 10 (1) | 10 (2) | 7 (1) | Ambiguous | Ambiguous |
| Vmax (nM s−1) | 11 (0) | 8 (0) | 2 (0) | 8 (0) | N.D. | N.D. |
| kcat (s−1) | 4 | 3 | 3 | 1 | N.D. | N.D. |
For each transporter, eight data points with varying substrate concentrations (0.5–100 µM) were acquired. Data represent the mean and SEM of n = 8 replicates. N.D., not determined.
Developmental Expression and Subcellular Localization.
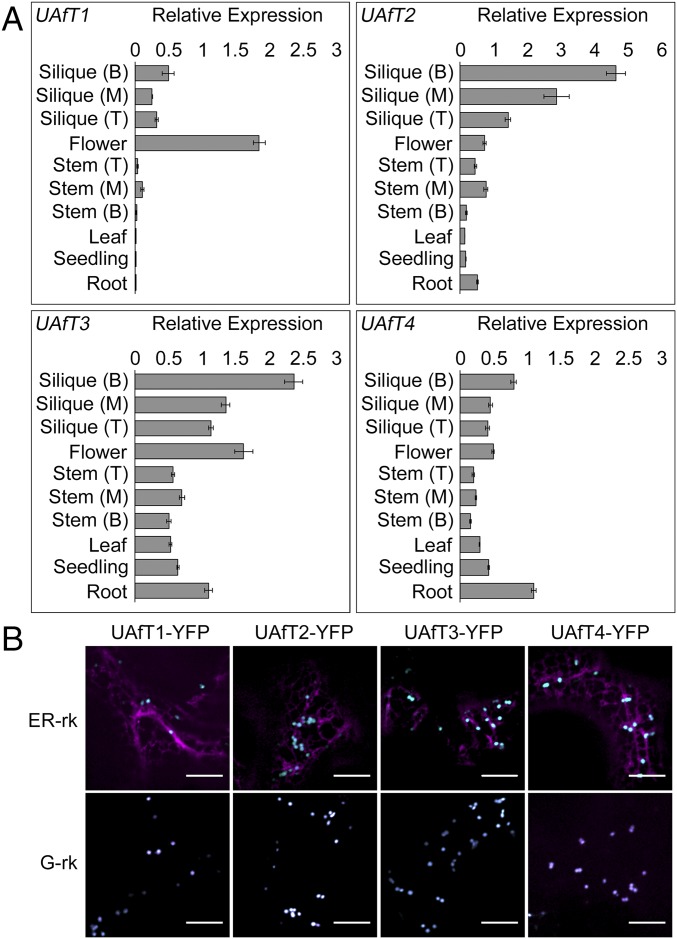
The expression patterns for the UAfTs were analyzed from a variety of organs using quantitative RT-PCR (qPCR) (Fig. 2A). All four members are expressed throughout plant development, although they are disproportionally expressed in flowers and during seed development (siliques), which are rich in Ara-containing polysaccharides. The expression patterns agree with those observed in publicly available microarray expression data (27). The subcellular locations were determined by transiently expressing C-terminal yellow fluorescent protein (YFP) fusions in Nicotiana benthamiana leaves and colocalizing the signal with the α-ManI mCherry Golgi-marker (G-rk) and a mCherry ER marker (ER-rk) (Fig. 2B). Simultaneous dual-channel live-cell imaging revealed localization of all UAfT–YFP fusions in the endomembrane system and resulted in punctate signals that highly colocalized with the Golgi marker. Subcellular proteome studies have identified all UAfTs, except for UAfT1, in the Golgi apparatus (28), supporting their roles as Golgi-localized UDP-Araf transporters.
Fig. 2.
Expression pattern and subcellular localizations. (A) Expression of UAfTs in different Arabidopsis organs was assessed by qPCR. Siliques and stem tissues were harvested from the bottom (B), middle (M) and top (T). Expression levels are the mean and SD of n = 3 expressed relative to three reference genes. (B) UAfTs were transiently coexpressed with the α-mannosidase I Golgi marker (G-rk) or the ER marker (ER-rk) in N. benthamiana leaves. All four UAfTs colocalize with the Golgi marker. (Scale bars, 10 μm.)
Determining in Planta Functions of UAfTs.
