Significance
Neisseria meningitidis (Nm) remains a leading cause of meningitis and rapidly fatal sepsis in otherwise healthy individuals. Historically, Nm is not recognized as a significant cause of urogenital infections. Since 2015, a significant increase of meningococcal urethritis primarily among heterosexual men has been reported in multiple US cities. We defined that a unique nonencapsulated Nm clade, which belonged to the cc11/ET-15 hyperinvasive lineage, was linked to these Nm urethritis clusters. The clade isolates causing urethritis clusters in the United States may have adapted to the urogenital environment with two unique molecular fingerprints: the insertion of IS1301 with associated deletion of capsule, enhancing mucosal adherence, and the acquisition of the gonococcal denitrification pathway by gene conversion, promoting anaerobic growth.
Keywords: Neisseria meningitidis, meningococcal urethritis, capsule, IS1301, denitrification
Abstract
Neisseria meningitidis (Nm) clonal complex 11 (cc11) lineage is a hypervirulent pathogen responsible for outbreaks of invasive meningococcal disease, including among men who have sex with men, and is increasingly associated with urogenital infections. Recently, clusters of Nm urethritis have emerged primarily among heterosexual males in the United States. We determined that nonencapsulated meningococcal isolates from an ongoing Nm urethritis outbreak among epidemiologically unrelated men in Columbus, Ohio, are linked to increased Nm urethritis cases in multiple US cities, including Atlanta and Indianapolis, and that they form a unique clade (the US Nm urethritis clade, US_NmUC). The isolates belonged to the cc11 lineage 11.2/ET-15 with fine type of PorA P1.5–1, 10–8; FetA F3-6; PorB 2–2 and express a unique FHbp allele. A common molecular fingerprint of US_NmUC isolates was an IS1301 element in the intergenic region separating the capsule ctr-css operons and adjacent deletion of cssA/B/C and a part of csc, encoding the serogroup C capsule polymerase. This resulted in the loss of encapsulation and intrinsic lipooligosaccharide sialylation that may promote adherence to mucosal surfaces. Furthermore, we detected an IS1301-mediated inversion of an ∼20-kb sequence near the cps locus. Surprisingly, these isolates had acquired by gene conversion the complete gonococcal denitrification norB-aniA gene cassette, and strains grow well anaerobically. The cc11 US_NmUC isolates causing urethritis clusters in the United States may have adapted to a urogenital environment by loss of capsule and gene conversion of the Neisseria gonorrheae norB-aniA cassette promoting anaerobic growth.
As an obligate human pathogen that can cause large epidemic outbreaks, Neisseria meningitidis remains a leading cause of meningitis and rapidly fatal sepsis in otherwise healthy individuals (1). Although new protein-capsular polysaccharide conjugate and serogroup B outer membrane protein vaccines provide protection against invasive meningococcal disease (IMD), ∼500,000 cases of IMD have occurred worldwide annually, with at least 50,000 deaths and as many survivors suffering neurological sequelae (2). The continued worldwide problem of IMD and the capacity of Nm to evolve quickly are well established.
Nm is asymptomatically carried in the nasopharynx of 5–10% of adults in nonepidemic periods, and transmission usually occurs by direct contact with oral or nasal secretions. Nm is also infrequently recovered from other mucosal sites, specifically the urogenital tract (cervix, vagina, urethra) and the rectum. Certain populations can have significantly higher (>30%) nasopharyngeal carriage (3). In particular, populations of men who have sex with men (MSM) can have >40% pharyngeal Nm carriage rates, urethral Nm carriage of 0.7%, and rectal carriage of up to 2% (4, 5). In addition, Neisseria gonorrheae (Ng) and Nm have been corecovered in MSM and other populations (6).
Historically, Nm is not documented as a significant cause of urogenital infections. However, case reports of meningococcal urethritis, cervicitis, vaginitis, proctitis, pelvic inflammatory disease, and postpartum endometritis date back to the 1940s (7–9). Since 2001, there have been several outbreaks (e.g., Toronto, Chicago, New York City, Los Angeles, Berlin, and Paris) of IMD among MSM (10, 11). Orogenital and anogenital contacts are postulated to be the sexual transmission route of invasive meningococcal infections observed in MSM (6). Members of the ST-11 clonal complex (cc11), a hyper-invasive meningococcal lineage (12, 13), have been linked to IMD in MSM.
Since January 2015 in the sexual health clinic of Columbus, Ohio, a significant portion (3–36%/month) of initially presumed Ng-symptomatic urethral infections, primarily among heterosexual men, have been determined to be meningococcal urethritis (now >100 cases) caused by nongroupable cc11 Nm (14). Smaller urethritis clusters, caused by Nm with the same genotype, have also been observed in Oakland County, Michigan (14), and later in multiple other US sites. In this study, we define genotypic properties of an emerging clade, termed the US Nm urethritis clade (US_NmUC), through whole-genome sequencing (WGS) analyses and biological characterization of key virulence factors that may contribute to Nm’s success in becoming a urogenital pathogen. This meningococcal cc11 clade, with novel genetic and phenotypic changes, has become competent for efficient transmission via sexual contact and can effectively colonize the urogenital tract to cause an unprecedented large US meningococcal urethritis clusters.
Results
Genotype and Genome Analyses.
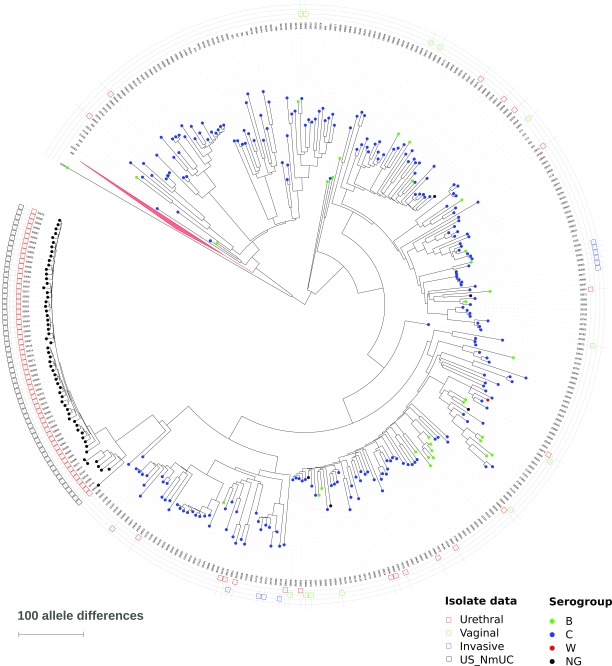
Multilocus sequence typing (MLST) extracted from WGS data confirmed that all 56 US_ NmUC isolates (52 from Columbus, 2 from Atlanta, and 2 from Indianapolis) belong to the ST-11 clonal complex (cc11). Meningococci of cc11 are further divided into two major sublineages, 11.1 and 11.2, with the ET-15 variant (15) found in lineage 11.2 (12). All 56 urethritis isolates were members of cc11 lineage 11.2/ET-15, contained IS1301, and were of fine-type PorA VR1 5–1, VR2 10–8, VR3 36–2, FetA F3-6, and PorB 2–2. Although the urethritis clusters caused by the US_NmUC isolates represent a clonal expansion event with no variation by conventional genotyping (MLST, fine-typing antigens), continued genomic alteration and evolution were evident within the genomes. For example, multiple genomic regions of the Columbus CNM26 isolate showed SNP differences in short nucleotide segments of a few hundred base pairs (mosaic nucleotide sequence variation) (16) from the corresponding regions of CNM10, which was isolated ∼1 1/2 mo earlier, indicating that homologous recombination is continuing to contribute to the evolution of this clade. Phylogenetic comparison of the US_NmUC isolates to lineage 11 isolates presented by Lucidarme et al. (12) and additional urogenital isolates identified in the PubMLST database using the Genome Comparator tool (17) clearly showed that the clade forms a distinct branch within the lineage 11.2 (Fig. S1).
