Significance
Human dietary exposures to aflatoxin in some of the most populated and underdeveloped portions of the world are a major contributing factor to the formation of human hepatocellular carcinomas (HCCs) that account for over 700,000 deaths annually. Although the genomic signatures for aflatoxin-driven carcinogenesis are G:C to T:A point mutations arising from bypass of aflatoxin-induced DNA adducts, maintenance of genome stability has been generally attributed to nucleotide excision repair. However, we present three lines of evidence that the DNA base excision repair pathway initiated by the DNA glycosylase NEIL1 is the major contributor in maintaining genomic stability following aflatoxin exposures. These findings suggest that inactivating NEIL1 polymorphic variants in the human population could affect susceptibility to aflatoxin-induced HCC.
Keywords: aflatoxin, base excision repair, ring-fragmented purines, liver cancer, environmental toxicant
Abstract
Global distribution of hepatocellular carcinomas (HCCs) is dominated by its incidence in developing countries, accounting for >700,000 estimated deaths per year, with dietary exposures to aflatoxin (AFB1) and subsequent DNA adduct formation being a significant driver. Genetic variants that increase individual susceptibility to AFB1-induced HCCs are poorly understood. Herein, it is shown that the DNA base excision repair (BER) enzyme, DNA glycosylase NEIL1, efficiently recognizes and excises the highly mutagenic imidazole ring-opened AFB1-deoxyguanosine adduct (AFB1-Fapy-dG). Consistent with this in vitro result, newborn mice injected with AFB1 show significant increases in the levels of AFB1-Fapy-dG in Neil1−/− vs. wild-type liver DNA. Further, Neil1−/− mice are highly susceptible to AFB1-induced HCCs relative to WT controls, with both the frequency and average size of hepatocellular carcinomas being elevated in Neil1−/−. The magnitude of this effect in Neil1−/− mice is greater than that previously measured in Xeroderma pigmentosum complementation group A (XPA) mice that are deficient in nucleotide excision repair (NER). Given that several human polymorphic variants of NEIL1 are catalytically inactive for their DNA glycosylase activity, these deficiencies may increase susceptibility to AFB1-associated HCCs.
Liver cancers pose an international public health concern as the second leading cause of cancer-related deaths worldwide, with >700,000 estimated deaths per year (1–3). This mortality approaches its annual incidence throughout the world, highlighting the need for development of effective treatments and early diagnostic tools. HCCs represent the major histological subtype among liver cancers. The global distribution of HCCs is dominated by its incidence in developing countries, especially in eastern Asia and Africa, where two major chronic etiological factors drive this disease: (i) routine dietary exposures to grains and nuts that are contaminated with molds, Aspergillus flavus and Aspergillus parasiticus, which produce aflatoxins, and (ii) extremely high rates of hepatitis B (HBV) and C viral infections. In geographical regions of China where aflatoxin contamination of human food products is highest, there is a large shift in not only the age of onset of HCCs, but also in the incidence rate. Within Qidong, a significant number of HCCs occur in males beginning in their early 20s, with the frequency of HCCs peaking between the ages of 40 and 50 (4). These data are in contrast to HCC frequencies in portions of China, such as Beijing, where aflatoxin exposures are minimal. The kinetics of HCC formation in aflatoxin-affected areas are similar to that observed in early onset breast and ovarian cancers in women who are carriers (heterozygotic) for inactivating mutations in BRCA1 or 2.
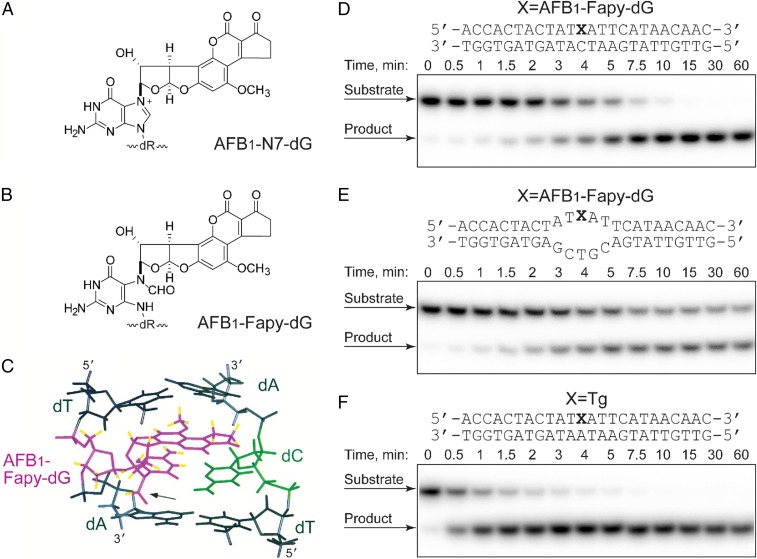
Although there are several different aflatoxin structures, AFB1 has been demonstrated to be the most potent hepatocarcinogen (5, 6). Following ingestion and activation by liver microsomal enzymes, the intermediate AFB1 epoxide can form a covalently bound adduct at N7 guanine in DNA, AFB1-N7-dG (Fig. 1A) (7–9). Kinetic analyses of the formation and disappearance of AFB1-DNA adducts revealed that ∼20% of the initial, quantitatively abundant, cationic AFB1-N7-dG lesions converts to the ring-opened trans-8,9-dihydro-8-(2,6-diamino-4-oxo-3,4-dihydropyrimid-5-yl-formamido)-9-hydroxy aflatoxin B1 (AFB1-Fapy-dG) adduct (Fig. 1B) within 24 h after a single dose. These adducts become the dominant DNA damage detected in cellular DNA 72 h postexposure, comprising up to 80% of all known AFB1-induced lesions (10). The AFB1-Fapy-dG adduct can exist in two interconvertible anomeric forms, α and β, that differ by orientation of the glycosyl bond relative to the deoxyribose (11). Although the α and β anomers equilibrate at an approximately equal ratio in single-stranded environments, the β form is strongly favored in duplex DNA where it intercalates within DNA (12) (Fig. 1C). Previously, we demonstrated that in primate cells, the AFB1-Fapy-dG adduct is more mutagenic than AFB1-N7-dG with mutation frequencies measured at 97% and 32%, respectively; the mutagenic spectrum for each was dominated by G-to-T transversions (13–15). Between the two anomers of AFB1-Fapy-dG, the α species was a severe block to replication, whereas the β species was implicated as a major contributor to mutagenesis (15). The spectra of mutations induced by AFB1-dG adducts correlate with the mutation signature observed in genomes of AFB1-associated HCCs (16, 17), thus underscoring a causal link between these lesions and genetic alterations associated with these cancers.
