Significance
The high-resolution structures presented herein explain how ERp44, a multifunctional chaperone cycling in the early secretory pathway, exploits the endoplasmic reticulum (ER)–Golgi pH gradient to bind clients in the acidic Golgi and release them into the neutral ER environment. Protonation of essential cysteine and histidine residues induces conformational changes that simultaneously expose Cys29 in the positively charged client-binding site and the C-terminal KDEL receptor-binding motif, making ERp44 a pH-sensitive molecular machine that controls fidelity of protein secretion.
Keywords: ERp44, protein quality control, retrograde transport, crystal structure, histidine protonation
Abstract
ERp44 retrieves some endoplasmic reticulum (ER)-resident enzymes and immature oligomers of secretory proteins from the Golgi. Association of ERp44 with its clients is regulated by pH-dependent mechanisms, but the molecular details are not fully understood. Here we report high-resolution crystal structures of human ERp44 at neutral and weakly acidic pH. These structures reveal key regions in the C-terminal tail (C tail) missing in the original crystal structure, including a regulatory histidine-rich region and a subsequent extended loop. The former region forms a short α-helix (α16), generating a histidine-clustered site (His cluster). At low pH, the three Trx-like domains of ERp44 (“a,” “b,” and “b′”) undergo significant rearrangements, likely induced by protonation of His157 located at the interface between the a and b domains. The α16-helix is partially unwound and the extended loop is disordered in weakly acidic conditions, probably due to electrostatic repulsion between the protonated histidines in the His cluster. Molecular dynamics simulations indicated that helix unwinding enhances the flexibility of the C tail, disrupting its normal hydrogen-bonding pattern. The observed pH-dependent conformational changes significantly enlarge the positively charged regions around the client-binding site of ERp44 at low pH. Mutational analyses showed that ERp44 forms mixed disulfides with specific cysteines residing on negatively charged loop regions of Ero1α. We propose that the protonation states of the essential histidines regulate the ERp44–client interaction by altering the C-tail dynamics and surface electrostatic potential of ERp44.
The early secretory pathway (ESP), which consists of the endoplasmic reticulum (ER), ER–Golgi intermediate compartment (ERGIC), and Golgi apparatus, has evolved elaborate and coordinated systems that facilitate the folding and maturation of secretory and membrane proteins (1). Nascent secretory proteins enter the ER and undergo folding, N-glycosylation, and disulfide-bond formation, in which a large number of chaperones, glycosylation enzymes, and oxidoreductases are involved (2, 3). For instance, BiP and calnexin/calreticulin function as stringent checkpoints that only allow natively folded proteins to be transported to the Golgi (4). In compartments downstream of the ER, ERp44, a member of the protein disulfide isomerase (PDI) family, serves as a second checkpoint that surveys the proper assembly of some secretory proteins (5, 6). ERp44 is mostly localized in the ERGIC and cis-Golgi. In concert with KDEL receptors, it mediates the transport of immature oligomeric proteins such as IgM (7, 8), adiponectin (9–11), and serotonin transporter (SERT) (12) from the Golgi to the ER. ERp44 also retains some ER-resident enzymes that lack KDEL-like motifs and are hence unable to return to the ER without assistance, including ER oxidoreductin 1 (Ero1) (7, 13, 14), peroxiredoxin 4 (Prx4) (15), sulfatase modifying factor 1 (Sumf1) (16), and ER aminopeptidase 1 (ERAP1) (17). ERp44 is composed of three thioredoxin (Trx)-like domains (“a,” “b,” and “b′”), followed by a C-terminal extension (C tail) composed of 52 residues (18). The a domain contains a rather unique CRFS motif (13). The cysteine in this motif (Cys29) is known to form mixed disulfide bonds with client proteins to promote their thiol-dependent retention (7).
We recently showed that interactions between ERp44 and client proteins are pH-dependent (19). At neutral pH (∼7.2) in the ER (20, 21), the C tail of ERp44 interacts with the a domain through a number of hydrogen bonds, including one between Cys29 and Thr369. These interactions shield the client-binding site around Cys29 (18). Once transported into the weakly acidic Golgi (pH 6.0 to 6.7), ERp44 undergoes conformational changes that open the C tail and facilitate association with its clients. Furthermore, conserved histidine residues located around the base of the C tail are important for the pH-dependent regulation of ERp44 (22). However, the original crystal structure was determined only at pH 7.5, and several important regions, including the histidine-rich region, were not defined in the electron density map. Thus, molecular details of the pH-dependent regulation of ERp44 were still unclear.