Fig. S1.
Core genome similarity dendrogram of US N. meningitidis urethritis clade (US_NmUC) isolates with respect to lineage 11 (ST-11 clonal complex) isolates presented by Lucidarme et al. (12), isolates identified in Table S1 as well as more recent urethral isolates from PubMLST. The dendrogram is rooted between sublineages 11.1 and 11.2. Isolates are marked with colored circles representing serogroup as documented in the PubMLST. The outer circle contains the PubMLST identifier and additional information about the sources of the isolates. The serogroup W clade in sublineage 11.1 is represented by a red triangle and contains 470 isolates, including one urethral isolate. A total of 812 isolates were included in the analysis.
The IS1301-Mediated Modification of the Capsular Polysaccharide Locus in the US_NmUC Isolates.
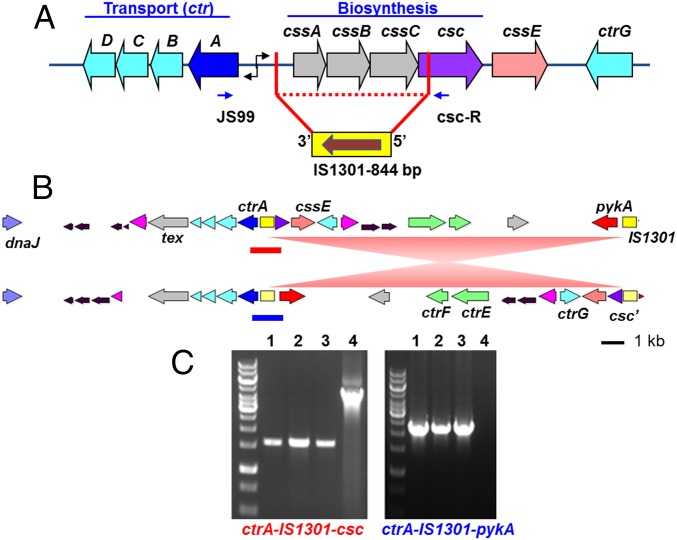
Characterization of the capsular polysaccharide (cps) locus of the US_NmUC isolates by serogrouping PCR (18) detected a product for the serogroup C csc gene. Subsequent overlapping PCR assays revealed a shorter-than-expected product size between csc and ctrA and an otherwise intact meningococcal cps locus. Sequencing of the csc to ctrA PCR product revealed a complete deletion of the sialic acid biosynthesis genes, cssA/B/C, and a deletion of 620 bp of csc with the insertion of an IS1301 element in a 3′-5′ orientation (Fig. 1A). This explained the nongroupable phenotype of US_NmUC isolates (14) and their corresponding susceptibility to killing by normal human serum in serum bactericidal assays. WGS of all 56 isolates confirmed the absence of cssA/B/C genes and the presence of IS1301, suggesting that this modification can serve as a unique genetic marker for this US Nm urethritis clade.
Fig. 1.
The cps locus of the US_NmUC isolates. (A) The IS1301 insertion and deletion of the capsule biosynthesis genes in the US Nm urethritis clade isolates. Based on overlapping PCR products and sequencing of the cps locus in clade isolates, an 844-bp insertion element (IS1301) has inserted into the IGR with a simultaneous 3,625-bp deletion of cssA/B/C and a partial csc deletion. When amplified across the deleted region using primers located in ctrA and csc (blue arrows), shorter PCR products (∼1.6 kb) were obtained from clade isolates compared with that (4.4 kb) of the serogroup C WT FAM18 strain (examples shown in C). The 1.6-kb PCR product was sequenced to determine the insertion/deletion junctions. (B) Genomic segments of ∼38 kb surrounding the cps locus of the US_NmUC isolates. A genome configuration between two IS1301 elements corresponding to the ctrA-IS1301-csc linkage in the US_NmUC isolates as determined by PCR and sequencing is shown on top, and the genome configuration of the single-contig PacBio genome of CNM10 is shown below. The ∼20-kb genome inversion between two IS1301 elements is depicted by red triangles. Genes are color-coded: ctrB/C/D and ctrG in cyan, ctrA in dark blue, ctrE/ctrG in green, truncated csc in purple, tex in gray, dnaJ in light blue, and pykA in red. The IS1301 elements are shown as yellow rectangles. (C) Gel pictures of the ctrA to csc (Left) and the ctrA to pykA (Right) PCR amplifications. The presence of both ctrA-IS1301-csc (red line in B) and ctrA-IS1301-pykA (blue line in B) orientations was determined by PCR amplification across IS1301 and sequencing of the resulting products. Lanes: 1, CNM3; 2, CNM10; 3, CNM26; 4, FAM18. Cell suspensions from single colonies were used as templates. The expected product sizes are 1,614 bp and 2,316 bp for ctrA-IS1301-csc and ctrA-IS1301-pykA, respectively.
The 3′ junction of IS1301 insertion within the intergenic region (IGR) has been previously described (but in the opposite orientation) in the hyper-encapsulated serogroup C cc11 invasive isolates from Spain that are highly resistant to conjugate vaccine-induced bactericidal antibodies and complement-mediated killing (19). The IS1301 insertion at this IGR site increases transcription of both css and ctr operons, resulting in increased capsule production and enhanced serum resistance of these hyper-encapsulated cc11 invasive isolates (19). In contrast, the IS1301 insertion in the cps locus of the US_NmUC isolates results in loss of the sialic acid capsule and intrinsic lipooligosaccharide (LOS) sialylation. The IS1301 insertion site within csc has not been reported previously, but the site does contain the IS1301 target consensus motif (5′-ATTAG-3′) (20).
Surprisingly, the single-contig CNM10 genome generated by PacBio showed that the IS1301 adjacent to ctrA was flanked by pykA, encoding a pyruvate kinase, whereas the partial csc gene was identified ∼20 kb downstream adjacent to another IS1301 element (Fig. 1B). This genome arrangement suggested an inversion of an ∼20-kb sequence between the two IS1301 elements. To determine if both the ctrA-IS1301-csc and the ctrA-IS1301-pykA configurations were present, we designed primers to amplify across IS1301 in both scenarios, using cell suspensions from a single colony as the template in colony PCR assays. Both PCR reactions were positive (Fig. 1C) and sequencing of the PCR products confirmed the expected sequences. The presence of two configurations between two IS1301 elements in a single colony population implied that a dynamic inversion process is occurring at the cps locus and that the PacBio genome assembly likely captured the dominant genome arrangement.
The Vaccine-Targeted Surface Protein Antigens.