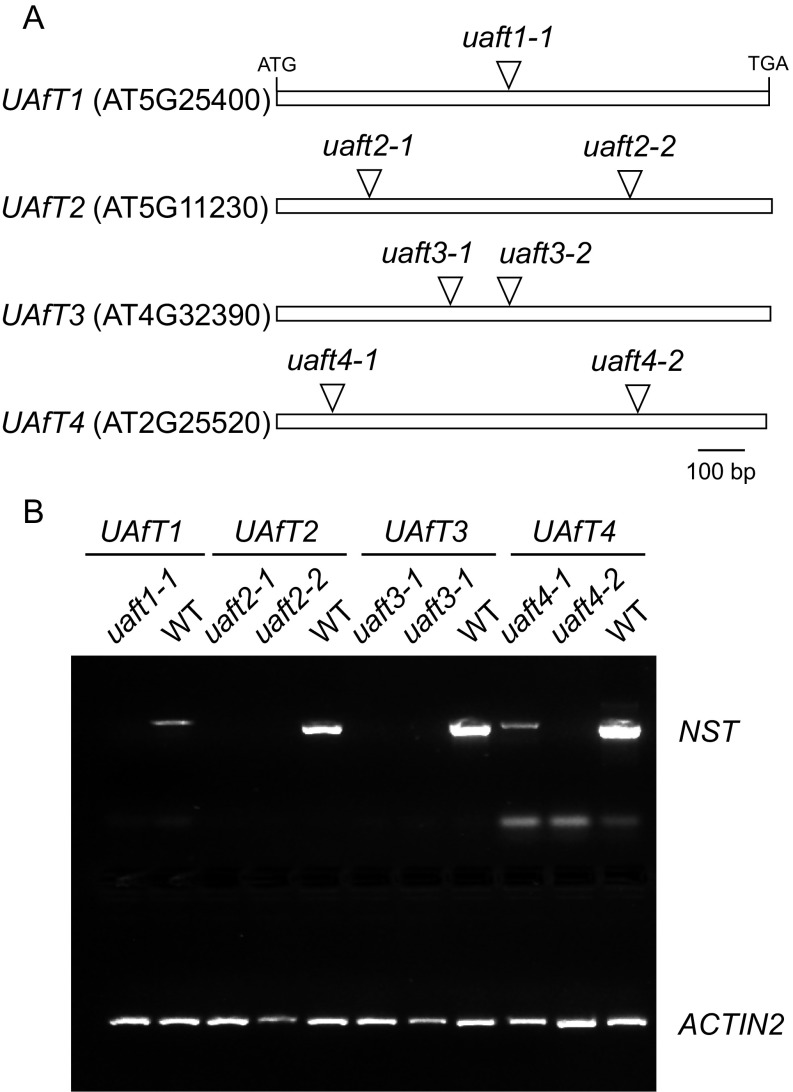
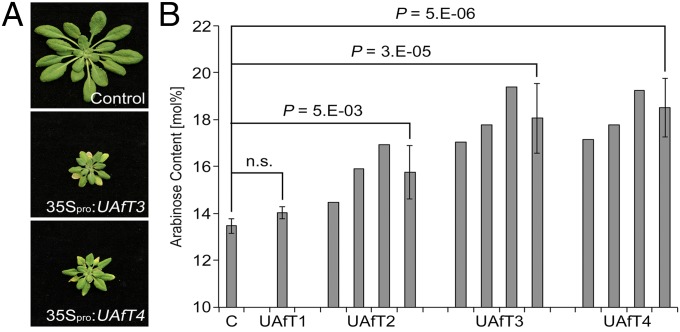
To confirm the in vitro biochemical characterization of the UAfTs, we obtained Arabidopsis mutant lines for each of the four members. For both, UAfT2 and UAfT3, we could confirm two independent knockout lines; for UAfT4 a knockout and knockdown line were verified and for UAfT1 only a single knockout line could be identified (Fig. S4). The analysis of homozygous UAfT knockout lines grown under standard growth conditions did not reveal any morphological alterations compared with wild-type plants. Cell wall preparations derived from leaves of 6-wk-old plants revealed a minor but significant reduction in Ara in both uaft4 lines (Table S2). Given the higher expression levels of UAfT2 and -3 in siliques (Fig. 2A) and of UAfT2, -3, and -4 in developing embryos (29), we sought to also analyze the cell wall monosaccharide composition of dry seeds derived from the corresponding knockout lines. As observed for leaf material, no major change in the abundance of monosaccharides was observed in dry seeds from these plants (Table S3). These results indicate that, at the examined resolution, some of the four UAfTs might function redundantly in Arabidopsis. Redundancy between NST family members in Arabidopsis seems to be relatively common (22), and we have previously found that the generation of NST overexpression (OE) lines results in attributable phenotypes. Consequently, we stably expressed the four UAfTs under the control of the CaMV 35S promoter and fused to YFP in Arabidopsis. For plants transformed with the 35Spro:UAfT1–YFP construct, YFP florescence could not be observed, indicating that this transgene had likely been silenced. For plants harboring the 35Spro:UAfT3–YFP or 35Spro:UAfT4–YFP constructs, we observed that lines possessing a strong YFP signal were retarded in their growth and showed early yellowing of leaves (Fig. 3A). An analysis of the monosaccharide content of leaf cell wall material indicated that UAfT2, -3, and -4 OE plant lines all contained a significant increase in the amounts of Ara (Fig. 3B). In particular, the overexpression of UAfT3 and UAfT4 led to a strong accumulation of Ara (up to 30% more compared with control plants) and concomitantly resulted in obvious phenotypic abnormalities (Fig. 3A). This result was observed predominantly for UAfT3 and UAfT4 OE lines, but only occasionally for UAfT2 overexpressors.
Fig. S4.
Characterization of UAfT insertion lines. (A) Schematic outlining insertion lines and locations in UAfTs. (B) Genotyping of UAfT insertion lines. uaft1-1 (SALK_039173C), uaft2-1 (SALK_011583), uaft2-2 (SALK_018646C), uaft3-1 (SALK_089917), uaft3-2 (SALK_148993), uaft4-1 (SALK_024802), and uaft4-2 (WiscDsLox355E01.0).
Fig. 3.
Overexpression of UAfTs result in morphological and biochemical phenotypes. (A) Representative phenotypes of plants overexpressing UAfT3 or UAfT4 compared with a control plant. (B) Arabinose content of rosette leaves. Three biological pools corresponding to the severity of the phenotype were analyzed. Arabinose levels are shown as molar percentage of total nonglucan carbohydrates ± SD (n = 3).
Glycome Profiling of UAfT3 and UAfT4 Overexpression Lines.