Fig. 1.
DNA adduct structures and repair of the AFB1-Fapy-dG adduct by NEIL1. (A) Structure of the AFB1-N7-dG adduct. (B) Structure of the AFB1-Fapy-dG adduct. (C) NMR solution structure of the AFB1-Fapy-dG adduct in a 5′-TXA-3′ sequence context where X is the AFB1-Fapy-dG adduct (12). (D–F) hNEIL1-catalyzed incision of lesion-containing DNA under single-turnover conditions. The 32P-labeled oligodeoxynucleotides (20 nM) containing AFB1-Fapy-dG either in fully duplex DNA (D) or a bubble structure (E) or Tg in fully duplex DNA (F) were incubated with hNEIL1 (230 nM). The aliquots were removed at the indicated times, and following separation by gel electrophoresis, DNA was visualized using a phosphor screen and Personal Molecular Imager System (Bio-Rad). The representative gel images are shown. The product formation was plotted as a function of time, and the kobs values were obtained from the best fit of the data to a single exponential equation using KaleidaGraph software.
The chemical stability of the AFB1-Fapy-dG adduct is consistent with the long half-lives of many ring-fragmented purines, and the persistence of the AFB1-Fapy-dG adduct in cells can be at least partially attributed to its intercalation into duplex DNA as detected by NMR (Fig. 1C) and consequent stabilization of the DNA as measured by increased melting temperature (12). As with many bulky DNA adducts, DNA repair of both AFB1-N7-dG and AFB1-Fapy-dG occurs via the NER pathway in Escherichia coli (18). The involvement of NER in the removal of AFB1-N7-dG in human cells is also evident because more adducts accumulate in NER-deficient XPA cells relative to NER-proficient fibroblasts 48 h posttreatment (19). In addition, XPA−/− mice are somewhat more susceptible to induction of HCC relative to WT mice following a single neonatal challenge with AFB1 (20). A potential role of the base excision repair (BER) pathway in removal of the AFB1-Fapy-dG lesion has previously been investigated. Chetsanga and Frenette (21) presented data suggesting that the E. coli formamidopyrimidine DNA glycosylase (Fpg) could incise a portion of the imidazole ring-opened guanines from DNAs that had been previously treated with aflatoxin. This activity could be inhibited using an Fpg inhibitor, suggesting an Fpg-catalyzed reaction. However, studies using E. coli cells deficient in Fpg did not support a role for this enzyme in aflatoxin adduct repair (18). A role for BER in the repair of AFB1-Fapy-dG in human cells has not been previously investigated. The human DNA glycosylase that is predominately responsible for the initiation of BER of Fapy adducts is hNEIL1, with substrates including the secondary oxidation products of 8-oxoG, Fapy-dA, Fapy-dG, methyl-Fapy-dG, and a subset of oxidized pyrimidines (22–30). In addition, this enzyme exhibits distinct structural preference for repairing lesions that are located within DNA bubble structures, modeling DNA intermediates formed during transcription and replication (29). Therefore, based on the broad substrate range of NEIL1, it was hypothesized that repair of AFB1-Fapy-dG could be initiated by NEIL1 as the first step of the BER pathway.
Results
NEIL1 Incision of DNA Containing a Site-Specific AFB1-Fapy-dG Adduct.
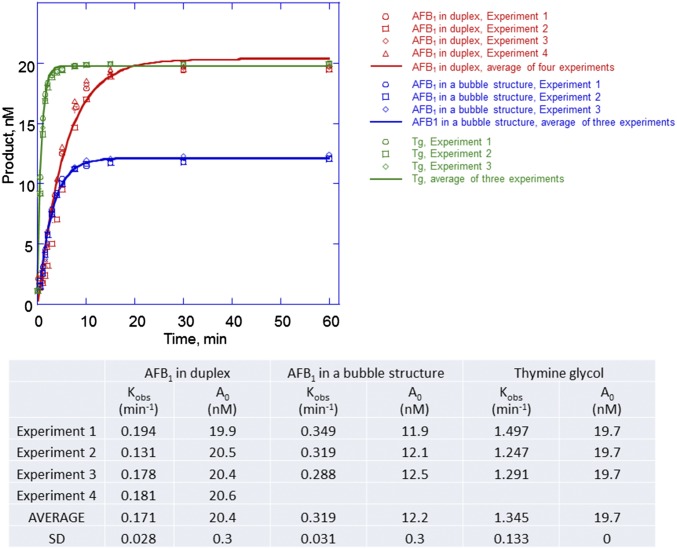
A site-specifically modified oligodeoxynucleotide (24 mer) was constructed with the adduct positioned at nucleotide position 12. Because the release of NEIL1 from its products is rate-limiting, conditions of single turnover kinetics were used, with NEIL1 in excess relative to the DNA substrate. The DNA substrates were either fully duplex or a 5-nucleotide single-stranded bubble structure (Fig. 1 D and E, respectively); DNA adducts in bubble structures have been identified as preferred substrates for NEIL1 (29). The product formation followed a single exponential rise with observed excision rate constants of 0.17 ± 0.03 and 0.32 ± 0.03 (average ± SD) min−1, respectively (Fig. 1 D and E and Fig. S1). Although the AFB1-Fapy-dG–containing duplex DNA was completely cleaved by NEIL1 (Fig. 1D), ∼40% of adducts in a bubble structure appeared to be resistant to cleavage (Fig. 1E). This observation suggests that only the β anomer is a substrate for NEIL1. Although these rate constants are close to previously reported rate constants for NEIL1 excision at 5-hydroxycytosine, 5-hydroxyuracil, and thymine glycol (Tg) (0.24, 0.14, and 1.3 min−1, respectively) (28, 30), we also confirmed these correlations by measuring incision kinetics on a duplex DNA containing a site-specific Tg adduct (Fig. 1F and Fig. S1). In excellent agreement with the prior literature, the observed rate constant for NEIL1-mediated incision of the Tg substrate was 1.35 ± 0.13 (average ± SD) min−1.