Factor H-binding protein (FHbp), a key virulence protein, is the main target of two new multicomponent protein-based meningococcal serogroup B-directed vaccines, 4CMenB and MenB-FHbp (21, 22). The 4CMenB protein vaccine antigens include FHbp, PorA, NHBA, and NadA. The fHbp gene of all US_NmUC isolates was allele 1127 and the FHbp peptide was allele 896, placing it in the variant 1 and subfamily B in the original Novartis (now GSK) and Pfizer FHbp classifications, respectively. The FHbp peptide allele, only found in isolates of the US_NmUC, is a hybrid of the subvariants 1.1 and 1.10, but with a unique residue 247 at the C terminus of the mature protein (23). The fHbp promoter sequence of the US_NmUC isolates belonged to the high FHbp-expressing promoter clade I defined by Biagini et al. (24). Preliminary data of flow analyses indicated high FHbp protein expression in US_NmUC isolates. This is in contrast to the urethritis cc11 Nm isolates described by Taha et al., all of which encoded a frame-shift mutation in fHbp (25).
The other three 4CMenB protein vaccine antigens, if not inactivated by IS1301 (see below), were conserved among all of the US_NmUC isolates including NadA peptide 2 (variant 2/3), NHBA peptide 20, and PorA P1.5–1, 10–8. An earlier study of 70 ET-15 strains found that 48 isolates (∼70%) contain the nadA::IS1301 insertion (26). Interestingly, the nadA gene was intact in 49 of the urethritis Nm isolates from Columbus, but was inactivated by IS1301 in the CNM45, CNM49, and CNM54 isolates (August–September 2015), the Indianapolis NM1 isolate (May 2015), and the Atlanta ATL2 isolate (August 2016), implying a recent movement of IS1301. The IS1301 appears to be shaping this clade.
The US Urethritis Clade Isolates Have Acquired the Gonococcal aniA and norB Genes by Gene Conversion and Grow Well Anaerobically.
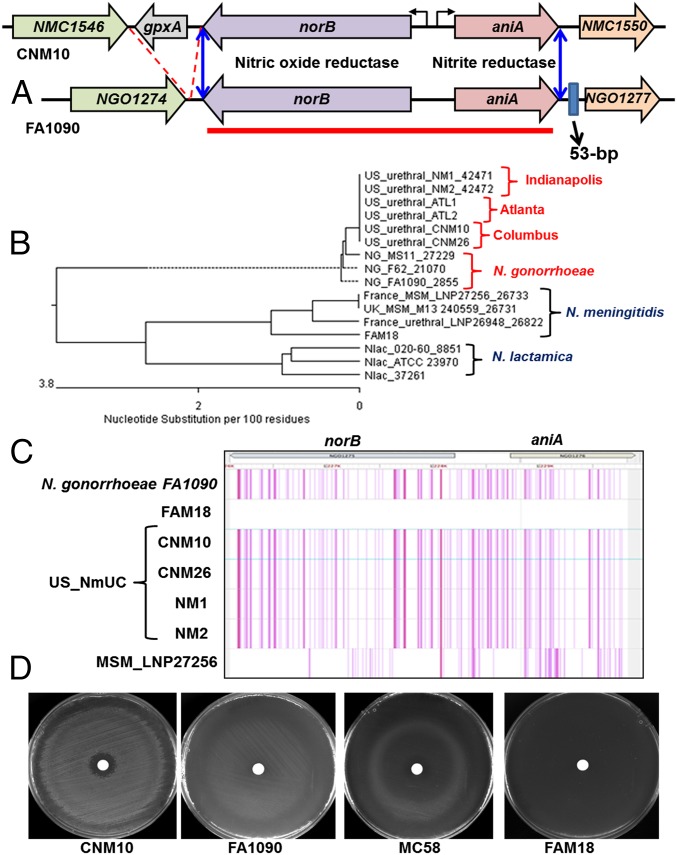
N. gonorrheae can grow anaerobically in the urogenital tract because it encodes an effective denitrification pathway composed of a nitrite reductase (AniA) and a nitric oxide reductase (NorB) (27) (Fig. 2A). In contrast, meningococci less commonly experience anaerobic conditions, and a functional denitrification pathway is not essential for their survival in the nasopharynx (28, 29). Many meningococcal isolates have frame-shift mutations in aniA (29) and cannot grow anaerobically using nitrite as the electron acceptor. Even in meningococcal isolates that retain intact aniA/norB, these alleles appear to be less efficient in supporting anaerobic growth (28).
Fig. 2.
Acquisition of the gonococcal norB-aniA gene cassette in the US Nm urethritis clade. (A) Schematics of the norB-aniA locus in the US_NmUC isolates and N. gonorrheae. Homologous genes are color-coded. A gpxA gene encoding a glutathione peroxidase adjacent to norB is present in the genomes of the US_NmUC clade isolates and other meningococci, but is absent in N. gonorrheae. On the 3′ side, a 53-bp sequence is present downstream of aniA in gonococci but absent in the US clade and other meningococci. The presence and the absence of these two genetic features (gpxA and the 53-bp element) in the US_NmUC genome indicated the recombination junctions (blue double-arrow lines). (B) The sequence between stop codons of norB and aniA (3808 bp) marked by the red line in A was used for the phylogenetic sequence analyses. Two isolates each from Columbus, Indianapolis, and Atlanta, together with three N. lactamica strains, two meningococcal urethral isolates, three meningococcal MSM isolates, and the FAM18 reference strain were included in the alignment by Clustal W. The US urethritis clade norB-aniA locus clustered with the gonococcal sequences. (C) A SNP density plot of the core genome alignment by Parsnp at the norB-aniA locus with FAM18 set as the reference genome. Each SNP that differed from FAM18 is shown as a single line, and multiple neighboring SNPs appear as thick lines. The sequence identity of four US_NmUC isolates to the gonococcal FA1090 sequence is compared with that of a meningococcal MSM isolate LNP27256 at this locus. The light-gray region indicates that sequence is absent in one or some of the aligned genomes (polymorphisms within the glycosylation motif of aniA). (D) Anaerobic growth of the US urethritis clade isolates on GC agar plates supplemented with sodium nitrite. The US clade isolate (CNM10) was compared with meningococcal strains with either an intact AniA (MC58) or a frame-shift inactivated AniA (FAM18) and the gonococcal reference strain FA1090. After incubating under anaerobic conditions for 24 h at 37 °C and another 24 h at room temperature, growth on the plates was recorded. Data shown are representative anaerobic growth experiments repeated five times. Additional CNM isolates showed anaerobic growth similar to that of CNM10.
Both norB and aniA were in-frame in all 56 US_NmUC isolates. In comparison, only 4 of 33 (12%) urogenital Nm isolates retrieved from the PubMLST database (Table S1) were predicted to encode functional AniA proteins, and the others had a frame-shift mutation. Unexpectedly, the divergently transcribed norB-aniA sequence of the US_NmUC isolates aligned almost perfectly with the homologous genes of N. gonorrheae, suggesting that the US_NmUC isolates had acquired the gonococcal version of the norB-aniA gene cassette, which was also distinct from those of Neisseria lactamica (Dataset S1). The phylogenetic tree based on the 3.8-kb norB-aniA gene sequence shows that the US_NmUC isolates clustered together with Ng (Fig. 2B), whereas they grouped with Nm at the core genome alignment (Fig. S1). The SNP density plot (Fig. 2C) demonstrated that the SNP signatures of the US_NmUC and Ng are near identical at this locus. Inspection of the norB-aniA locus in additional cc11 Nm urogenital isolates and MSM invasive isolates in the PubMLST database confirmed that all were of meningococcal sequence and that none had such a gene conversion event at this locus, whereas WGS data of all 56 US_NmUC isolates from three cities confirmed the identical gonococcal norB-aniA gene conversion event.