To elucidate the mechanisms underlying the pH-dependent conformational changes of ERp44, we determined the crystal structures of ERp44 in neutral and weakly acidic conditions at higher resolutions (2.00- and 2.05-Å resolution, respectively) than that of the original crystal structure (2.6-Å resolution). The updated crystal structures reveal that protonation of several conserved histidines (His157, His299, His328, His332, and His333) induces pH-dependent movements of the three Trx-like domains and triggers local helix unwinding, leading to enhanced dynamics of the C tail. These conformational changes expose a wider positively charged molecular surface, promoting the electrostatic interactions with client proteins. These findings provide a basic framework for understanding the ERp44-mediated protein quality control cycle at the ER–Golgi interface.
Results
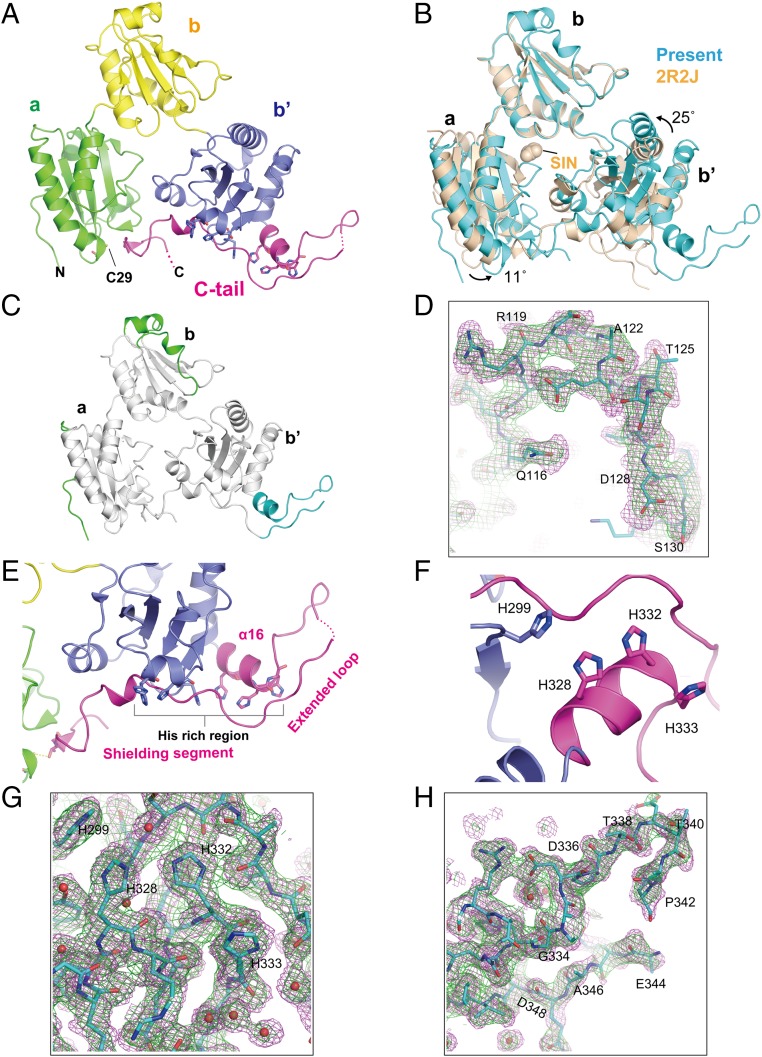
Higher-Resolution Crystal Structures of ERp44.