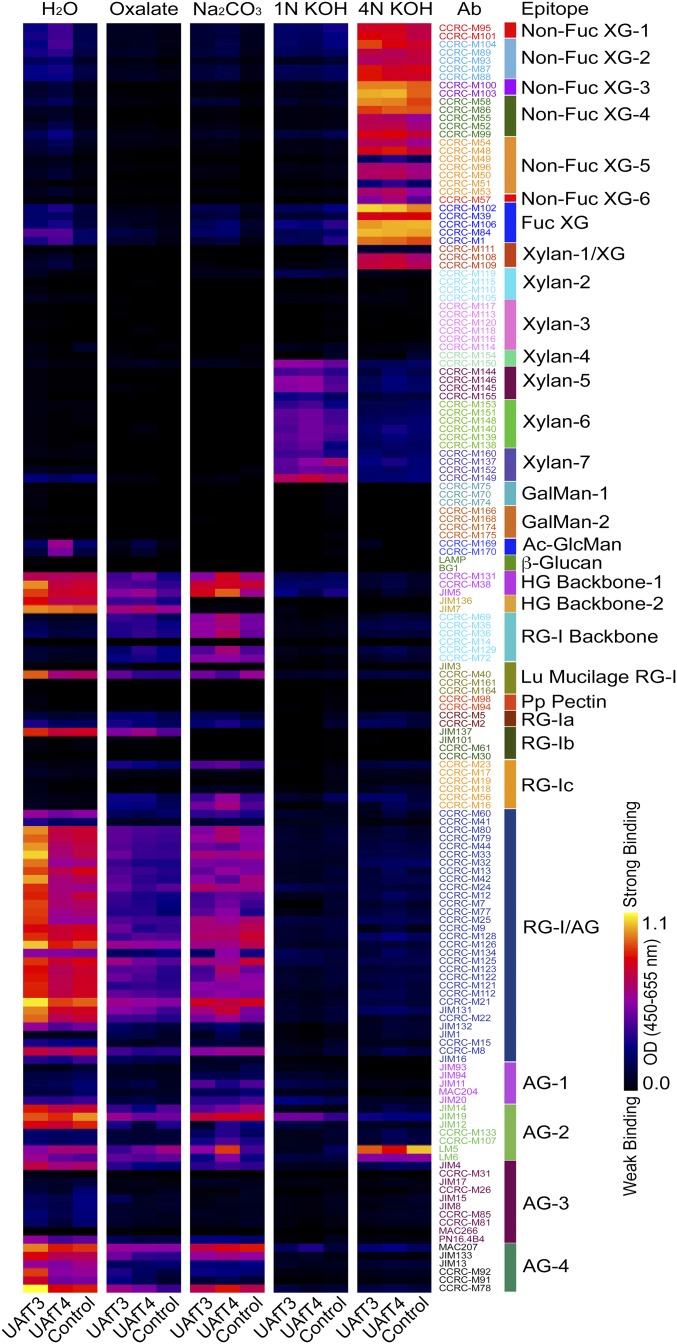
To investigate where the additional Ara in plants overexpressing UAfT3 or UAfT4 was incorporated, we performed glycome profiling using an ELISA-based screen with a collection of 155 plant cell wall glycan-directed monoclonal antibodies (30). Interestingly, the glycome profiles of wall extracts from UAfT3 and UAfT4 OE lines were different (Fig. 4). The most pronounced differences between UAfT3 OE lines and the control were observed in water-soluble cell wall material, particularly for antibodies recognizing arabinogalactan epitopes in RG-I and AGPs (RG-I/AG and AG-4 antibody groups). In contrast, the most pronounced differences between cell wall extracts from UAfT4 OE lines and control plants were observed in the sodium carbonate extracts (enriched for pectic polymers). Similar to the results observed for UAfT3 OE lines, the difference in the recognition pattern between the control and UAfT4 OE lines was predominantly due to antibodies recognizing pectic backbones, such as JIM5, which recognizes homogalacturonan (31), or CCRC-M36, which is specific for the RG-I backbone (32), or arabinogalactan epitopes in RG-I and AGP (RG-I/AG and AG-4). The observation that the UAfT3 and UAfT4 OE lines vary between different extracts is further supported by the data for the LM6 antibody, which is specific to 1,5-α-arabinan (33). An increased signal for LM6 was observed in the water-extracted material derived from UAfT3 OE plants, whereas the increase in binding intensity in UAfT4 OE lines is clearly visible in the sodium carbonate extract. Water-soluble extracts of UAfT4 OE lines showed stronger binding to acetylated glucomannan-directed antibodies than that observed for the UAfT3 OE or control lines. The 4 N KOH extracts (enriching hemicelluloses) showed decreased binding by LM5, which is specific for a β-1,4-galactan epitope (34) and could indicate that an increase in arabinose content leads to increased 1,5-α-arabinan decoration and consequently reduced unsubstituted galactan epitopes in RG-I in the OE lines. Antibodies belonging to the groups of nonfucosylated and fucosylated xyloglucan also show subtle differences in their binding to wall extracts between the control and both the UAfT3 and UAfT4 OE lines. Another interesting difference was the marginal increase in the abundances of 1 N KOH extractable xylan epitopes (especially epitopes recognized by xylan-4 through xylan-6 groups of antibodies) in the two OE lines.
Fig. 4.
Glycome profiling. Sequentially extracted cell wall material derived from plants overexpressing UAfT3, UAfT4, and control plants was subjected to glycome profiling using 155 plant cell wall glycan-directed monoclonal antibodies. A black–yellow scale indicates signal intensity in the ELISA, with black corresponding to no binding and yellow to strongest binding. Labels at the Top specify the different reagents used to extract the cell wall material.