Table S1.
Comparison of the US_NmUC isolates (CNM, NM1, NM2, ATL1, ATL2) and previously isolated urogenital isolates and MSM-invasive isolates in PubMLST database
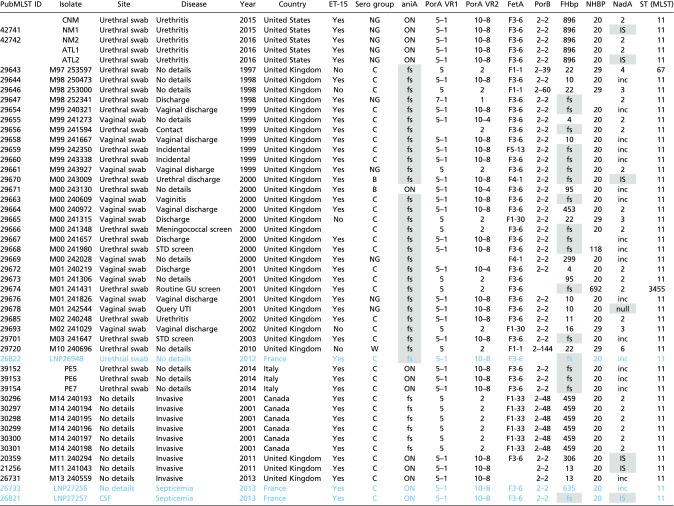 |
For the aniA column: ON, in-frame; fs, frame-shift inactivation. For the NadA column: inc, incomplete sequence; IS, IS element insertion; null, the nadA locus is missing. No data are available for the blank cells. Isolates included in the study of Taha et al. (25) are in blue, and the inactivated genes are shaded in gray.
To identify the recombination junction, the sequence alignment was extended upstream and downstream of norB-aniA (Dataset S1). In meningococci, a 534-bp ORF encoding a glutathione peroxidase (gpxA) is 178 bp upstream of the norB stop codon (Fig. 2A and Dataset S1). The gpxA gene and the intergenic sequence between gpxA and norB were absent in N. gonorrheae (a 672-bp deletion), but were present in the US_NmUC isolates. Thus, the recombination event likely occurred very close to the norB stop codon. On the other side, a 53-bp gonococcal sequence beginning 103 bp downstream of the aniA stop codon was absent in all meningococci including the US_NmUC isolates, indicating that recombination happened upstream of this 53-bp sequence (Fig. 2A and Dataset S1). Thus, the norB-aniA gene conversion appeared to be a precise transformation/recombination event.
There were 107 SNPs scattered throughout the norB coding sequence (2253 bp), and only six SNPs were nonsynonymous between Nm and Ng (and US_NmUC) (Fig. S2). The aniA coding sequences of Ng and Nm are 1,179 and 1,146 bp, respectively, and differed by 62 polymorphic sites. The AniA proteins of Ng and US_NmUC were identical. Compared with the meningococcal protein, both have a four-residue extension, one residue change in the N-terminal repeat region, two residue changes in the reductase domain, four amino acid changes, and seven extra residues within the C-terminal glycosylation region (29) (Fig. S3). Finally, norB and aniA are divergently transcribed and are separated by an IGR of 370 bp in Nm, 373 bp in Ng, and 372 bp in the US_NmUC. The IGR of the US_NmUC was identical to Ng with only one difference: a C6 vs. a C5 tract (absent in Nm) within the untranslated region of aniA in Ng and the clade, respectively (Fig. S4).
Fig. S2.
NorB protein sequence alignment by clustalW. Three N. gonorrheae (Ng) strains (FA1090, F62, and MS11), two US_NmUC isolates (CNM10 and NM1), one Nm urethral isolate, one Nm MSM isolate, and one N. lactamica (Nl) strain are included in the alignment. Residues different from FA1090 are shaded in black.
Fig. S3.
AniA protein sequence alignment. The US urethritis clade isolate CNM10 is aligned to the gonococcal strain FA1090, the N. lactamica strain 020–60, the meningococcal urethral isolate PE5, and the MSM invasive isolate LNP27256 with intact AniA. Residues different from FA1090 are highlighted in yellow. Residues important for activity are marked with an asterisk.
Fig. S4.
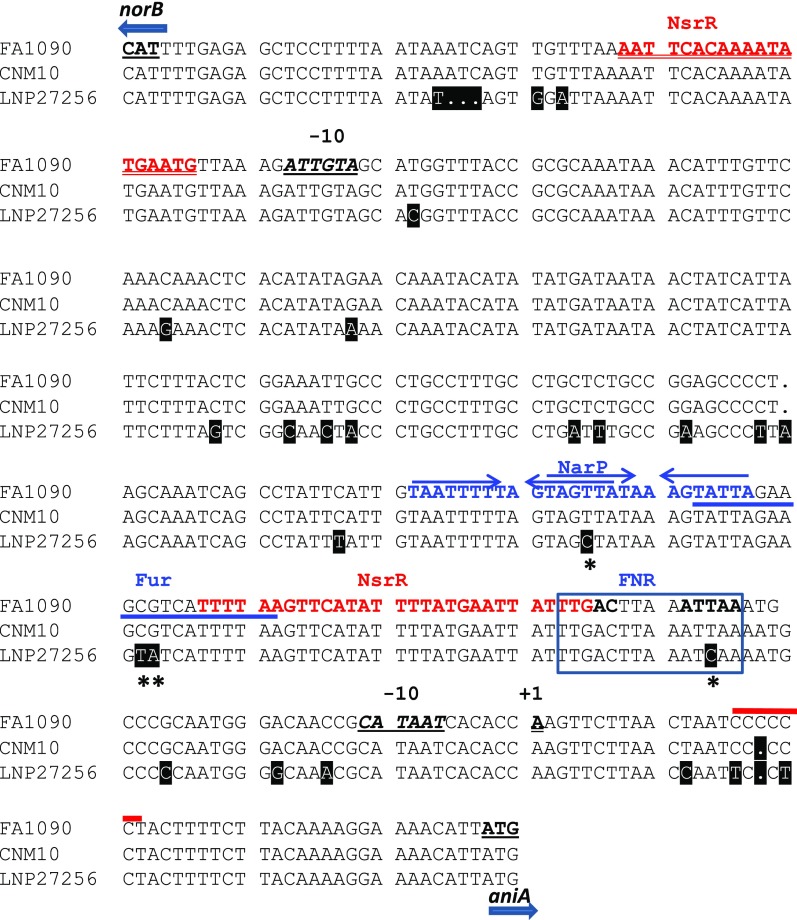
The intergenic region between start codons of norB and aniA of CNM10 was aligned to gonococcal (FA1090) and meningococcal (LNP27256) sequences. The nucleotide different from that of strain FA1090 is shaded in black. The ATG start codons, the −10 element, and the transcriptional start site (+1) are underlined. The NsrR-binding motifs of norB and aniA are in red. Two NarP-binding inverted repeat sequences are marked with two sets of arrows and are in blue. The Fur-binding site is labeled with blue underline and the FNR-binding motif is boxed. The location of the poly-C tract is shown with a thick red line above. The SNPs shown to affect regulatory activities of FNR and NarP (44) are marked with asterisks.