Previous studies indicated that the pH gradient between the ER and the Golgi regulates the interaction of ERp44 with its clients (19). To reveal the structural and mechanistic basis of the pH-dependent regulation of ERp44, we attempted to solve high-resolution crystal structures of ERp44 in the range from pH 6.5 (Golgi) to pH 7.2 (ER). By optimizing precipitant and cryoprotectant conditions as well as deleting the C-terminal RDEL motif, we successfully determined the crystal structure of ERp44 at neutral pH (pH 7.2) at 2.00-Å resolution (E form; Table S1). As previously shown (18), the overall structure of ERp44 consists of three Trx-like domains (a, b, and b′ domains) and a closed C tail, adopting a clover-shaped structure (Fig. 1A). However, the domain arrangement in the present structure differs, the a and b′ domains rotating by ∼11° and 25°, respectively, with respect to their positions in the original structure (Fig. 1B). This difference is presumably due to the small molecule (succinic acid) bound to ERp44 in the original structure (Fig. 1B). In the updated structure, almost all of the residues, including the C tail and other regions missing in the original structure, are clearly resolved in the electron density map (Fig. 1 C, D, G, and H). Residues Arg119 to Ser130 and Ser170 to Gly177 in the flexible loops of the b domain are found to assume 310- and α-helices, respectively (shown in green, Fig. 1C). The crystal structure further reveals that the C tail of ERp44 consists of three regions: an α-helix (α16), extended loop, and shielding segment (Fig. 1E). The histidine-rich region (residues 326 to 333) containing three conserved histidine residues (His328, His332, and His333) forms the α16-helix. Notably, His328 and His332 are in close proximity to His299, forming a histidine-clustered site (His cluster) (Fig. 1F). This site may facilitate metal ion binding to ERp44. Residues Gly334 to Ala350 form the long extended loop, connecting the α16-helix to the shielding segment (Fig. 1 E and G). The shielding segment (residues 351 to 378) covers a hydrophobic patch in the b′ domain and interacts with the a domain through several hydrogen bonds, as observed in the original structure (see also the next section).
Table S1.
Data collection and refinement statistics
| E form (pH 7.2) | G form (pH 6.5) | |
| Data collection | ||
| Space group | P3121 | P3121 |
| Cell dimensions | ||
| a = b, Å | 81.63 | 84.15 |
| c, Å | 139.91 | 133.18 |
| Resolution,* Å | 19.61 to 2.00 (2.07 to 2.00) | 37.9 to 2.05 (2.10 to 2.05) |
| Reflections, total/unique | 672,732/36,963 | 595,593/34,847 |
| Completeness,* % | 99.7 (99.1) | 99.9 (100) |
| Redundancy | 18.2 | 17.1 |
| I/σ(I)* | 22.5 (2.2) | 26.0 (2.3) |
| Rmeas,*,† % | 7.7 (120) | 6.5 (123) |
| CC1/2* | 0.998 (0.759) | 0.999 (0.764) |
| Refinement | ||
| Rwork/Rfree, % | 20.40/24.05 | 20.0/23.5 |
| No. of atoms | ||
| Protein | 3,087 | 3,005 |
| Ion | 1 | |
| Water | 111 | 146 |
| Average B factor | ||
| Protein | 69.83 | 62.23 |
| Water | 60.78 | 56.99 |
| Rms deviations | ||
| Bond length, Å | 0.004 | 0.007 |
| Bond angles, ° | 0.67 | 0.85 |
| Ramachandran plot, % | ||
| Favored | 98.2 | 99.5 |
| Allowed | 1.8 | 0.5 |
| Outliers | 0.0 | 0.0 |
Values in parentheses are for the highest-resolution shell.
Rmeas = Σhkl{n/[n − 1]}1/2Σi|Ii(hkl) − <I(hkl)>|/ΣhklΣIi(hkl), where Ii(hkl) is the ith observed intensity and <I(hkl)> is the average intensity from multiple observations.
Fig. 1.
Higher-resolution crystal structure of ERp44 at neutral pH. (A) Overall structure of ERp44 at 2.0-Å resolution. The a, b, and b′ domains and the C tail are shown in green, yellow, blue, and magenta, respectively. (B) Superposition of the present (cyan) and original (wheat) structures of ERp44. A succinic acid (SIN) molecule bound to the original structure is shown in sphere representation. (C) Regions that are built into the present structure but absent in the original structure (2.6-Å resolution) are highlighted in green (residues Arg119 to Ser130 and Ser170 to Gly177) and cyan (residues Lys326 to Ala350). (D) 2mFo − DFc sigma-weighted map (green; 1σ) and feature-enhanced map (magenta; 1σ) (51) of the region around residues 119 to 130 (cyan). (E) Close-up view of the C tail of ERp44. Conserved histidine residues are shown as sticks. A dotted line represents a missing region in the electron density map. (F) Close-up view of the histidine-clustered site in the C tail. (G and H) 2mFo − DFc sigma-weighted map (green; 1σ) and feature-enhanced map (magenta; 1σ) of the region around the His cluster (G) or the extended loop (H).