Discussion
The ubiquitous presence of Ara in an assortment of cell wall-associated polymers, its presence in flavonoids, and its association with signaling molecules highlight the important role of this monosaccharide as both a structural component of walls and in regulating growth and development. The majority of Ara is found in cell wall-associated polymers where it is mainly present in its furanose form. Whereas luminal nucleotide sugar interconversion results in the biosynthesis of an activated Ara donor, it is in the pyranose form. The characterization of the UDP-Ara mutases and their localization to the cytosol resulted in a biosynthetic dilemma, because UDP-Araf is required within the Golgi lumen necessary for the biosynthesis of cell wall-associated polysaccharides. The characterization of a family of four Arabidopsis NSTs with substrate specificity for UDP-Araf reveals the final step in the delivery pathway for Araf-containing compounds synthesized in the lumen and supports the current biosynthetic model of nucleotide sugar interconversion (Fig. S1).
The Partitioning of UDP-Ara Within the Plant Cell.
The biosynthesis of Ara-containing compounds within the Golgi requires UDP-Ara be transported twice across the Golgi membrane. A likely explanation for this process is the thermodynamic equilibrium of UDP-Araf biosynthesis by the UDP-Ara mutases. The in vitro characterization of those enzymes from either rice or Arabidopsis indicates that only about 10% of UDP-Arap is converted to UDP-Araf (1, 35). This inefficient conversion is a probable reason for the partitioning of UDP-Araf biosynthesis in the cytosol and its incorporation within the Golgi lumen, enabling the efficient and noncompetitive transfer of Araf to a nucleophilic acceptor. The ability to modify the composition of Ara in cell wall-associated material through the up-regulation of UAfTs indicates that the supply of UDP-Araf within the Golgi lumen is likely limiting. The increased availability of this nucleotide sugar within the Golgi and resultant increase in Ara in the cell wall also provides information about the distribution of UDP-Araf and UDP-Arap within the cell. We previously determined that the amount of UDP-Arap within a variety of Arabidopsis organs is 40–105 pmol mg−1 dry weight, whereas the amount of UDP-Araf ranges from 5 to 15 pmol mg−1 dry weight (22). Considering the thermodynamic equilibrium of UDP-Ara mutases with these concentrations and given the limited availability of UDP-Araf for incorporation into luminal polysaccharides, it is likely that the biochemical pools of both UDP-Arap and UDP-Araf exist predominantly within the cytosol, even with the former actively made by the Golgi-resident UDP-Xyl 4-epimerase 1 (UXE1). The location of UDP-Araf in the cytosol is supported by the investigation of a GT47 family member, ARABINAN DEFICIENT 1 (ARAD1), which is implicated in pectic arabinan biosynthesis (36). Plants overexpressing ARAD1 did not show an altered arabinan content (36), potentially indicating limitations in substrate availability (37). In contrast, evidence for a distinct cytosolic pool of UDP-Arap is supported by the recently characterized Golgi-resident UDP-xylose transporters (UXTs) and their role in xylan biosynthesis (24) as well as the fact that the Golgi-localized UDP-xylose synthases (UXS) have a limited role in supplying substrates for xylose-containing polymers within the Golgi lumen (38). Both findings indicate that the fate of luminal synthesized UDP-Xyl is mainly to provide the substrate for UXE1 for the biosynthesis of UDP-Arap. This latter observation is supported by a UXS triple mutant in Arabidopsis (uxs3uxs5uxs6) that eliminated cytosolic-derived UDP-Xyl (38), and resulted in an increased Ara content in cell wall extracts. The removal of the cytosolic UDP-Xyl pool likely results in an increased flux through the luminal interconversion pathway, driving the biosynthesis of UDP-Arap thus explaining the increased levels of Ara. Moreover, because the reported equilibrium between UDP-Xyl and UDP-Arap by UXE1 is around 1:1 (14), the transport of UDP-Arap outside of the Golgi (by an unidentified NST) would provide the necessary pull on the luminal interconversion pathway to ensure sufficient supply of UDP-Araf for luminal polysaccharide biosynthesis. This model is also supported by studies on the mur4 mutant and wild-type plants, where the addition of exogenous Ara increased polysaccharide arabinosylation (17), supporting a limited delivery of UDP-Araf into the Golgi lumen.
The Role of Ara in the Plant Cell Wall.
Ara comprises a significant proportion of the plants cell wall. In Arabidopsis, the modulation of Ara quantity results in a severe growth phenotype. Both UAfT3 and UAfT4 OE lines displayed up to ∼30% increase in Ara content in cell wall extracts, which correlated with reduced growth and early senescing leaves (Fig. 3A). Glycome profiling indicated that epitopes corresponding to RG-I, RG-I-associated arabinogalactans, and AGPs increased in both UAfT3 and UAfT4 OE lines. Similarly, cytosolic UXS triple mutant plants (uxs3uxs5uxs6), which likely result in an increased flux through the luminal interconversion pathway, contain a twofold increase in cell wall-derived Ara and display a reduced growth phenotype (38). Analysis of stem wall material from the uxs3uxs5uxs6 triple mutant indicated that increased Ara was mainly present as 1,5-Ara(f) linkage and represents an increase in length or quantity of α-1,5-arabinan sidechains in the pectic polymer RG-I. Thus, the increased Ara content observed here can likely be attributed to increases in RG-I arabinan.