We examined whether the US meningococcal urethritis clade isolates with the gonococcal denitrification pathway differed in nitrite-dependent anaerobic growth compared with the reference strain MC58 with a meningococcal pathway. FAM18 was a negative control (frame-shift in aniA) whereas the gonococcal strain FA1090 served as a positive control. No growth was detected for FAM18, consistent with the presence of the frame-shift mutation (Fig. 2D). Results of anaerobic nitrite-dependent growth assays correlated with the differences between meningococci and gonococci in the efficiency of the AniA/NorB systems (28). The US_NmUC isolates yielded substantial growth around the nitrite disk similar to that observed for Ng FA1090 (a representative CNM10 shown in Fig. 2D), whereas the meningococcal MC58 strain with a less efficient anaerobic system demonstrated scanty growth (Fig. 2D).
Discussion
A limited number of virulent clonal complexes cause most of the invasive meningococcal diseases. Encapsulated cc11 strains are hyper-invasive and have caused multiple outbreaks and epidemics, such as epidemics in North America, Europe, and Australia in the 1990s/2000s, Hajj-associated global outbreaks, epidemics in the African meningitis belt, and more recently, endemic disease in South Africa, Europe, and Brazil (12). The serogroup C encapsulated cc11/ET-15 sublineage has been associated with a higher case-fatality rate and a high proportion of sequelae (30). Members of cc11 have been linked to IMD in MSM (10, 11) and now to Nm urethritis in heterosexual men. Our data indicate that a new nonencapsulated meningococcal clade of cc11 lineage 11.2/ET-15 (IS1301 containing), fine-type P1.5–1, 10–8, F3-6, 2–2, which is monophylogenic within cc11 (Fig. S1), is causing unprecedented clusters of urethritis in the United States (14). In addition to the large cluster observed in Columbus, Ohio, other sexually transmitted infection clinics in Oakland County, Michigan, Indianapolis, Atlanta, and other cities have also reported increased cases of cc11 Nm causing urethritis. Since 2015, this clade was the exclusive cause of a significant portion (∼20%) of symptomatic urethral infections (with urethral Gram-negative intracellular diplococci and growth of oxidase-positive Gram-negative diplococci) in Columbus (14). In Indianapolis, rates of discordant nongonococcal urethritis were 2.8, 1.4, and 6.9% in 2013, 2014, and 2015, respectively, indicating a significant increase in meningococcal urethritis (31). In addition to the isolates characterized in this study, other isolates from Indianapolis, Atlanta, and other cities also appear to have the same detailed molecular profile and characteristics as the US_NmUC isolates. These data represented a clonal expansion within the cc11 lineage and that the US_NmUC has acquired a capacity to better adapt to the urogenital (e.g., urethra) tract and perhaps to other mucosal environments. The specific signatures of the US_NmUC include (i) an IS1301-mediated specific deletion of the sialic acid biosynthesis genes with associated genome inversion near the cps locus, (ii) expression of a unique fHBP protein, and (iii) the acquisition of gonococcal NorB-AniA denitrification apparatus. These unique features differ from the cc11 Nm urethritis isolates described by Taha et al., which express a serogroup C capsule, encode a frame-shifted fHbp allele, and have a meningococcal denitrification pathway (25).
The US_NmUC isolates displayed active genomic inversion events mediated by IS1301 elements. Budroni et al. has previously recognized an inversion of ∼15 kb localized within the cps locus to occur within related lineages, i.e., between cc11 and cc8 and between cc41/44 and cc269, likely mediated by the duplicated regions flanking the cps locus (32). We have also detected such an inversion because both cc11 and cc8 orientations of the cps locus were identified in the cc11 US_NmUC isolates. Additionally, we discovered a unique and likely independent inversion mediated by IS1301 elements. The inversion may be drifting toward the major configuration detected by PacBio WGS that links the functional capsule transport operon to the intact pyruvate kinase pykA instead of the remnant of an inactivated biosynthesis gene. Whether this arrangement achieved any biological benefits awaits further studies. The loss of both capsule and LOS sialylation due to deletion of the sialic acid biosynthesis genes should enhance close adherence at mucosal surfaces (33).
The US_NmUC isolates contain a novel fHbp gene coding for a subvariant 1 peptide, not found in other meningococcal isolates. The ability of a strain to bind factor H (fH) is influenced by the expression level of FHbp protein and amino acid differences in the subvariant sequences (34). The fH is present at mucosal surfaces with concentrations expected to be ∼10% of serum levels (35). Hence, it is proposed that the different levels of fH binding by subvariants may be more relevant in different niches with low fH levels during colonization or disease, rather than during disease in the blood where fH is sufficiently abundant to saturate FHBP binding (34). An additional role of FHbp is to provide resistance to killing by the antimicrobial peptide LL-37 as the ΔfHbp mutants (both subvariants 1.1 and 1.10) have been shown to be more sensitive to LL37 (36). Nm and Ng have adopted different strategies to bind fH to their surface to prevent complement activation. The fHbp homolog encoded by Ng is not surface-expressed and does not bind fH; gonococci instead use abundant porin proteins to recruit fH (37). Without capsule, Ng and the nonencapsulated Nm urethritis clade isolates depend on alternative factors to confer complement resistance. The highly expressed, unique fHBP variant in the US_NmUC isolates may afford immuno-protective effects in the urogenital tract that has lower levels of fH. Compared with a related MSM invasive isolate expressing functional fHBP, the Nm urethritis isolates characterized by Taha et al., which has a frame-shifted inactivated fHbp, has reduced survival in a transgenic mouse model expressing human factor H (25). Based on the predicted protein sequence, both MenB-FHbp and 4CMenB vaccines might provide protection against this clade, but the effectiveness of MenB vaccines at a mucosal surface has not been demonstrated. The NadA adhesion, another major component of the 4CMenB vaccine that was absent in Ng (21), was inactivated by IS1301 in a few of the later isolates. Thus, IS1301 insertion and possible movement and transformation/homologous recombination continue to shape the pathogenic potential of Nm and provide genetic diversities that facilitate new niche adaptation.
Gene conversion events have been previously documented between meningococcal genomes and are more likely to happen within related lineages defined by MLST (e.g., cc11 and cc8) and to involve longer DNA transformations (3.89 kb vs. 0.68 kb) (32). These differences coincide with the distributions of restriction modification systems (32). In addition, replacement of the capsule biosynthesis genes, in particular the capsule polymerase, by gene conversion results in “capsule switching” and immune escape (38). Ecological separation of Ng and Nm within the human host is proposed as an explanation for the lower frequency of interspecies recombination noted between these species (39). However, a meningococcal vaginal isolate with a gonococcal PIB class porin gene (40) and urethral isolates carrying a 16s rRNA copy from Ng (41) have been reported. The norB-aniA exchange of the US_NmUC reported here is a large (3.8 kb), complete, and precise gene conversion event from Ng that may provide an advantageous metabolic function to survive the anaerobic urogenital environment.