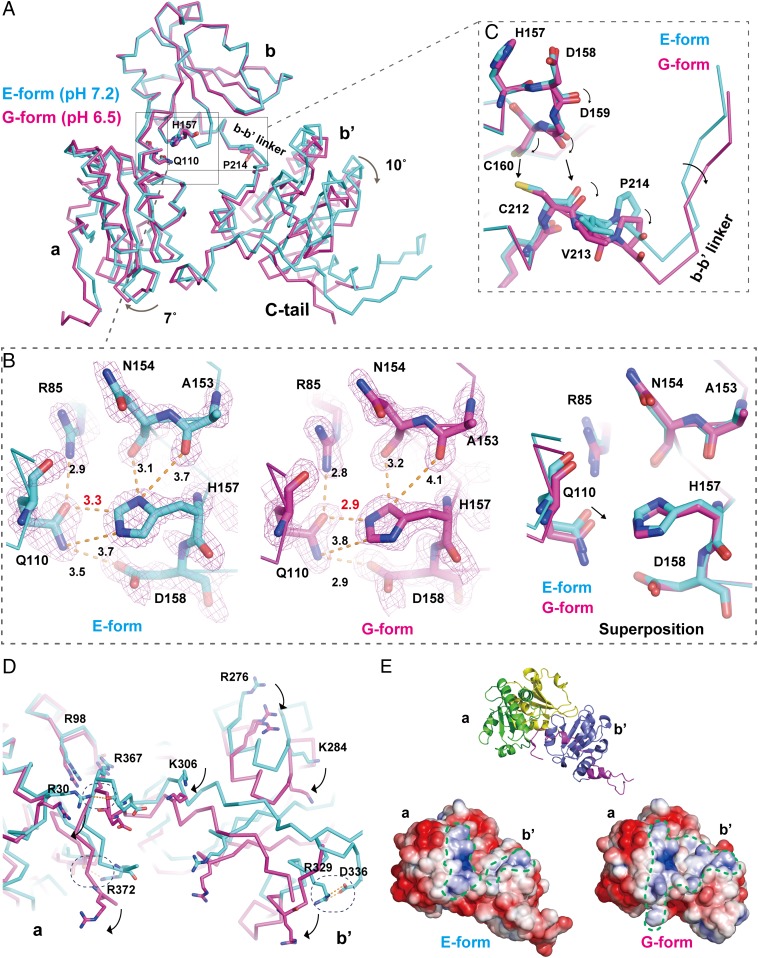
Histidine Protonation Induces the pH-Dependent Domain Rearrangement and Changes in Surface Potential.
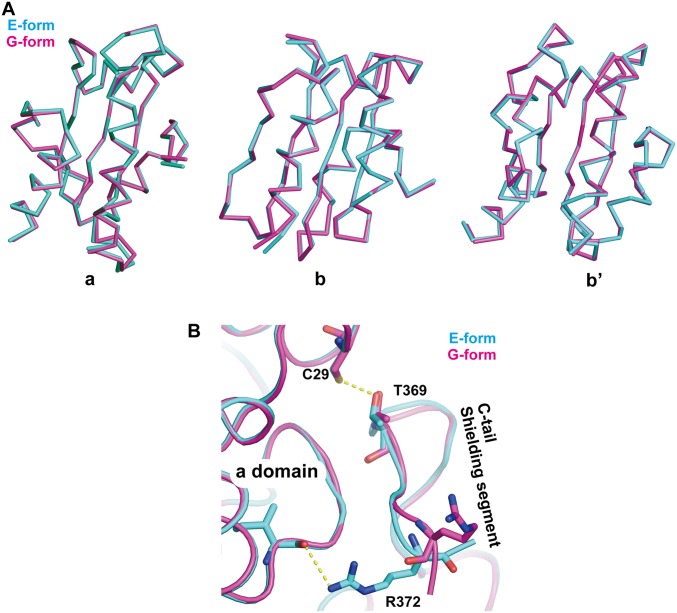
To elucidate the pH-dependent conformational changes of ERp44, we also determined the crystal structure at weakly acidic pH (pH 6.5) at 2.05-Å resolution (G form; Table S1). Both E and G forms belong to the space group P3121, with similar unit-cell dimensions and crystal packing. Therefore, the observed structural differences between the two forms should not be due to the difference in crystal packing but most likely to pH-dependent conformational changes. Each Trx-like domain in the G form is nearly identical to its corresponding domain in the E form (Fig. S1A). However, superposition of the b domain of the two forms shows that the a and b′ domains in the G form are rotated by ∼7° and ∼10°, respectively, with respect to their counterparts in the E form, leading to a significant separation between the b′ and b domains (Fig. 2A). These movements appear to be induced in part by the different modes of interaction between His157 (b domain) and Gln110 (a domain) (Fig. 2B). In the E form, a configuration of His157 protonated at Nδ1 produced the best fit in the electron density map. The Nδ1 of His157 is within hydrogen-bonding distance (3.1 Å) of the carbonyl oxygen of Asn154 and also forms a weak hydrogen bond with the main-chain carbonyl oxygen of Asn153. The Oε of Gln110 makes van der Waals contact with the Cε of His157 at a distance of 3.3 Å. In the G form, the flipped imidazole ring of His157 gives a better fit, suggesting that the Nε1 of His157 is protonated and forms a hydrogen bond with the Oε of Gln110. As a result, Gln110 gets closer to His157, leading to the ∼7° rotation of the a domain. These local conformational changes surrounding His157 are also transmitted to the linker region between the b and b′ domains through van der Waals contacts between Cys160 and Cys212, resulting in the ∼10° rotation of the b′ domain (Fig. 2C). These findings indicate that His157 protonation significantly influences the overall conformation of ERp44.