Fig. S1.
hNEIL1-catalyzed incision of the AFB1-Fapy-dG–containing DNA under single-turnover conditions. The 32P-labeled oligodeoxynucleotides (20 nM) containing AFB1-Fapy-dG either in full duplex or bubble structure or Tg were incubated with hNEIL1 (230 nM). The aliquots were removed in the course of reaction, and following separation by gel electrophoresis, DNA was visualized using a phosphor screen and Personal Molecular Imager System (Bio-Rad). The substrate structures and representative gel images are shown in Fig. 1. The product formation was plotted as a function of time, and the kobs and A0 values were obtained from the best fit of the data to a single exponential equation [P]t = A0[1 − exp(kobst)] using KaleidaGraph software. The data of individual experiments and the average values with the corresponding SDs are shown in the table below. For ease of visualization, the averages of individual time points were calculated from four or three experiments, and the resulting curves are shown as solid lines.
Increased AFB1-Fapy-dG Adduct Accumulation in Livers from Neil1−/− Relative to WT Mice.
The high efficiency with which NEIL1 catalyzed the release of the AFB1-Fapy-dG adducts was unanticipated because the base modification stabilizes the melting temperature of a 12-mer duplex DNA by 15 °C (12), and this increase would make the modified nucleotide resistant to extrahelical flipping as a prerequisite of glycosyl bond scission. Sterically, this adduct also represents the largest DNA base modification reported as a substrate for any DNA glycosylase. To extend the in vitro biochemical analyses to an in vivo repair assay, DNA adduct levels were measured in the livers of newborn WT and Neil1−/− mice (31, 32). We hypothesized that levels of AFB1-Fapy-dG would be significantly lower in WT vs. Neil1−/− mice because the WT mice could repair these adducts via both the NER and BER pathways, whereas the Neil1−/− mice would only repair this lesion via NER.
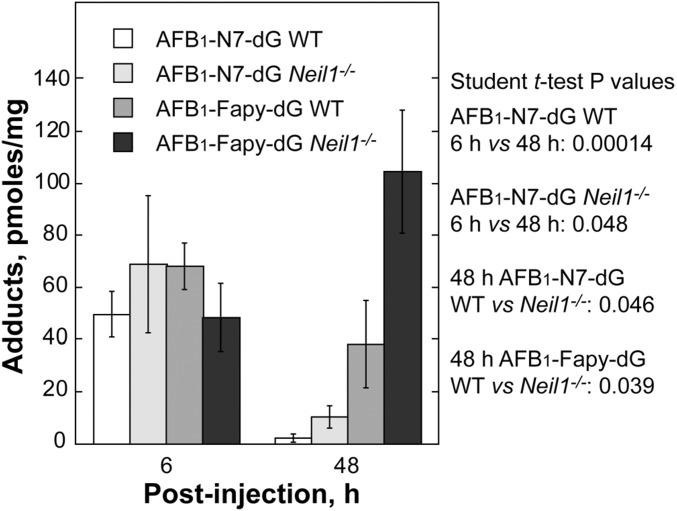
Newborn WT and Neil1−/− mice (∼6 d old) were i.p. injected with ∼3.5 mg/kg AFB1 in DMSO. Livers were harvested at 6 and 48 h postinjection and DNA was extracted and analyzed by mass spectrometry. At the 6-h time point, both genotypes had approximately equal levels of the AFB1-N7-dG and AFB1-Fapy-dG adducts (Fig. 2). At 48-h, the levels of AFB1-N7-dG were significantly decreased in both genotypes, with these decreases attributable to a combination of spontaneous depurination, repair by NER, and conversion to the AFB1-Fapy-dG adduct. As anticipated from the hypothesis that NEIL1 would significantly contribute to the repair of AFB1-Fapy-dG adducts, there was a significant difference in the amount of these adducts at 48 h in WT vs. Neil1−/− mice (38.2 vs. 104.5 pmol/mg DNA, P = 0.039, respectively). These data implicate NEIL1 as a major contributor to repair of this lesion.
Fig. 2.
Levels of AFB1-induced DNA adducts in liver DNA from WT and Neil1−/− mice following AFB1 i.p. injection. Neil1−/− and control WT mice (6-d-old pups) were injected with 10 mM AFB1 in DMSO at a dose of 3.5 mg/kg. Livers were harvested at 6 and 48 h postinjection, and AFB1-induced DNA adducts were measured.
Neil1−/− Mice Develop AFB1-Induced HCCs at Significantly Higher Frequencies Relative to WT Mice.
Because both the biochemical and in vivo adduct accumulation data showed that NEIL1-initiated BER can contribute to the overall repair of the mutagenic AFB1-Fapy-dG adduct, it was hypothesized that Neil1−/− mice would be more susceptible to AFB1-induced carcinogenesis relative to WT C57Bl6J mice. Newborn pups (<7 d old) from matings of Neil1−/− × Neil1−/− and C57Bl6/J × C57Bl6/J (Neil1+/+) were challenged with a single i.p. injection of AFB1 in DMSO at 1.0 or 7.5 mg/kg. Injections of DMSO alone were used as the control. All mice were immediately returned to their litters and monitored daily for adverse health effects. No injection-associated lethality was observed for either the DMSO controls or the 1.0 mg/kg AFB1 doses; for mice challenged with 7.5 mg/kg AFB1, there was significant lethality observed for both genotypes, with a greater toxicity observed in WT relative to Neil1−/− (Fig. S2). Although the mechanism underlying the decreased lethality in knockout mice is not known, the deficiency in BER-initiated repair may reduce the number of DNA strand breaks in affected tissues, with a concomitant decrease in apoptosis, but at the expense of increased mutagenesis.
Fig. S2.
Kaplan–Meier estimated survival curves for WT (solid black) and Neil1−/− (dashed gray) pups given 7.5 mg/kg AFB1. Survival was censored at 30 d (vertical tick mark on each curve) with the proportion surviving beyond 30 d noted for each group.