It has been speculated that RG-I sidechains act as spacers and regulate interactions with homogalacturonan, thus controlling the flexibility of the cell wall (39). The plasticity of the wall relies on the mobility of RG-I sidechains, especially arabinan (40), and the modulation of sidechain lengths and branching seems to have a deleterious effect on the physical properties of the wall. Such conclusions have been made examining transgenic potato plants expressing a Golgi-localized α-1,5-arabinanase (41). This contrasts with modulations to the galactan sidechains of RG-I because plants overexpressing the galactan synthase GALS1 or the UDP-Gal/UDP-Rha transporter URGT1 contain up to 50% more cell wall galactose specifically in the form of pectic β-1,4-galactan but plant growth and development are not affected (22, 42). These contrasting observations highlight the special importance of arabinan sidechains for proper RG-I function.
Conclusion
Ara plays an essential role in the growth and development of plants. Here we have characterized a family of Golgi nucleotide sugar transporters (UAfTs) responsible for the delivery of UDP-Araf for a range of Golgi luminal arabinosylation reactions in Arabidopsis. This study represents the only example of UDP-Araf transport in biology and the characterization of this family supports the curious model for the incorporation of UDP-Araf in plants.
Materials and Methods
Nucleotide Sugar Substrates.
Nucleotide/nucleotide sugar substrates used in this study are listed in Table S1. UDP-β–L-rhamnose and GDP-α–L-galactose were synthesized enzymatically as previously described (22).
Heterologous Expression of NSTs in Yeast and Transport Assays.
Heterologous expression was undertaken in Saccharomyces cerevisiae strain INVSc1 using the expression vector pYES-DEST52 (Thermo Fisher Scientific). Reconstitution of NSTs and transport activity assays were conducted as previously described (22). Kinetic parameters were determined using nonlinear regression with Prism 7 software (GraphPad Software). Polyacrylamide gel electrophoreses and immunoblot analyses were carried out as previously described (22).
Analysis of Nucleotide Sugars by Tandem Mass Spectrometry.
Nucleotide sugars were separated on a Hypercarb porous graphitic carbon column (Thermo Fisher Scientific) with a 1100 series HPLC system (Agilent Technologies) and analyzed by LC-MS/MS and multiple reaction monitoring (MRM) using a 4000 QTRAP system (Sciex) with conditions and parameters as previously outlined (22).
Plant Material.
A. thaliana seeds were acquired from the Arabidopsis Biological Resource Center (SI Materials and Methods).
Fractionation of Cell Wall Material and Glycome Profiling.
Glycome profiling was performed with a comprehensive suite of plant cell wall glycan-directed monoclonal antibodies (30) as described (43) (SI Materials and Methods).
SI Materials and Methods
Phylogenetic Analysis.
Sequences were obtained from The Arabidopsis Information Resource (TAIR) (44). Alignments and phylogenetic trees were generated as previously described (22) using the Molecular Evolutionary Genetics Analysis (MEGA) software package version 6.0 (45).
Quantitative PCR.
RNA was extracted using the RNEasy RNA Plant Kit (Qiagen) and reverse transcribed using SuperScriptII reverse transcriptase and d(T)15 oligomers (Thermo Fisher Scientific). Quantitative PCR (qPCR) was performed using SYBR Select Master Mix (Applied Biosystems) as previously described (24). For normalization, the housekeeping genes UBQ10 (AT4G05320), PP2A (AT1G13320), and MON1 (AT2G28390) were amplified (primers are listed in Table S4). Expression levels were calculated using the comparative CT method and normalized against the geometric mean of the three housekeeping genes (46).
Quantitation of Expressed NST Proteins.
Quantitation of expressed NSTs in yeast proteoliposomes was undertaken by MRM of a shared tryptic peptide (SRGPFEGKPIPNPLLGLDSTR) derived from the V5-tag region of constructs as previously described (24). Quantitation of expressed NSTs was estimated by linear regression using a standard curve (0.5–10 pmol) of a synthetic peptide with the above sequence. Data were analyzed using Skyline (v2.5.0.6157) (47). The amount of expressed NST was estimated by integrating the total signal peak area from Skyline for two transitions on the 563.560 [M+4H]4+ precursor ion, namely L [y7] 761.452 [M+H]1+ and G [y6] 648.3311 [M+H]1+. The UAfTs comprised around 0.1–1.5% of total proteins in proteoliposome preparations (Table S5).
Plant Material, Growth, Transformation, and Selection.
Arabidopsis thaliana (Col-0) seeds were acquired from the Arabidopsis Biological Resource Center (https://abrc.osu.edu/). T-DNA insertion mutants were obtained from the SIGnAL Salk (48) and WiscDsLox collections (49) (Table S4 and Fig. S4). Seeds were sown on soil (PRO-MIX, Premier Horticulture, Inc.) and grown in a growth chamber (Percival Scientific) under short-day light conditions [10 h of fluorescent light (120 μmol m−2 s−1) at 22 °C and 60% relative humidity (RH)/14 h of dark at 22 °C and 60% RH]. After 4 wk, plants were transferred to long-day conditions [16 h of fluorescent light (120 μmol m−2 s−1) at 22 °C and 60% RH/8 h of dark at 22 °C and 60% RH]. To generate plants overexpressing the UAfTs, A. thaliana (Col-0) plants were transformed with Agrobacterium tumefaciens strain GV3101 using the floral dip method (50). Selection of transgenic plants was done with glufosinate ammonium as outlined in ref. 1.