Fig. S1.
Comparison of the crystal structures of ERp44 at neutral and weakly acidic pHs. (A) Superposition of the a (Left), b (Center), and b′ domains (Right) of the E form (pH 7.2) in cyan and G form (pH 6.5) in magenta. (B) Close-up view of the superposition of the interfaces between the a domain and the shielding segment.
Fig. 2.
pH-dependent domain movements in ERp44. (A) Comparison of the overall structure of the E form (pH 7.2; cyan) and G form (pH 6.5; magenta). The b domains of the two structures are superposed onto each other. (B) Close-up view of the interaction between His157 and Gln110 (Left, E form; Center, G form; Right, superposition). Hydrogen bonds and van der Waals contacts are represented by yellow broken lines, with distances in angstroms. Feature-enhanced maps at 2.5σ are shown in pink. (C) Close-up view of a communication between His157 and Pro213 that leads to the rotation in the b′ domain. (D) Comparison of the positions of arginine and lysine residues of the two structures. Arginine and lysine residues are represented by sticks. Hydrogen bonds and a salt bridge are indicated with dashed circles. (E) Electrostatic potential of ERp44 in the E form at pH 7.2 (Lower Left) and G form at pH 6.5 (Lower Right), viewed from the same direction as the ribbon diagram (Upper). Positively and negatively charged regions are denoted in blue [+3 Boltzmann constant (kB)T/e] and red (−3 kBT/e), respectively. The positively charged regions of the a and b′ domains encircled by green dashed lines bury ∼1,800 and ∼1,000 Å2 of accessible surface in the G and E forms, respectively.
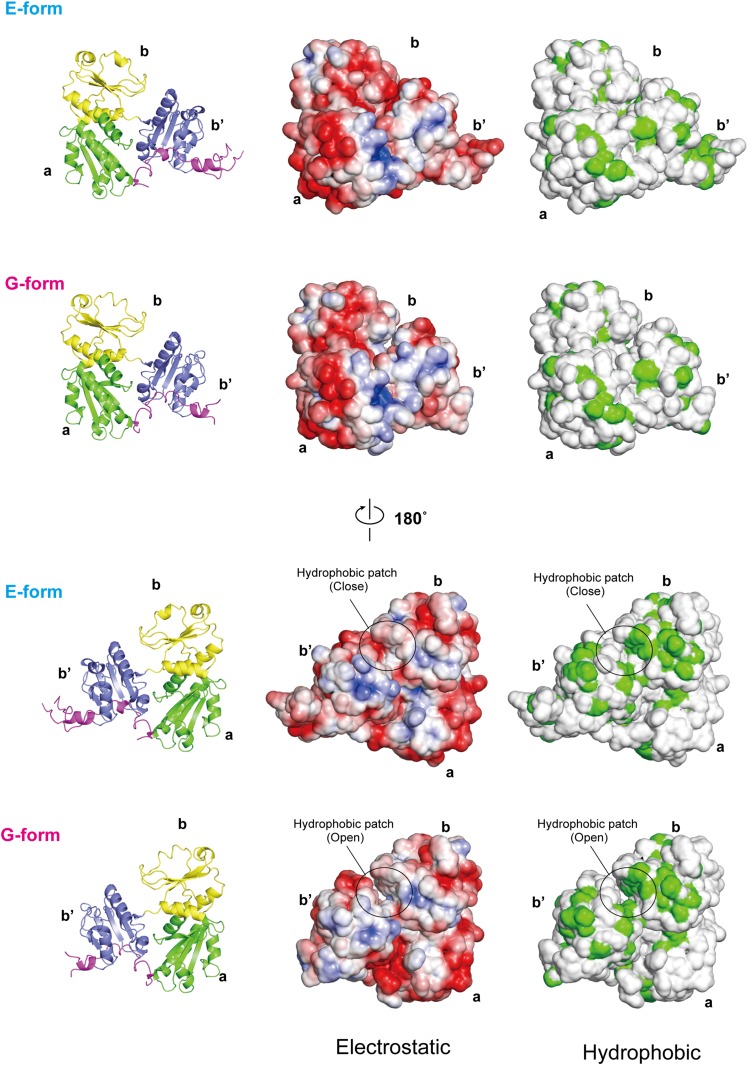
Importantly, the observed domain movements result in substantial changes in the electrostatic surface potential. In the G form, the relative positions of several arginine and lysine residues are altered relative to those in the E form, and two hydrogen bonds and a salt bridge formed between Arg329 and Asp336 in the E form are disrupted (Fig. 2D). As a result, the