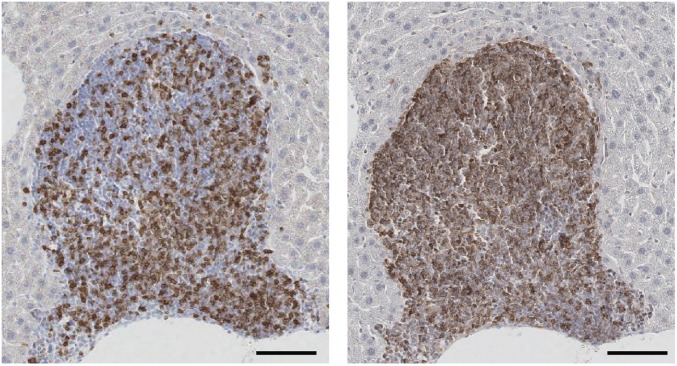
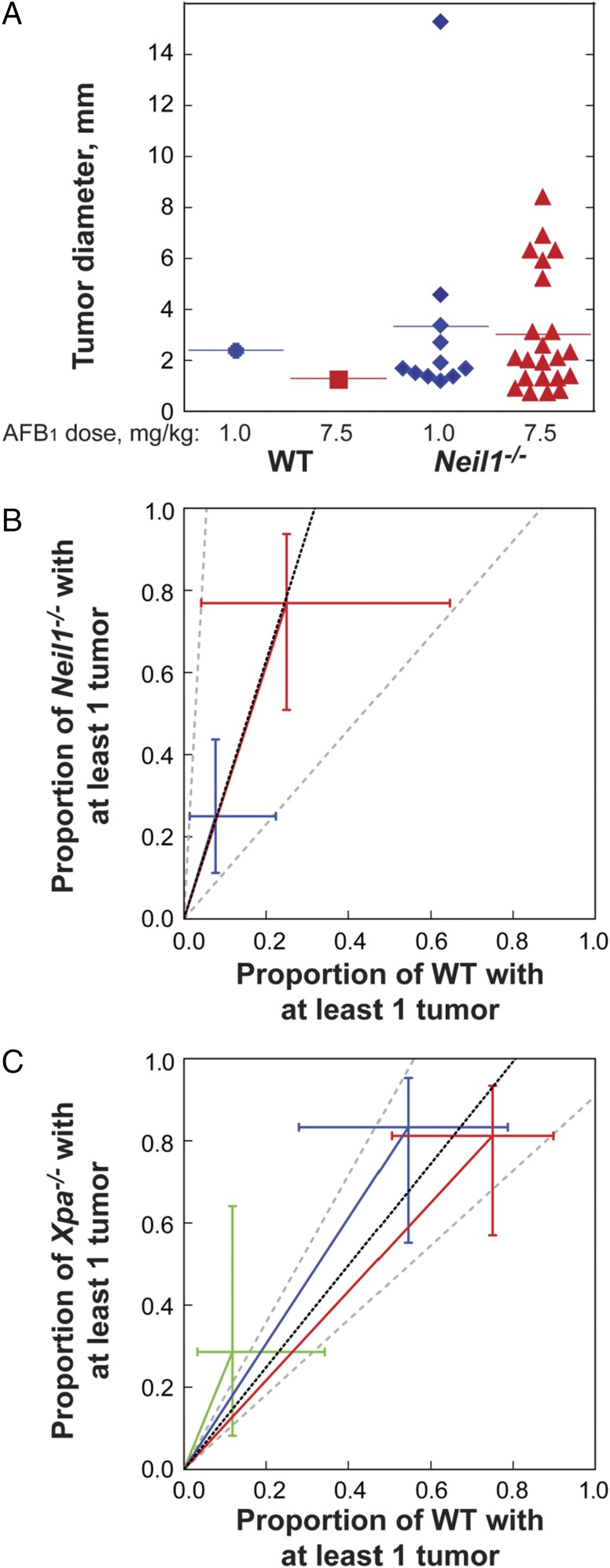
At 15 mo postexposure, all mice were euthanized and examined for macroscopic tumors of the liver. All livers were fixed and prepared for serial sectioning and staining to determine tumor number and size. No tumors were detected in any of the DMSO-injected controls. Consistent with prior literature (33), female mice were highly resistant to liver tumor induction; specifically, only 2 of 32 Neil1−/− females developed tumors, whereas no tumors were observed in any WT females. The mechanistic basis for this strong gender effect is not known, but does not appear to be associated with differences in the number of mutations generated in male vs. female mice, even at 10 wk postexposure (34). All remaining analyses are restricted to tumors developing in male mice, and the quantitation of these is given below. Representative H&E and glypican stains of both small (∼2 mm) and large (>12 mm) tumors are shown in Fig. 3. Based on glypican and reticulin stains, all tumors were classified as carcinomas. During routine analyses of H&E-stained slides, it was observed that there were frequent pockets of lymphoid cell infiltration as an indication of significant inflammation. To distinguish these sites from lymphoid tumors, liver sections were stained with antibodies against CD3 and CD20 (Fig. S3). Analyses of these data revealed that all lymphoid aggregates were complex mixtures of T and B cells, indicating that these were likely to be regions of the liver that were undergoing significant tissue injury. Further evaluation of serial H&E sections through tumor-bearing tissues allowed for the maximum diameter to be measured, with the size of each tumor shown (Fig. 4A). Liver cancers were observed in both the Neil1−/− and WT males, albeit at greatly different frequencies and size distributions at both doses (Fig. 4A and Table 1). For Neil1−/− mice, the risk ratio of developing at least one tumor was calculated to be 3.15-fold greater than the corresponding risk of tumor development for WT mice (95% CI: 1.15–18.17; P = 0.020; Fig. 4B). It is interesting to note that the effect due to dose (7.5 mg/kg AFB1 vs. 1.0 mg/kg AFB1) was comparable in size (risk ratio = 3.09; 95% CI: 1.57–7.06, P = 0.001) relative to the increase in tumor risk associated with genotype. The mean number of tumors per Neil1−/− mouse vs. WT was estimated to be 4.76-fold greater (95% CI: 1.36–30.03; P = 0.012). Similarly, the associated effect due to dose was comparable in the Neil1−/− mice, with the mean number of tumors at 7.5 vs. 1 mg/kg AFB1 calculated to be 4.14 (95% CI: 1.77–10.56, P = 0.001). Due to the large number of animals with no tumors (diameter of 0 mm) and the somewhat irregular distribution of diameters for those with tumors, a permutation test was used to assess whether the observed allocation of tumor diameters in WT vs. Neil1−/− mice for both doses was likely the result of chance (statistical analyses presented in Materials and Methods). These data suggested that observed differences in the distribution of tumor size between WT and Neil1−/− mice could not be attributed to chance (permutation P = 0.0286), but with no evidence to suggest systematic differences due to dose (permutation P = 0.1692) or the interaction between genotype and dose (permutation P = 0.2534). Even though there was a strong dependence on both dose and genotype for the frequencies of tumors, the lack of either a dose or genotype effect on the size of the tumor was anticipated because tumor induction is an extremely rare event relative to the total number of cells exposed to AFB1.