Genotyping of Mutant Lines by PCR.
To identify homozygous T-DNA insertion lines, genomic DNA was prepared from leaf material using the cetyltrimethylammonium bromide (CTAB) method and assessed by PCR on cDNA prepared from leaf material using primers listed in Table S4. The Arabidopsis ACTIN2 gene (AT3G18780) was amplified as a control for equal loading.
Cloning Procedures.
Coding sequences without native stop codons were PCR amplified from leaf cDNA using primers listed in Table S4. The resultant PCR products were introduced into the pENTR/SD/d-TOPO cloning vector (AT1G48230) or the pCR8/GW/TOPO cloning vector (UAfT1–4, and AT3G17430) (Thermo Fisher Scientific). C-terminal YFP fusions were created by introduction of the constructs into the 35S promoter carrying pEarleyGate101 plant transformation vector (51) using the LR Clonase II reaction (Thermo Fisher Scientific).
Subcellular Localization and Microscopy.
Agrobacteria strain AGL1 carrying the respective vectors for UAfT C-terminal YFP fusions, the αManI-mCherry Golgi (G-rk) and the mCherry ER marker (ER-rk) (52) were grown overnight in lysogeny broth media at 30 °C with constant shaking. Cultures were centrifuged, the supernatant was discarded and the cell pellets were resuspended in infiltration buffer (10 mM Mes, 10 mM MgCl2, and 100 μM acetosyringone to an OD600 = 0.01–0.15). Epidermal cells of leaves from 4-wk-old N. benthamina plants were transiently cotransformed with the NST–YFP fusions and the respective marker and imaged 72 h after infiltration. Leaf segments were imaged using a 100× total internal reflection fluorescence (N.A. 1.49) lens on Nikon Ti-E inverted microscope equipped with an Andor Revolution CSU-W1 spinning disk unit, a Borealis homogeneous illumination unit, and two Andor iXon Ultra 888BV-EMCCDs in dual camera mode. YFP and mCherry were simultaneously excited with a 488-nm and a 561-nm laser. Emission was collected simultaneously for both fluorophores on two EMCCDs using a 535/30-nm emission filter for YFP and a 610/40-nm emission filter for mCherry.
Cell Wall Monosaccharide Composition.
Cell wall monosaccharide compositions were determined following the protocol outlined in ref. 25. Alcohol insoluble residues (AIRs) were prepared, extracts were hydrolyzed in 2 N trifluoroacetic acid (TFA) for 1 h at 120 °C, and subsequently High Performance Anion Exchange Chromatography with Pulsed Amperometric Detection (HPAEC-PAD) was performed on an ICS 3000 (Thermo Fisher Scientific) using a CarboPac PA20 (3× 150 mm) anion exchange column.
Fractionation of Cell Wall Material and Glycome Profiling.
Plants were grown in short-day conditions. Whole rosettes of 6-wk-old plants were harvested and AIR was prepared. A total of 10 mg was sequentially extracted with water, ammonium oxalate (50 mM, pH 5), 50 mM Na2CO3/0.5% NaBH4, 1 N KOH/1.0% NaBH4, and 4 N KOH/1.0% NaBH4. Extractions were carried out overnight at 4 °C under constant shaking. Samples were pelleted and the resultant supernatant was transferred into a fresh tube before subjecting it to the next extraction step. The pellet was washed once with water. Fractions were dialyzed. The KOH fractions were neutralized with acetic acid before dialysis. All samples were lyophilized and glycome profiling was performed as described in ref. 43. Samples were dissolved in water and coated onto ELISA plates on an equal-weight-per-well basis (0.3 μg per well). The samples were then subjected to ELISA screening with a comprehensive suite of cell wall glycan-directed monoclonal antibodies (Table S6) as described (30). Assays were done in technical duplicates and data represent the average. Cell wall glycan-directed antibodies and CBMs were obtained from laboratory stocks (CCRC, JIM, and MAC series) at the Complex Carbohydrate Research Center and from PlantProbes (LM5 and LM6).
Supplementary Material
Acknowledgments
J.L.H., B.E., and S. Persson are supported by Australian Research Council (ARC) Future Fellowships (FT130101165, FT160100276, and FT160100218). H.E.M. is supported by an ARC Discovery Early Career Researcher Award (DE170100054). Microscopy was supported by an ARC Linkage Infrastructure, Equipment, and Facilities (LE150100011) and Biological Optical Microscopy Platform (The University of Melbourne). The work was supported by the US Department of Energy through contract DE-AC02-05CH11231 with Lawrence Berkeley National Laboratory and contract DE-AC05-00OR22725 with Oak Ridge National Laboratory. A.O. is supported by Fondecyt 1151335, Fondo de Areas Prioritarias-Centro de Regulacion del Genoma 15090007, Committee of Evaluation and Direction of the Scientific Cooperation-CONICYT C14B02, and Basal Program PB-16. S.S.-A. is supported by Fondecyt 11160787. The generation of the Complex Carbohydrate Research Center series of plant cell wall glycan-directed antibodies was supported by the NSF Plant Genome Program (DBI-0421683 and IOS-0923992). Substrates from CarboSource Services (Athens, GA) were supported by NSF-Research Coordination Networks Grant 0090281.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1701894114/-/DCSupplemental.