AniA catalyzes the conversion of nitrite to nitric oxide (NO), which is subsequently reduced to nitrous oxide by NorB (27). In addition to supporting microaerophilic and anaerobic growth, the denitrification pathway plays a significant role in protection against NO-mediated toxicity. Gonococcal AniA is a glycosylated surface-exposed outer membrane lipoprotein (29), is the major protein induced anaerobically, and has been shown to confer enhanced serum resistance in gonococci (42). Patients with gonococcal infection produced antibodies to AniA, indicating that it is expressed during disease (43). Thus, the denitrification pathway plays a crucial role in gonococcal biology, and gonococcal isolates universally have highly functional AniA and NorB proteins. In contrast, meningococci less commonly experience an anaerobic or oxygen-limited environment at the nasopharynx, and a functional denitrification pathway is not essential for meningococcal survival (28, 29). Many meningococcal isolates have various mutations in aniA or completely lack the aniA gene (27–29), which seemingly points to a selection against expression of a functional AniA in meningococci. A recent report indicated that the invasive meningococcal MSM isolates and related urethritis isolates, but not other invasive meningococci, maintained a functional meningococcal AniA (25). This trait was proposed to reflect an adaptation and selection, which allows meningococci to colonize at this uncommon etiological site (25). However, even an intact meningococcal AniA/NorB system is less efficient than the gonococcal system. For example, the relative in vitro specific activity of meningococcal AniA is lower than that of the corresponding enzyme from Ng (28). Thus, the acquisition of a gonococcal norB-aniA cassette through homologous recombination in the US_NmUC isolates could be a major contributor to the success of this clade in adapting to the urethra, and, like gonococci, causing urethritis clusters. The clade indeed displayed a robust anaerobic growth phenotype over meningococci with an active Nm system. The finding of a gonococcal denitrification system also suggests that US_NmUC evolved in the presence of gonococcal DNA for recombination when in this new niche.
The expression of both meningococcal and gonococcal AniA proteins is under the control of a complex regulatory network involving multiple transcriptional regulators (27), but with subtle differences (44). Expression of aniA is induced by fumarate and nitrate reduction (FNR) regulator in response to oxygen limitation, induced by nitrite via the NarQ/NarP two-component system, activated by the iron uptake regulator Fur, and repressed by the NO-sensitive repressor NsrR (45). Meningococcal IGR was quite distinct with ∼30 polymorphic sites (Fig. S4) compared with that of Ng. The critical SNP differences noted in a previous promoter comparison study that distinguished aniA cis-regulation of Ng (44) from that of Nm were present in the US_NmUC. Further studies of regulatory network mediated by global regulators are needed to determine if the effects of gonococcal promoter regulation in the US_NmUC genetic background are similar to those observed in Ng (44) and to better delineate the advantage afforded by the gonococcal apparatus in meningococci.
The unprecedented large US meningococcal urethritis clusters suggest that a meningococcal cc11 lineage 11.2 clade, with novel genetic and phenotypic changes, appears to have become competent for efficient transmission via sexual contact and can effectively colonize the urogenital tract. An IS1301 insertion/deletion resulted in the loss of both encapsulation and intrinsic LOS sialylation, which is recognized to facilitate adherence at mucosal surfaces and provide a molecular fingerprint for this clade. A key finding of this study was the discovery of the gonococcal AniA/NorB denitrification system in the US meningococcal urethritis clade that enabled robust anaerobic growth. Considering the uniformity of housekeeping genes, N. gonorrheae may have evolved from a subset of meningococci that could colonize the genital tract (39). The further speciation of the gonococcus and meningococcus is suggested to be a consequence of the adaptation to different environments (46). The deletion of capsule genes, the conversion to the gonococcal denitrification pathway, and potentially other phenotypic changes suggest that the emergence of this new cc11 lineage 11.2 meningococcal clade through multiple independent evolutionary events has occurred and been selected for to better assimilate into the same niche first adopted by gonococci. In summary, a N. meningitidis cc11 clade causing the US Nm urethritis clusters appears to be newly emergent as a successful urogenital pathogen. Further epidemiologic and laboratory investigations are needed to determine the full extent of spread of this clade within the US population.
Materials and Methods
Bacterial Isolates and Growth Conditions.
The collection and demographic characterization of 52 Nm urethritis isolates from nonepidemiologically related cases between January and September 2015 at the sexual health clinic at Columbus Public Health, Columbus, Ohio, has been described earlier (14). The isolate collection was conducted as part of the Gonococcal Isolate Surveillance Project (GISP) that has been determined by the CDC as nonhuman subject research and exempt from Institutional Review Board (IRB) approval. The outbreak investigation, including molecular characterization of the isolates as described in this report, was approved by the Ohio State University IRB (protocol 2015H0388). The isolates were named CNM for Columbus N. meningitidis. In addition, two US_NmUC isolates each from Indianapolis (NM1 and NM2) and from Atlanta (ATL1 and ATL2) were included in this study. Gonococcal reference strain FA1090 was kindly provided by W. Shafer, Emory University, Atlanta. The strain M7 is a nonencapsulated meningococcal control derived from the NMB strain (B:2B:P1.2,5:L2, CDC8201085) (47). Meningococcal and gonococcal strains were grown with 5% CO2 at 37 °C on gonococcal base agar (GC, BD) plates or GC broth as previously described (48).
WGS.
WGS of all 52 CNM isolates was performed by Illumina at the Centers for Disease Control and Prevention (CDC), and relevant genotypes were extracted. Two Columbus isolates, CNM10 (April 2015) and CNM26 (May 2015), were sequenced by Pacific Biosciences (PacBio) technology, yielding ∼200× coverage, and assembled with Hierarchical Genome Assembly Process v3 (49), which resulted in a single contig and two contigs, respectively. Nm urethritis isolates from Indianapolis, NM1 (May 2015) and NM2 (February 2016), and isolates from Atlanta, ATL1 and ATL2 (August 2016), were sequenced by MiSeq, yielding ∼600× coverage of paired-end 250-bp reads, assembled using SPAdes (50) and annotated by RAST (51). Draft genome assemblies of urogenital sourced Nm and closed reference genomes of N. meningitidis, N. gonorrheae, and N. lactamica were downloaded from PubMLST (https://pubmlst.org/neisseria/) (52). Genome assemblies were compared using Harvest (v1.1.2); a reference-independent core genome alignment and maximum-likelihood phylogeny were constructed using the Parsnp aligner and then visualized using Gingr (53). The MegAlign program in the DNASTAR Lasergene 13 suite was used for multialignments of short DNA and protein sequences by Clustal W. Allele assignments of MLST, fine-typing antigens, and Bexsero Antigen Sequence Typing (54) were retrieved from the PubMLST database.
Genome assemblies were uploaded to PubMLST, and the Genome Comparator tool was used to calculate allele distances according to the cgMLST scheme (v1.0) (17). A neighbor-joining dendrogram was constructed using SplitsTree v4 (55) and displayed on the interactive Tree of Life website (56). The genomes of the US_NmUC isolates were also compared with ST-11 urethritis and invasive isolates in the PubMLST database (Table S1).
Nitrite-Dependent Anaerobic Growth and Serum Bactericidal Assays.
The nitrite-dependent growth assays were performed as previously described with minor modifications (57). GC broth cultures at midlog phase were adjusted to a density of 0.3 at OD550, and 100-µL aliquots of suspension were then spread on GC agar plates. A 6-mm AA disk was placed on the agar, and 10 µL of 20% NaNO2 was spotted onto the disk to provide nitrite. The plates were incubated at 37 °C for 24 h in an anaerobic jar (Oxoid) with the GasPak EZ anaerobe container system with anaerobic indicator (BD) and allowed to grow at room temperature for an additional 24 h before imaging their growth. Serum bactericidal assays were conducted in 96-well microtiter plates with 5% commercial pooled normal human serum (Fisher) as previously described (58) except that the RPMI medium was used for the assays.