positively charged regions on the surface of the a and b′ domains (about 1,800 Å2) are significantly enlarged in the G form compared with those in the E form (about 1,000 Å2) (Fig. 2E). Likewise, the overall electrostatic surface potential is affected (Fig. S2). A hydrophobic patch between the b and b′ domains is more solvent-exposed in the G form (Fig. S2), which is ascribed to the significant separation between these two domains mentioned above (Fig. 2A). These differences in the surface charge distribution between the E and G forms are consistent with our previous finding that ERp44 binds to and dissociates from client proteins in a pH-dependent manner (19) (see also Discussion).
Fig. S2.
Comparison of the molecular surface properties of ERp44 at pH 7.2 and 6.5. (Left) Ribbon representations of the molecules as a reference. (Center) Electrostatic potential of ERp44, in which positively and negatively charged regions are shown in blue and red, respectively. A circle highlights a hydrophobic patch formed at the interface of the b and b′ domains. (Right) Hydrophobicity of the molecular surface of ERp44. Hydrophobic residues (except main-chain oxygen and nitrogen atoms) are shown in green.
The pH-Dependent Dynamics of the C Tail.
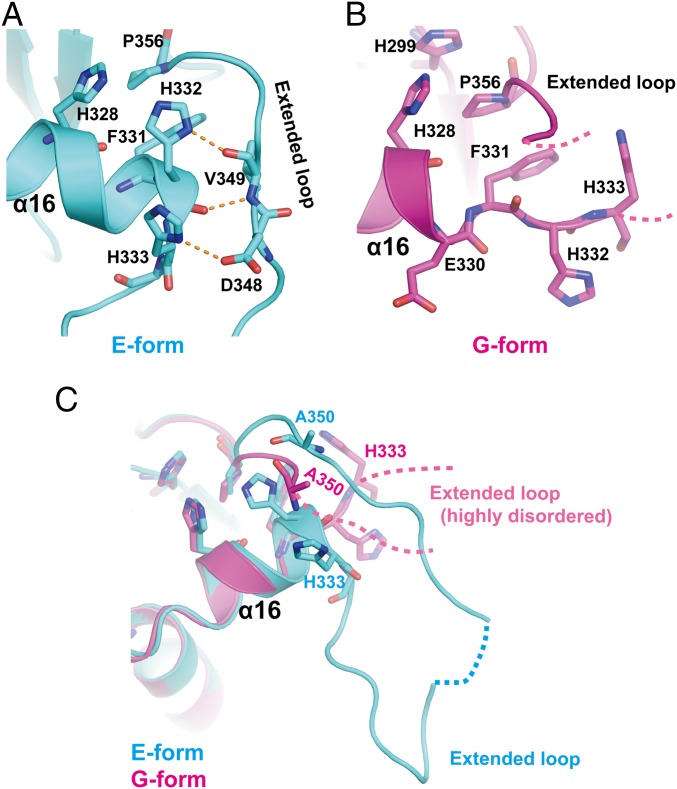
A change in pH also affects the conformation and dynamics of the C tail (Fig. 3A). In the E form, the histidine-rich region in the C tail (residues 326 to 333) adopts an α-helical conformation (α16), in which His332 and His333 form hydrogen bonds with the main-chain atoms of Val349 and the side chain of Asp348 in the extended loop, respectively (Fig. 3A). Notably, a part of the α16-helix (residues Glu330 to His333) is unwound in the G form (Fig. 3B). His332 and His333 assume an extended configuration in the G form, leading to disruption of the hydrogen bonds at the interface of the α16-helix and the extended loop (Fig. 3B). As a result, the extended loop is highly disordered in the G form (Fig. 3C, magenta dotted line).