Fig. 3.
Representative AFB1-induced HCCs in Neil1−/− mice. (A) Representative images of H&E-stained tumors. (Scale bars: Left, 1 mm; Right, 100 μm.) (B). Representative images of serial sections of HCCs and peripheral tissues stained with H&E (Left) and glypican (Right). (Scale bars: 100 µm.)
Fig. S3.
Representative immunohistochemistry of large lymphoid aggregates in liver tissues of Neil1−/− mice. Liver tissues were collected 15 mo following AFB1 challenge, fixed, and serial sections processed for immunohistochemistry. (Left) CD3 antibody stain revealing T-cell infiltration. (Right) CD20 antibody stain revealing B-cell infiltration. Costaining is indicative of tissue injury. (Scale bars: 100 μm.)
Fig. 4.
AFB1-induced carcinogenesis in Neil1−/− and XPA−/− mice. (A) The individual diameter of liver tumors observed in AFB1-injected WT and Neil1−/− mice. Relative AFB1-induced tumor risk analysis in (B) Neil1−/− mice, with data illustrated by blue and red symbols representing AFB1 doses of 1.0 and 7.5 mg/kg respectively, and (C) XPA−/− mice, with data illustrated by green, blue, and red symbols representing AFB1 doses of 0, 0.6, and 1.5 mg/kg, respectively.
Table 1.
Tumor frequency in AFB1-challenged male mice
| Dose, mg/kg | Genotype | n | Proportion with at least one tumor | Mean number of tumors per mouse |
| 1.0 | WT | 13 | 0.0770.079 (0.014, 0.224) | 0.0770.088 (0.013, 0.318) |
| Neil1−/− | 24 | 0.2500.249 (0.112, 0.437) | 0.4580.417 (0.180, 0.831) | |
| 7.5 | WT | 4 | 0.2500.244 (0.042, 0.646) | 0.2500.362 (0.057, 1.260) |
| Neil1−/− | 13 | 0.7690.770 (0.509, 0.937) | 1.6921.724 (0.984, 2.855) |
Subscribed values are the estimates from a model based on main effects of dosage and genotype; confidence intervals (95% coverage) are given in parentheses.
NEIL1 Deficiency Confers Increased AFB1-Mediated HCC Formation Relative to XPA-Deficient Mice.
Because these data demonstrated that NEIL1-initiated repair contributed significantly to the reduction in AFB1-induced HCCs in mice, we sought to compare these data with a previous investigation that examined HCC formation in NER-deficient (XPA−/−) mice following AFB1 challenges (20). In contrast to our study in which all mice were extensively backcrossed into the C57BL/6 background, the XPA−/− carcinogenesis study used mice that had been backcrossed 10 times into inbred C3H/HeN mice, and we recognize that these strain differences could lead to differences in the magnitude of tumor formation. Further, due to the XPA−/− background, these mice developed high levels of spontaneous liver tumors, such that at 16 mo of age, with no exogenous challenges, 79% of these mice developed liver cancers, with 57% being classified as HCCs. Given this high background at 16 mo of age, the AFB1 carcinogenesis study was carried out for only 11 mo, and these data revealed that a single dose of AFB1 at 0, 0.6, and 1.5 mg/kg into 7-d-old mice resulted in 14, 50, and 38% HCCs, respectively. Based on the reported numbers of mice with at least one tumor (tables III–V in ref. 20), the overall common risk ratio (adjusted for dose) was estimated to be 1.24 (95% CI: 0.91–1.79, P = 0.174; Fig. 3C). Relative to the current study investigating the risk ratio associated with the Neil1−/− genotype in which the risk ratio was 3.15, the effect of the NER deficiency in XPA−/− mice was considerably less.
Discussion
The constellation of events that contribute to an individual developing early onset HCC is multifactorial, with potential contributions from the following: (i) the robustness of DNA repair to complete removal of stable aflatoxin-induced DNA adducts before replication; (ii) the mutagenic burden sustained by cells in the target tissues; (iii) the balance of bioactivation and detoxication pathways; (iv) the extent of in utero and dietary aflatoxin exposures; (v) other dietary and lifestyle choices (i.e., alcohol consumption and smoking that promote inflammation); and (vi) the specific strain of HBV with which the individual is infected, such that infection with HBV carrying the 1762T/1764A double mutation significantly increases susceptibility vs. infection with the more common strain of HBV. Genetic polymorphisms that tip the balance in favor of compromised vs. efficient DNA repair, bioactivation over detoxication, and the specifics of the HBV strain will increase the odds ratio for early onset disease (35, 36). However, despite more than 50 years of aflatoxin research, the specific genetic variants that influence or drive this early onset disease have not been elucidated.