Supplementary Material
Acknowledgments
We thank the Epidemiology Team of the Meningitis and Vaccine Preventable Disease Branch, Division of Bacterial Diseases, National Center for Immunizations and Respiratory Diseases, CDC, in particular Elizabeth Briere, MD, and Jessica MacNeil, MPH; the CDC Gonococcal Isolate Surveillance Program and Division of STD Prevention of the National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention, CDC, in particular Dr. Robert Kirkcaldy; Dr. Jay Lucidarme and Dr. Ray Borrow at the Meningococcal Reference Unit, Public Health England, for providing urogenital-sourced meningococcal isolates and associated epidemiological information; and Baderinwa Offutt for technical assistance. This work was supported in part by NIH Grants R01AI107116 and R21AI128313 (to Y-L.T.), Grant R21AI121860 (to T.D.R.), Grant R01AI116706 (to D.E.N.), and CDC Grant 5 NH25PS004311 (to C.D.R.). This publication made use of the Neisseria Multi Locus Sequence Typing website (https://pubmlst.org/neisseria/) developed by Keith Jolley at the University of Oxford. The development of this site has been funded by the Wellcome Trust and European Union. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the BioProject database (accession nos. PRJNA324131 and PRJNA319252).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620971114/-/DCSupplemental.
References
- 1.Rouphael NG, Stephens DS. Neisseria meningitidis: Biology, microbiology, and epidemiology. Methods Mol Biol. 2012;799:1–20. doi: 10.1007/978-1-61779-346-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilder-Smith A. Meningococcal vaccine in travelers. Curr Opin Infect Dis. 2007;20:454–460. doi: 10.1097/QCO.0b013e3282a64700. [DOI] [PubMed] [Google Scholar]
- 3.William DC, Schapiro CM, Felman YM. Pharyngeal carriage of Neisseria meningitidis and anogenital gonorrhea: Evidence for their relationship. Sex Transm Dis. 1980;7:175–177. doi: 10.1097/00007435-198010000-00006. [DOI] [PubMed] [Google Scholar]
- 4.Janda WM, Bohnoff M, Morello JA, Lerner SA. Prevalence and site-pathogen studies of Neisseria meningitidis and N gonorrhoeae in homosexual men. JAMA. 1980;244:2060–2064. [PubMed] [Google Scholar]
- 5.Salit IE, Frasch CE. Seroepidemiologic aspects of Neisseria meningitidis in homosexual men. Can Med Assoc J. 1982;126:38–41. [PMC free article] [PubMed] [Google Scholar]
- 6.Janda WM, Morello JA, Lerner SA, Bohnhoff M. Characteristics of pathogenic Neisseria spp. isolated from homosexual men. J Clin Microbiol. 1983;17:85–91. doi: 10.1128/jcm.17.1.85-91.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Givan KF, Thomas BW, Johnston AG. Isolation of Neisseria meningitidis from the urethra, cervix, and anal canal: Further observations. Br J Vener Dis. 1977;53:109–112. doi: 10.1136/sti.53.2.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conde-Glez CJ, Calderón E. Urogenital infection due to meningococcus in men and women. Sex Transm Dis. 1991;18:72–75. doi: 10.1097/00007435-199118020-00003. [DOI] [PubMed] [Google Scholar]
- 9.Maini M, French P, Prince M, Bingham JS. Urethritis due to Neisseria meningitidis in a London genitourinary medicine clinic population. Int J STD AIDS. 1992;3:423–425. doi: 10.1177/095646249200300604. [DOI] [PubMed] [Google Scholar]
- 10.Kratz MM, et al. Community-based outbreak of Neisseria meningitidis serogroup C infection in men who have sex with men, New York City, New York, USA, 2010-2013. Emerg Infect Dis. 2015;21:1379–1386. doi: 10.3201/eid2108.141837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nanduri S, et al. Outbreak of Serogroup C meningococcal disease primarily affecting men who have sex with men: Southern California, 2016. MMWR Morb Mortal Wkly Rep. 2016;65:939–940. doi: 10.15585/mmwr.mm6535e1. [DOI] [PubMed] [Google Scholar]
- 12.Lucidarme J, et al. Genomic resolution of an aggressive, widespread, diverse and expanding meningococcal serogroup B, C and W lineage. J Infect. 2015;71:544–552. doi: 10.1016/j.jinf.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caugant DA, Maiden MC. Meningococcal carriage and disease: Population biology and evolution. Vaccine. 2009;27:B64–B70. doi: 10.1016/j.vaccine.2009.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bazan JA, et al. Notes from the field: Increase in Neisseria meningitidis-associated urethritis among men at two sentinel clinics: Columbus, Ohio, and Oakland County, Michigan, 2015. MMWR Morb Mortal Wkly Rep. 2016;65:550–552. doi: 10.15585/mmwr.mm6521a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vogel U, Claus H, Frosch M, Caugant DA. Molecular basis for distinction of the ET-15 clone within the ET-37 complex of Neisseria meningitidis. J Clin Microbiol. 2000;38:941–942. doi: 10.1128/jcm.38.2.941-942.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong Y, et al. Homologous recombination drives both sequence diversity and gene content variation in Neisseria meningitidis. Genome Biol Evol. 2013;5:1611–1627. doi: 10.1093/gbe/evt116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bratcher HB, Corton C, Jolley KA, Parkhill J, Maiden MC. A gene-by-gene population genomics platform: De novo assembly, annotation and genealogical analysis of 108 representative Neisseria meningitidis genomes. BMC Genomics. 2014;15:1138. doi: 10.1186/1471-2164-15-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennett DE, Cafferkey MT. Consecutive use of two multiplex PCR-based assays for simultaneous identification and determination of capsular status of nine common Neisseria meningitidis serogroups associated with invasive disease. J Clin Microbiol. 2006;44:1127–1131. doi: 10.1128/JCM.44.3.1127-1131.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uria MJ, et al. A generic mechanism in Neisseria meningitidis for enhanced resistance against bactericidal antibodies. J Exp Med. 2008;205:1423–1434. doi: 10.1084/jem.20072577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hilse R, Hammerschmidt S, Bautsch W, Frosch M. Site-specific insertion of IS1301 and distribution in Neisseria meningitidis strains. J Bacteriol. 1996;178:2527–2532. doi: 10.1128/jb.178.9.2527-2532.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Serruto D, Bottomley MJ, Ram S, Giuliani MM, Rappuoli R. The new multicomponent vaccine against meningococcal serogroup B, 4CMenB: Immunological, functional and structural characterization of the antigens. Vaccine. 2012;30:B87–B97. doi: 10.1016/j.vaccine.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seib KL, Scarselli M, Comanducci M, Toneatto D, Masignani V. Neisseria meningitidis factor H-binding protein fHbp: A key virulence factor and vaccine antigen. Expert Rev Vaccines. 2015;14:841–859. doi: 10.1586/14760584.2015.1016915. [DOI] [PubMed] [Google Scholar]
- 23.Brunelli B, et al. Influence of sequence variability on bactericidal activity sera induced by Factor H binding protein variant 1.1. Vaccine. 2011;29:1072–1081. doi: 10.1016/j.vaccine.2010.11.064. [DOI] [PubMed] [Google Scholar]