References
- 1.Rautengarten C, et al. The interconversion of UDP-arabinopyranose and UDP-arabinofuranose is indispensable for plant development in Arabidopsis. Plant Cell. 2011;23:1373–1390. doi: 10.1105/tpc.111.083931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bar-Peled M, O’Neill MA. Plant nucleotide sugar formation, interconversion, and salvage by sugar recycling. Annu Rev Plant Biol. 2011;62:127–155. doi: 10.1146/annurev-arplant-042110-103918. [DOI] [PubMed] [Google Scholar]
- 3.Showalter AM, Basu D. Extensin and arabinogalactan-protein biosynthesis: Glycosyltransferases, research challenges, and biosensors. Front Plant Sci. 2016;7:814. doi: 10.3389/fpls.2016.00814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seifert GJ, Roberts K. The biology of arabinogalactan proteins. Annu Rev Plant Biol. 2007;58:137–161. doi: 10.1146/annurev.arplant.58.032806.103801. [DOI] [PubMed] [Google Scholar]
- 5.Willats WG, McCartney L, Mackie W, Knox JP. Pectin: Cell biology and prospects for functional analysis. Plant Mol Biol. 2001;47:9–27. [PubMed] [Google Scholar]
- 6.Bar-Peled M, Urbanowicz BR, O’Neill MA. The synthesis and origin of the pectic polysaccharide rhamnogalacturonan II: Insights from nucleotide sugar formation and diversity. Front Plant Sci. 2012;3:92. doi: 10.3389/fpls.2012.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheller HV, Ulvskov P. Hemicelluloses. Annu Rev Plant Biol. 2010;61:263–289. doi: 10.1146/annurev-arplant-042809-112315. [DOI] [PubMed] [Google Scholar]
- 8.Tan L, et al. An Arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. Plant Cell. 2013;25:270–287. doi: 10.1105/tpc.112.107334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peña MJ, Darvill AG, Eberhard S, York WS, O’Neill MA. Moss and liverwort xyloglucans contain galacturonic acid and are structurally distinct from the xyloglucans synthesized by hornworts and vascular plants. Glycobiology. 2008;18:891–904. doi: 10.1093/glycob/cwn078. [DOI] [PubMed] [Google Scholar]
- 10.Schultink A, Cheng K, Park YB, Cosgrove DJ, Pauly M. The identification of two arabinosyltransferases from tomato reveals functional equivalency of xyloglucan side chain substituents. Plant Physiol. 2013;163:86–94. doi: 10.1104/pp.113.221788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y. A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat Chem Biol. 2009;5:578–580. doi: 10.1038/nchembio.182. [DOI] [PubMed] [Google Scholar]
- 12.Halabalaki M, et al. Quercetin and kaempferol 3-O-[α-L-rhamnopyranosyl-(1→2)-α-L-arabinopyranoside]-7-O-α-L-rhamnopyranosides from Anthyllis hermanniae: Structure determination and conformational studies. J Nat Prod. 2011;74:1939–1945. doi: 10.1021/np200444n. [DOI] [PubMed] [Google Scholar]
- 13.Torres-Mendoza D, et al. Weakly antimalarial flavonol arabinofuranosides from Calycolpus warszewiczianus. J Nat Prod. 2006;69:826–828. doi: 10.1021/np050484i. [DOI] [PubMed] [Google Scholar]
- 14.Burget EG, Verma R, Mølhøj M, Reiter WD. The biosynthesis of L-arabinose in plants: Molecular cloning and characterization of a Golgi-localized UDP-D-xylose 4-epimerase encoded by the MUR4 gene of Arabidopsis. Plant Cell. 2003;15:523–531. doi: 10.1105/tpc.008425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geserick C, Tenhaken R. UDP-sugar pyrophosphorylase is essential for arabinose and xylose recycling, and is required during vegetative and reproductive growth in Arabidopsis. Plant J. 2013;74:239–247. doi: 10.1111/tpj.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kotake T, et al. Properties and physiological functions of UDP-sugar pyrophosphorylase in Arabidopsis. Biosci Biotechnol Biochem. 2007;71:761–771. doi: 10.1271/bbb.60605. [DOI] [PubMed] [Google Scholar]
- 17.Burget EG, Reiter WD. The mur4 mutant of arabidopsis is partially defective in the de novo synthesis of uridine diphospho L-arabinose. Plant Physiol. 1999;121:383–389. doi: 10.1104/pp.121.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rösti J, et al. UDP-glucose 4-epimerase isoforms UGE2 and UGE4 cooperate in providing UDP-galactose for cell wall biosynthesis and growth of Arabidopsis thaliana. Plant Cell. 2007;19:1565–1579. doi: 10.1105/tpc.106.049619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dolezal O, Cobbett CS. Arabinose kinase-deficient mutant of Arabidopsis thaliana. Plant Physiol. 1991;96:1255–1260. doi: 10.1104/pp.96.4.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konishi T, et al. A plant mutase that interconverts UDP-arabinofuranose and UDP-arabinopyranose. Glycobiology. 2007;17:345–354. doi: 10.1093/glycob/cwl081. [DOI] [PubMed] [Google Scholar]
- 21.Oikawa A, Lund CH, Sakuragi Y, Scheller HV. Golgi-localized enzyme complexes for plant cell wall biosynthesis. Trends Plant Sci. 2013;18:49–58. doi: 10.1016/j.tplants.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Rautengarten C, et al. The Golgi localized bifunctional UDP-rhamnose/UDP-galactose transporter family of Arabidopsis. Proc Natl Acad Sci USA. 2014;111:11563–11568. doi: 10.1073/pnas.1406073111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orellana A, Moraga C, Araya M, Moreno A. Overview of nucleotide sugar transporter gene family functions across multiple species. J Mol Biol. 2016;428:3150–3165. doi: 10.1016/j.jmb.2016.05.021. [DOI] [PubMed] [Google Scholar]