To better understand the genetic landscape that could modulate individual sensitivities to dietary aflatoxin exposures, it is critical to know the biochemical pathways that contribute to the repair of the AFB1-Fapy-dG adduct. Herein, we have presented three sets of data, all implicating NEIL1-initiated BER as a significant modulator of AFB1-induced carcinogenesis. First, we have shown the ability of NEIL1 to catalyze the release of the AFB1-Fapy-dG adduct from synthetic oligodeoxynucleotides. Second, we demonstrated that Neil1−/− mice accumulate approximately threefold more AFB1-Fapy-dG adducts relative to WT. Third, data were presented showing that Neil1−/− mice were considerably more susceptible to AFB1-induced HCCs. Overall, these data suggest that the majority of repair of AFB1-Fapy-dG adducts may be catalyzed by NEIL1-initiated BER. If these mouse carcinogenesis studies can be extrapolated to the human populations exposed to various aflatoxins, then deficiencies in the enzymatic activities of NEIL1 may reduce the efficiency of repair of these highly mutagenic adducts. In this regard, others and we have previously carried out biochemical characterization of several NEIL1 polymorphic variants (37, 38). When expressed as purified proteins, two of the variants, G83D and C136R, showed little to no DNA glycosylase activity. We hypothesize that humans who are heterozygous for a glycosylase-defective form of NEIL1 or possess mutations that effect (i) the stabilization or localization of the enzyme; (ii) the protein domains responsible for other important protein–protein interactions such as PARP1, XRCC1, hnRNP-U, and the 9-1-1 complex (reviewed in ref. 39) or (iii) regulation of transcription/translation processes may be at an elevated risk for increased steady-state levels of AFB1-Fapy-dG adducts; this in turn would affect mutation load and the potential loss of heterozygosity in NEIL1 by a secondary mutagenic event. These data could be used in early diagnosis/disease prevention strategies.
Materials and Methods
Materials.
The following materials and reagents were purchased: T4 polynucleotide kinase and BSA (New England BioLabs); [γ-32P]ATP (PerkinElmer Life Sciences); Micro Bio-Spin 6 column (Bio-Rad); all other chemicals were from Sigma-Aldrich or Fisher Scientific. hNEIL1 was purified as previously described (37) and the total concentration of the protein was determined by the Bradford assay, using BSA as the standard. Aflatoxin B1 was purchased from Sigma Aldrich; it was dissolved in DMSO and used immediately after preparation. Aflatoxin B1 is a very potent carcinogen and thus great care should be exercised to avoid personnel exposure. Further, the crystalline material presents an inhalation hazard.
DNA Substrate Preparation.
The AFB1-Fapy-dG containing oligodeoxynucleotide was synthesized as previously described (40). The nonadducted and Tg-containing oligodeoxynucleotides were obtained from Integrated DNA Technologies, Inc. The adducted oligodeoxynucleotide was labeled at the 5′ terminus using [γ-32P]ATP and T4 polynucleotide kinase and annealed to the complementary strand at molar ratio of 1:1.2 by heating the mixture at 90 °C for 2 min and gradual cooling to room temperature.
DNA Glycosylase Assays.
To evaluate the glycosylase activity of hNEIL1, the rate constants were measured under single-turnover conditions. Reactions were initiated by the addition of 230 nM active enzyme into a total volume of 80 μL in the presence of 20 mM Tris⋅HCl, pH 7.4, 100 mM KCl, and 100 μg/mL BSA and incubated at 37 °C with a final DNA concentration of 20 nM. The aliquots (5 μL) were removed in the course of reactions, and following addition of equal volume of 0.5 N NaOH, incubated at 90 °C for 5 min. By converting the newly formed abasic sites into DNA strand breaks, the treatment with NaOH allowed for the measurement of the rates of glycosylase activity separately from the lyase activity. DNA was denatured by addition of 20 μL of solution containing 95% formamide, 20 mM EDTA, 0.02% xylene cyanol, and 0.02% bromophenol blue followed by incubation at 90 °C for 2 min, and resolved by electrophoresis in a 20% denaturing polyacrylamide gel containing 8 M urea in Tris-borate-EDTA buffer. The reaction products were visualized using Personal Molecular Imager System (Bio-Rad). The product [P] was quantified from a phosphor screen image by the Quantity One Software (Bio-Rad) and plotted as a function of time (t) using KaleidaGraph software. The first-order rate constant (kobs) and extrapolated maximal product (A0) were obtained from the best fit of the data to equation [P]t = A0[1 − exp(kobst)]. Control reactions without enzyme were performed under identical conditions showing ∼5% of a nonenzymatic cleavage that was not affected by the duration of incubation in the reaction buffer at 37 °C. The extension of incubation with NaOH up to 20 min also did not change the level of this cleavage. Thus, the AFB1-Fapy-dG–modified oligodeoxynucleotides were stable under used experimental conditions.
Animals.
Construction and characterization of Neil1 knockout mice was previously described (31, 32). The Neil1 genotype has been backcrossed >15 generations onto a C57Bl/6J background. Control mating pairs of WT C57Bl6/J were obtained from Jackson Laboratories. All animal protocols were preapproved through the Oregon Health & Science University Institutional Animal Care and Use Committee and monitored by the Department of Comparative Medicine.
Measurement of AFB1-Induced DNA Adducts in Liver of Exposed Mice.
Separate mating of Neil1−/− and control WT mice were set up, and 6-d-old pups were injected with freshly reconstituted 10 mM AFB1 in DMSO. The pups were weighed, i.p. injected with AFB1 at a dose of 3.5 mg/kg, and returned to their original cages. The livers were harvested at 6 and 48 h postinjection and immediately frozen in liquid nitrogen. DNA for AFB1 adduct measurement was isolated using the previously described method (10). AFB1-induced DNA adducts were hydrolyzed by treatment with 0.1 N HCl at 95 °C for 15 min, and following hydrolysis, internal 15N5-guanine–derived standards were added to permit quantitative analysis by isotope dilution mass spectrometry for both AFB1-N7-dG and AFB1-Fapy-dG. Adducts were separated by ultra-performance liquid-chromatography mass spectrometry. The protonated parent ion of the AFB1-N7-Gua adduct (m/z 480.1) was selected and subjected to collision-induced fragmentation producing a m/z 152 product ion that was monitored to quantify adduct levels. The AFB1-Fapy-Gua adduct was measured by selection of the m/z 498 parent ion and monitoring the collision-induced product ion m/z 452.29 (41).
Mouse Carcinogenesis.