- 24.Biagini M, et al. Expression of factor H binding protein in meningococcal strains can vary at least 15-fold and is genetically determined. Proc Natl Acad Sci USA. 2016;113:2714–2719. doi: 10.1073/pnas.1521142113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taha MK, et al. Evolutionary events associated with an outbreak of meningococcal disease in men who have sex with men. PLoS One. 2016;11:e0154047. doi: 10.1371/journal.pone.0154047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elias J, Vogel U. IS1301 fingerprint analysis of Neisseria meningitidis strains belonging to the ET-15 clone. J Clin Microbiol. 2007;45:159–167. doi: 10.1128/JCM.01322-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barth KR, Isabella VM, Clark VL. Biochemical and genomic analysis of the denitrification pathway within the genus Neisseria. Microbiology. 2009;155:4093–4103. doi: 10.1099/mic.0.032961-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stefanelli P, et al. Molecular characterization of nitrite reductase gene (aniA) and gene product in Neisseria meningitidis isolates: Is aniA essential for meningococcal survival? IUBMB Life. 2008;60:629–636. doi: 10.1002/iub.95. [DOI] [PubMed] [Google Scholar]
- 29.Ku SC, Schulz BL, Power PM, Jennings MP. The pilin O-glycosylation pathway of pathogenic Neisseria is a general system that glycosylates AniA, an outer membrane nitrite reductase. Biochem Biophys Res Commun. 2009;378:84–89. doi: 10.1016/j.bbrc.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 30.Yazdankhah SP, et al. Distribution of serogroups and genotypes among disease-associated and carried isolates of Neisseria meningitidis from the Czech Republic, Greece, and Norway. J Clin Microbiol. 2004;42:5146–5153. doi: 10.1128/JCM.42.11.5146-5153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toh E, et al. Neisseria meningitidis ST11 complex isolates associated with nongonococcal urethritis, Indiana, USA, 2015-2016. Emerg Infect Dis. 2017;23:336–339. doi: 10.3201/eid2302.161434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Budroni S, et al. Neisseria meningitidis is structured in clades associated with restriction modification systems that modulate homologous recombination. Proc Natl Acad Sci USA. 2011;108:4494–4499. doi: 10.1073/pnas.1019751108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bartley SN, et al. Attachment and invasion of Neisseria meningitidis to host cells is related to surface hydrophobicity, bacterial cell size and capsule. PLoS One. 2013;8:e55798. doi: 10.1371/journal.pone.0055798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seib KL, et al. Characterization of diverse subvariants of the meningococcal factor H (fH) binding protein for their ability to bind fH, to mediate serum resistance, and to induce bactericidal antibodies. Infect Immun. 2011;79:970–981. doi: 10.1128/IAI.00891-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Persson CG, et al. Plasma-derived proteins in airway defence, disease and repair of epithelial injury. Eur Respir J. 1998;11:958–970. doi: 10.1183/09031936.98.11040958. [DOI] [PubMed] [Google Scholar]
- 36.Seib KL, et al. Factor H-binding protein is important for meningococcal survival in human whole blood and serum and in the presence of the antimicrobial peptide LL-37. Infect Immun. 2009;77:292–299. doi: 10.1128/IAI.01071-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jongerius I, et al. Distinct binding and immunogenic properties of the gonococcal homologue of meningococcal factor h binding protein. PLoS Pathog. 2013;9:e1003528. doi: 10.1371/journal.ppat.1003528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swartley JS, et al. Capsule switching of Neisseria meningitidis. Proc Natl Acad Sci USA. 1997;94:271–276. doi: 10.1073/pnas.94.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vázquez JA, et al. Ecological separation and genetic isolation of Neisseria gonorrhoeae and Neisseria meningitidis. Curr Biol. 1993;3:567–572. doi: 10.1016/0960-9822(93)90001-5. [DOI] [PubMed] [Google Scholar]
- 40.Vázquez JA, et al. Interspecies recombination in nature: A meningococcus that has acquired a gonococcal PIB porin. Mol Microbiol. 1995;15:1001–1007. doi: 10.1111/j.1365-2958.1995.tb02275.x. [DOI] [PubMed] [Google Scholar]
- 41.Walcher M, Skvoretz R, Montgomery-Fullerton M, Jonas V, Brentano S. Description of an unusual Neisseria meningitidis isolate containing and expressing Neisseria gonorrhoeae-specific 16S rRNA gene sequences. J Clin Microbiol. 2013;51:3199–3206. doi: 10.1128/JCM.00309-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cardinale JA, Clark VL. Expression of AniA, the major anaerobically induced outer membrane protein of Neisseria gonorrhoeae, provides protection against killing by normal human sera. Infect Immun. 2000;68:4368–4369. doi: 10.1128/iai.68.7.4368-4369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clark VL, Knapp JS, Thompson S, Klimpel KW. Presence of antibodies to the major anaerobically induced gonococcal outer membrane protein in sera from patients with gonococcal infections. Microb Pathog. 1988;5:381–390. doi: 10.1016/0882-4010(88)90038-1. [DOI] [PubMed] [Google Scholar]
- 44.Edwards J, et al. Neisseria meningitidis and Neisseria gonorrhoeae are differently adapted in the regulation of denitrification: Single nucleotide polymorphisms that enable species-specific tuning of the aerobic-anaerobic switch. Biochem J. 2012;445:69–79. doi: 10.1042/BJ20111984. [DOI] [PubMed] [Google Scholar]
- 45.Rock JD, Thomson MJ, Read RC, Moir JW. Regulation of denitrification genes in Neisseria meningitidis by nitric oxide and the repressor NsrR. J Bacteriol. 2007;189:1138–1144. doi: 10.1128/JB.01368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feavers IM, Maiden MC. A gonococcal porA pseudogene: Implications for understanding the evolution and pathogenicity of Neisseria gonorrhoeae. Mol Microbiol. 1998;30:647–656. doi: 10.1046/j.1365-2958.1998.01101.x. [DOI] [PubMed] [Google Scholar]
- 47.Stephens DS, Swartley JS, Kathariou S, Morse SA. Insertion of Tn916 in Neisseria meningitidis resulting in loss of group B capsular polysaccharide. Infect Immun. 1991;59:4097–4102. doi: 10.1128/iai.59.11.4097-4102.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Piek S, et al. The role of oxidoreductases in determining the function of the neisserial lipid A phosphoethanolamine transferase required for resistance to polymyxin. PLoS One. 2014;9:e106513. doi: 10.1371/journal.pone.0106513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chin CS, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 50.Bankevich A, et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aziz RK, et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jolley KA, Maiden MC. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brehony C, et al. Distribution of Bexsero® antigen sequence types (BASTs) in invasive meningococcal disease isolates: Implications for immunisation. Vaccine. 2016;34:4690–4697. doi: 10.1016/j.vaccine.2016.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 56.Letunic I, Bork P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Householder TC, Fozo EM, Cardinale JA, Clark VL. Gonococcal nitric oxide reductase is encoded by a single gene, norB, which is required for anaerobic growth and is induced by nitric oxide. Infect Immun. 2000;68:5241–5246. doi: 10.1128/iai.68.9.5241-5246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kahler CM, et al. The (α2→8)-linked polysialic acid capsule and lipooligosaccharide structure both contribute to the ability of serogroup B Neisseria meningitidis to resist the bactericidal activity of normal human serum. Infect Immun. 1998;66:5939–5947. doi: 10.1128/iai.66.12.5939-5947.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.