Fig. 3.
pH-dependent conformational changes around the histidine-rich region. (A) Close-up view of interactions between the histidine residues in the α16-helix and their contact residues located in the extended loop in the E form. Hydrogen bonds are shown as yellow dashed lines. (B) Close-up view of the partially unwound α16-helix in the G form. Note that electron density was invisible for most of the extended loop in the G form, probably due to the highly disordered structure. Only the tentatively placed N-terminal and C-terminal short segments of the loop are represented by dashed lines. (C) Superposition of the α16-helices of the two structures. The dotted lines indicate the unstructured regions whose electron density is invisible even in the present higher-resolution crystal structures.
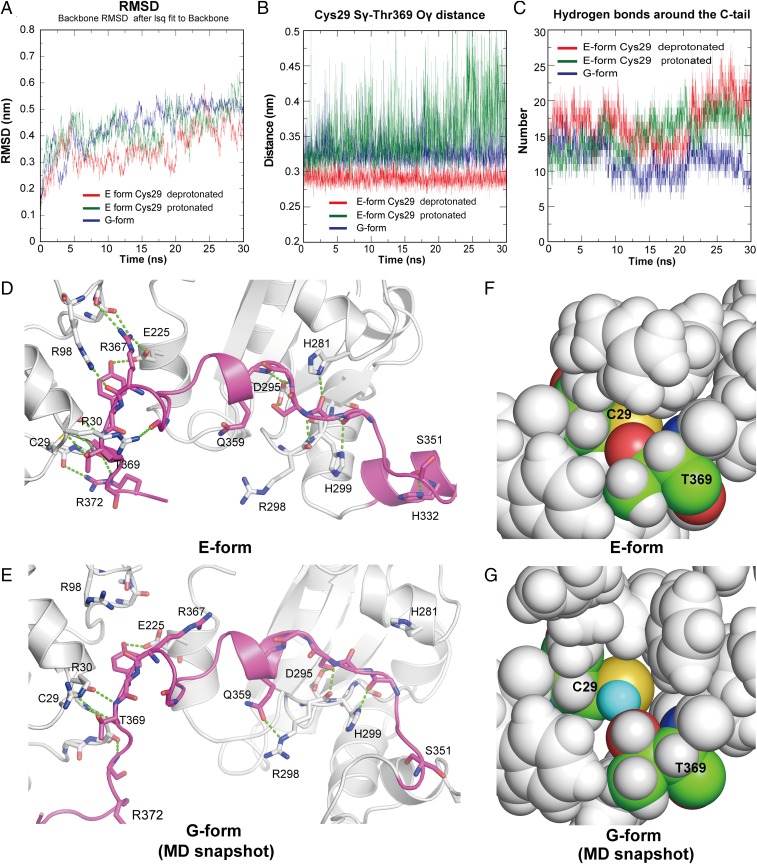
Compared with the extended loop, the shielding segment of the C tail in the G form undergoes little displacement from the position in the E form upon a 7.2 to 6.5 pH shift (Fig. S1B), despite previous studies that suggested a significant opening of the C tail at weakly acidic pH (19). This observation may be ascribed to the crystal packing restricting the dynamics of the shielding segment. To model the dynamics of the C tail, we carried out molecular dynamics (MD) simulations using the higher-resolution crystal structures determined in the present work. The overall structures were stable over the entire simulation duration, with an average backbone rmsd of ∼3 to ∼5 Å from the starting structure (Fig. 4A). In the MD simulation of the E form with deprotonated Cys29, the distance between Sγ of Cys29 and Oγ of Thr369 remained almost constant throughout (Fig. 4B, red line and Movie S1). Protonation of Cys29 in the E form markedly increased the fluctuation of the Sγ-to-Oγ distance (Fig. 4B, green line). However, the number of hydrogen bonds between the C tail and the a or b′ domain did not change appreciably throughout the simulation (Fig. 4 C and D and Movie S2). In contrast, the number of hydrogen bonds formed around the C tail in the G form decreased significantly during the simulation (Fig. 4 C and E and Movie S3), resulting in the transient exposure of the Sγ of Cys29 (Fig. 4 F and G). These results strongly suggest that unwinding of the α16-helix at weakly acidic pH increases the dynamic properties of the extended loop and shielding segment through significant disruption of hydrogen bonds around the C tail.