The pups were obtained as described above and, at <7 d of age, injected with AFB1 in DMSO at a dose of either 1.0 or 7.5 mg/kg. DMSO alone was used as a vehicle control. Postinjection, pups were immediately returned to the litter and weaned at 21 d of age. Litters were monitored on a daily basis for mortality. At weaning, genders were determined and pups were group housed up to five per box. Both males and females were routinely monitored for developmental abnormalities and changes in weight throughout the 15-mo study. Any mouse that either experienced weight loss of >15% or ceased grooming or became severely lethargic was euthanized before death. For the remaining majority of mice, these were euthanized at 15 mo of age and the livers and lungs and associated tissues examined for morphological changes, including tumors. Harvested tissues were fixed in aqueous buffered zinc formalin [4% (wt/vol) formaldehyde and 600 ppm zinc], and after 2 d transferred into 70% (vol/vol) ethanol. Tissues were paraffin embedded and sections cut for H&E staining. Selected tissues were also analyzed by immunohistochemistry, including detection of glypican (Abcam) and reticulin (American MasterTech) for tumor classification and CD3 (Spring Bioscience) and CD20 (Invitrogen/Life Technologies) for lymphoid aggregate analyses.
Statistical Analyses.
Survival curves for WT and Neil1−/− pups were produced using Kaplan–Meier estimates, with censoring of survival times beyond 30 d. Curves were compared using both the Peto–Peto test, which places more weight/influence on the earlier part of the survival curves, and the log-rank test that treats all data equally, making it susceptible to large differences at the end of the curves. The proportion of surviving pups, as well as the proportion of mice that develop at least one tumor, were compared using log-binomial regression, and the number of tumors was estimated and tested using a negative binomial model with constant dispersion. Due to the limited sample sizes, likelihood ratio tests (instead of asymptotic Wald tests) were used to determine significance for each of the above models (log binomial or negative binomial) and confidence intervals were constructed by inverting the test. Tumor diameter was compared between genotypes and doses using analysis of variance, with P values determined through 5,000 random permutations of the data. Analyses were conducted using Stata (ver. 13.1; StataCorp) and R (R Core Team).
Acknowledgments
This work was supported by NIH Grants R01 CA 055678 (to M.P.S. and R.S.L.); R01 ES 016313, P30 ES002109, and R01 CA080024 (to J.M.E.); and R01 CA190610 and P30 CA006973 (to J.D.G.). S.C. is supported by a Schlumberger Foundation Faculty for the Future Grant.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620932114/-/DCSupplemental.
References
- 1.Blonski W, Kotlyar DS, Forde KA. Non-viral causes of hepatocellular carcinoma. World J Gastroenterol. 2010;16:3603–3615. doi: 10.3748/wjg.v16.i29.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F, Ren JS, Masuyer E, Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer. 2013;132:1133–1145. doi: 10.1002/ijc.27711. [DOI] [PubMed] [Google Scholar]
- 3.Chitapanarux T, Phornphutkul K. Risk factors for the development of hepatocellular carcinoma in Thailand. J Clin Transl Hepatol. 2015;3:182–188. doi: 10.14218/JCTH.2015.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kensler TW, Roebuck BD, Wogan GN, Groopman JD. Aflatoxin: A 50-year odyssey of mechanistic and translational toxicology. Toxicol Sci. 2011;120:S28–S48. doi: 10.1093/toxsci/kfq283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ayres JL, Lee DJ, Wales JH, Sinnhuber RO. Aflatoxin structure and hepatocarcinogenicity in rainbow trout (Salmo gairdneri) J Natl Cancer Inst. 1971;46:561–564. [PubMed] [Google Scholar]
- 6.Wogan GN, Edwards GS, Newberne PM. Structure-activity relationships in toxicity and carcinogenicity of aflatoxins and analogs. Cancer Res. 1971;31:1936–1942. [PubMed] [Google Scholar]
- 7.Croy RG, Essigmann JM, Reinhold VN, Wogan GN. Identification of the principal aflatoxin B1-DNA adduct formed in vivo in rat liver. Proc Natl Acad Sci USA. 1978;75:1745–1749. doi: 10.1073/pnas.75.4.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Essigmann JM, et al. Structural identification of the major DNA adduct formed by aflatoxin B1 in vitro. Proc Natl Acad Sci USA. 1977;74:1870–1874. doi: 10.1073/pnas.74.5.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin CN, Garner RC. Aflatoxin B-oxide generated by chemical or enzymic oxidation of aflatoxin B1 causes guanine substitution in nucleic acids. Nature. 1977;267:863–865. doi: 10.1038/267863a0. [DOI] [PubMed] [Google Scholar]
- 10.Croy RG, Wogan GN. Temporal patterns of covalent DNA adducts in rat liver after single and multiple doses of aflatoxin B1. Cancer Res. 1981;41:197–203. [PubMed] [Google Scholar]
- 11.Brown KL, et al. Unraveling the aflatoxin-FAPY conundrum: Structural basis for differential replicative processing of isomeric forms of the formamidopyrimidine-type DNA adduct of aflatoxin B1. J Am Chem Soc. 2006;128:15188–15199. doi: 10.1021/ja063781y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mao H, Deng Z, Wang F, Harris TM, Stone MP. An intercalated and thermally stable FAPY adduct of aflatoxin B1 in a DNA duplex: Structural refinement from 1H NMR. Biochemistry. 1998;37:4374–4387. doi: 10.1021/bi9718292. [DOI] [PubMed] [Google Scholar]
- 13.Lin YC, et al. Molecular basis of aflatoxin-induced mutagenesis-role of the aflatoxin B1-formamidopyrimidine adduct. Carcinogenesis. 2014;35:1461–1468. doi: 10.1093/carcin/bgu003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin YC, et al. Error-prone replication bypass of the primary aflatoxin B1 DNA adduct, AFB1-N7-Gua. J Biol Chem. 2014;289:18497–18506. doi: 10.1074/jbc.M114.561563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smela ME, et al. The aflatoxin B1 formamidopyrimidine adduct plays a major role in causing the types of mutations observed in human hepatocellular carcinoma. Proc Natl Acad Sci USA. 2002;99:6655–6660. doi: 10.1073/pnas.102167699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bressac B, Kew M, Wands J, Ozturk M. Selective G to T mutations of p53 gene in hepatocellular carcinoma from southern Africa. Nature. 1991;350:429–431. doi: 10.1038/350429a0. [DOI] [PubMed] [Google Scholar]