- 24.Ebert B, et al. Identification and characterization of a Golgi-localized UDP-xylose transporter family from Arabidopsis. Plant Cell. 2015;27:1218–1227. doi: 10.1105/tpc.114.133827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rautengarten C, et al. The Arabidopsis Golgi-localized GDP-L-fucose transporter is required for plant development. Nat Commun. 2016;7:12119. doi: 10.1038/ncomms12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saez-Aguayo S, et al. UUAT1 is a Golgi-localized UDP-uronic acid transporter that modulates the polysaccharide composition of Arabidopsis seed mucilage. Plant Cell. 2017;29:129–143. doi: 10.1105/tpc.16.00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmid M, et al. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 28.Nikolovski N, et al. Putative glycosyltransferases and other plant Golgi apparatus proteins are revealed by LOPIT proteomics. Plant Physiol. 2012;160:1037–1051. doi: 10.1104/pp.112.204263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le BH, et al. Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci USA. 2010;107:8063–8070. doi: 10.1073/pnas.1003530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pattathil S, et al. A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiol. 2010;153:514–525. doi: 10.1104/pp.109.151985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Casero PJ, Knox JP. The monoclonal-antibody Jim5 indicates patterns of pectin deposition in relation to pit fields at the plasma-membrane-face of tomato pericarp cell-walls. Protoplasma. 1995;188:133–137. [Google Scholar]
- 32.Young RE, et al. Analysis of the Golgi apparatus in Arabidopsis seed coat cells during polarized secretion of pectin-rich mucilage. Plant Cell. 2008;20:1623–1638. doi: 10.1105/tpc.108.058842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willats WGT, Marcus SE, Knox JP. Generation of monoclonal antibody specific to (1–>5)-alpha-L-arabinan. Carbohydr Res. 1998;308:149–152. doi: 10.1016/s0008-6215(98)00070-6. [DOI] [PubMed] [Google Scholar]
- 34.Jones L, Seymour GB, Knox JP. Localization of pectic galactan in tomato cell walls using a monoclonal antibody specific to (1->4)-beta-D-galactan. Plant Physiol. 1997;113:1405–1412. doi: 10.1104/pp.113.4.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Konishi T, et al. Purification and biochemical characterization of recombinant rice UDP-arabinopyranose mutase generated in insect cells. Biosci Biotechnol Biochem. 2010;74:191–194. doi: 10.1271/bbb.90619. [DOI] [PubMed] [Google Scholar]
- 36.Harholt J, et al. ARABINAN DEFICIENT 1 is a putative arabinosyltransferase involved in biosynthesis of pectic arabinan in Arabidopsis. Plant Physiol. 2006;140:49–58. doi: 10.1104/pp.105.072744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geshi N, Harholt J, Sakuragi Y, Krüger Jensen J, Scheller HV. In: Glycosyltransferases of the GT47 Family. Plant Polysaccharides, Biosynthesis and Bioengineering, Annual Plant Reviews. Ulvskov P, editor. Vol 41. Wiley-Blackwell; New Jersey: 2010. pp. 265–283. [Google Scholar]
- 38.Kuang B, et al. The role of UDP-glucuronic acid decarboxylase(s) (UXS) in xylan biosynthesis in Arabidopsis. Mol Plant. 2016;9:1119–1131. doi: 10.1016/j.molp.2016.04.013. [DOI] [PubMed] [Google Scholar]
- 39.Harholt J, Suttangkakul A, Vibe Scheller H. Biosynthesis of pectin. Plant Physiol. 2010;153:384–395. doi: 10.1104/pp.110.156588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ha MA, Viëtor RJ, Jardine GD, Apperley DC, Jarvis MC. Conformation and mobility of the arabinan and galactan side-chains of pectin. Phytochemistry. 2005;66:1817–1824. doi: 10.1016/j.phytochem.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 41.Ulvskov P, et al. Biophysical consequences of remodeling the neutral side chains of rhamnogalacturonan I in tubers of transgenic potatoes. Planta. 2005;220:609–620. doi: 10.1007/s00425-004-1373-8. [DOI] [PubMed] [Google Scholar]
- 42.Liwanag AJ, et al. Pectin biosynthesis: GALS1 in Arabidopsis thaliana is a β-1,4-galactan β-1,4-galactosyltransferase. Plant Cell. 2012;24:5024–5036. doi: 10.1105/tpc.112.106625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pattathil S, Avci U, Miller JS, Hahn MG. Immunological approaches to plant cell wall and biomass characterization: Glycome Profiling. Methods Mol Biol. 2012;908:61–72. doi: 10.1007/978-1-61779-956-3_6. [DOI] [PubMed] [Google Scholar]
- 44.Lamesch P, et al. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 47.MacLean B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 49.Woody ST, Austin-Phillips S, Amasino RM, Krysan PJ. The WiscDsLox T-DNA collection: An arabidopsis community resource generated by using an improved high-throughput T-DNA sequencing pipeline. J Plant Res. 2007;120:157–165. doi: 10.1007/s10265-006-0048-x. [DOI] [PubMed] [Google Scholar]
- 50.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 51.Earley KW, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 52.Nelson BK, Cai X, Nebenführ A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007;51:1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- 53.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2011;39:D32–D37. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.