- 17.Hsu IC, et al. Mutational hotspot in the p53 gene in human hepatocellular carcinomas. Nature. 1991;350:427–428. doi: 10.1038/350427a0. [DOI] [PubMed] [Google Scholar]
- 18.Alekseyev YO, Hamm ML, Essigmann JM. Aflatoxin B1 formamidopyrimidine adducts are preferentially repaired by the nucleotide excision repair pathway in vivo. Carcinogenesis. 2004;25:1045–1051. doi: 10.1093/carcin/bgh098. [DOI] [PubMed] [Google Scholar]
- 19.Leadon SA, Tyrrell RM, Cerutti PA. Excision repair of aflatoxin B1-DNA adducts in human fibroblasts. Cancer Res. 1981;41:5125–5129. [PubMed] [Google Scholar]
- 20.Takahashi Y, et al. Enhanced spontaneous and aflatoxin-induced liver tumorigenesis in Xeroderma pigmentosum group A gene-deficient mice. Carcinogenesis. 2002;23:627–633. doi: 10.1093/carcin/23.4.627. [DOI] [PubMed] [Google Scholar]
- 21.Chetsanga CJ, Frenette GP. Excision of aflatoxin B1-imidazole ring opened guanine adducts from DNA by formamidopyrimidine-DNA glycosylase. Carcinogenesis. 1983;4:997–1000. doi: 10.1093/carcin/4.8.997. [DOI] [PubMed] [Google Scholar]
- 22.Dizdaroglu M. Oxidatively induced DNA damage and its repair in cancer. Mutat Res Rev Mutat Res. 2015;763:212–245. doi: 10.1016/j.mrrev.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 23.Dizdaroglu M, Kirkali G, Jaruga P. Formamidopyrimidines in DNA: Mechanisms of formation, repair, and biological effects. Free Radic Biol Med. 2008;45:1610–1621. doi: 10.1016/j.freeradbiomed.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 24.Hazra TK, et al. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc Natl Acad Sci USA. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu J, et al. Repair of formamidopyrimidines in DNA involves different glycosylases: Role of the OGG1, NTH1, and NEIL1 enzymes. J Biol Chem. 2005;280:40544–40551. doi: 10.1074/jbc.M508772200. [DOI] [PubMed] [Google Scholar]
- 26.Morland I, et al. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: An alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res. 2002;30:4926–4936. doi: 10.1093/nar/gkf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wallace SS, Bandaru V, Kathe SD, Bond JP. The enigma of endonuclease VIII. DNA Repair (Amst) 2003;2:441–453. doi: 10.1016/s1568-7864(02)00182-9. [DOI] [PubMed] [Google Scholar]
- 28.Vik ES, et al. Biochemical mapping of human NEIL1 DNA glycosylase and AP lyase activities. DNA Repair (Amst) 2012;11:766–773. doi: 10.1016/j.dnarep.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 29.Dou H, Mitra S, Hazra TK. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J Biol Chem. 2003;278:49679–49684. doi: 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- 30.Krishnamurthy N, Zhao X, Burrows CJ, David SS. Superior removal of hydantoin lesions relative to other oxidized bases by the human DNA glycosylase hNEIL1. Biochemistry. 2008;47:7137–7146. doi: 10.1021/bi800160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vartanian V, et al. The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase. Proc Natl Acad Sci USA. 2006;103:1864–1869. doi: 10.1073/pnas.0507444103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sampath H, et al. Variable penetrance of metabolic phenotypes and development of high-fat diet-induced adiposity in NEIL1-deficient mice. Am J Physiol Endocrinol Metab. 2011;300:E724–E734. doi: 10.1152/ajpendo.00387.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vesselinovitch SD, Mihailovich N, Wogan GN, Lombard LS, Rao KV. Aflatoxin B1, a hepatocarcinogen in the infant mouse. Cancer Res. 1972;32:2289–2291. [PubMed] [Google Scholar]
- 34.Wattanawaraporn R, et al. A single neonatal exposure to aflatoxin b1 induces prolonged genetic damage in two loci of mouse liver. Toxicol Sci. 2012;128:326–333. doi: 10.1093/toxsci/kfs151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuang SY, et al. Specific mutations of hepatitis B virus in plasma predict liver cancer development. Proc Natl Acad Sci USA. 2004;101:3575–3580. doi: 10.1073/pnas.0308232100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuang SY, et al. Hepatitis B 1762T/1764A mutations, hepatitis C infection, and codon 249 p53 mutations in hepatocellular carcinomas from Thailand. Cancer Epidemiol Biomarkers Prev. 2005;14:380–384. doi: 10.1158/1055-9965.EPI-04-0380. [DOI] [PubMed] [Google Scholar]
- 37.Roy LM, et al. Human polymorphic variants of the NEIL1 DNA glycosylase. J Biol Chem. 2007;282:15790–15798. doi: 10.1074/jbc.M610626200. [DOI] [PubMed] [Google Scholar]
- 38.Prakash A, Carroll BL, Sweasy JB, Wallace SS, Doublié S. Genome and cancer single nucleotide polymorphisms of the human NEIL1 DNA glycosylase: Activity, structure, and the effect of editing. DNA Repair (Amst) 2014;14:17–26. doi: 10.1016/j.dnarep.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dutta A, Yang C, Sengupta S, Mitra S, Hegde ML. New paradigms in the repair of oxidative damage in human genome: Mechanisms ensuring repair of mutagenic base lesions during replication and involvement of accessory proteins. Cell Mol Life Sci. 2015;72:1679–1698. doi: 10.1007/s00018-014-1820-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Banerjee S, Brown KL, Egli M, Stone MP. Bypass of aflatoxin B1 adducts by the Sulfolobus solfataricus DNA polymerase IV. J Am Chem Soc. 2011;133:12556–12568. doi: 10.1021/ja2015668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Egner PA, Groopman JD, Wang JS, Kensler TW, Friesen MD. Quantification of aflatoxin-B1-N7-guanine in human urine by high-performance liquid chromatography and isotope dilution tandem mass spectrometry. Chem Res Toxicol. 2006;19:1191–1195. doi: 10.1021/tx060108d. [DOI] [PubMed] [Google Scholar]