Fig. 4.
pH-sensitive dynamics of the C tail. (A) Root-mean-square deviation plots of the backbone atoms (N, Cα, and C) from the energy-minimized starting structures. (B) Distances between the Sγ of Cys29 and the Oγ of Thr369 during the MD simulations. (C) Number of hydrogen bonds between the C tail (residues 350 to 378) and the a and b′ domains (residues 10 to 340) during the MD simulations. (D and E) Hydrogen bonds around the C tail in the crystal structure of the E form (D) and those in a snapshot of the MD simulation of the G form (E) are represented by green dashed lines. (F) Close-up view around Cys29 (shown in sphere representation) in the initial structure of the E form with Cys29 modeled as the deprotonated state. All atoms except Cys29 and Thr369 are colored in gray. The carbon, oxygen, nitrogen, and sulfur atoms of Cys29 and Thr369 are colored green, blue, red, and yellow, respectively. (G) Close-up view around Cys29 from an MD simulation snapshot of the G form. The Hγ of Cys29 is colored cyan.
ERp44 Recognizes Multiple Ero1α Cysteines Through Electrostatic Interactions.
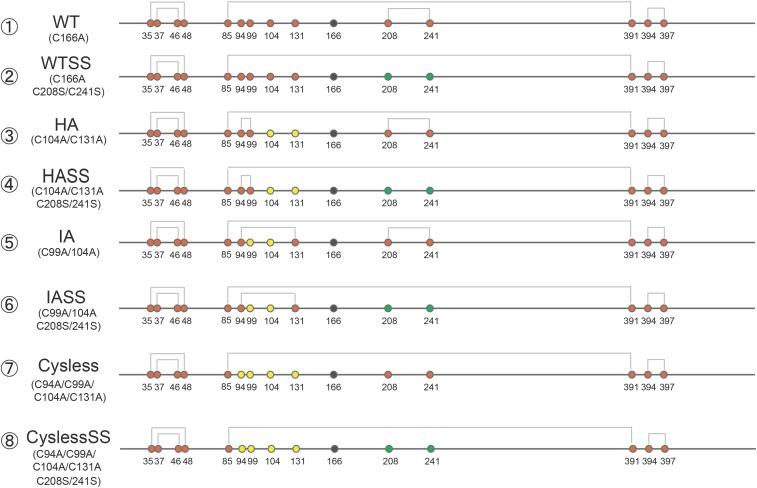
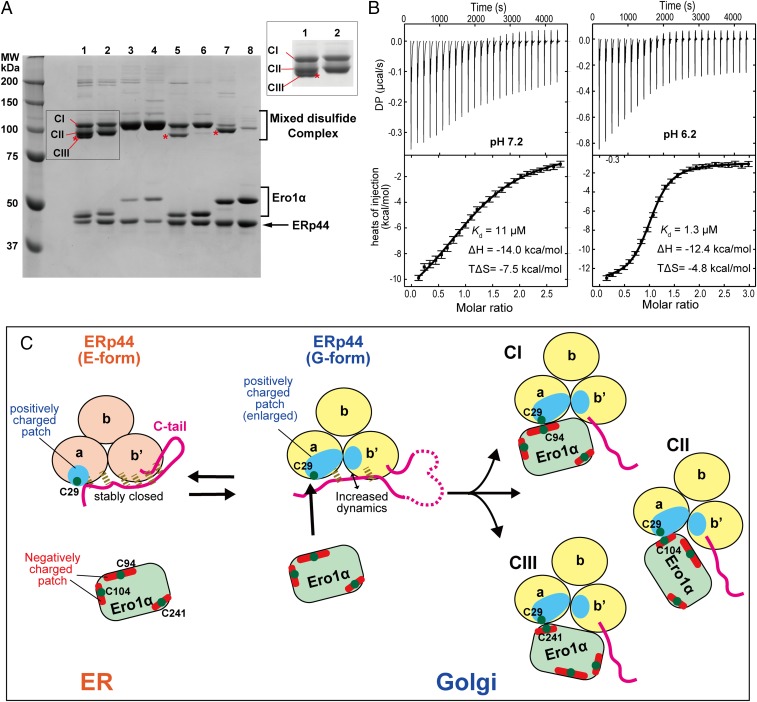
The present finding that the putative client-binding surface around Cys29 is more positively charged at low pH (Fig. 2E) suggests that ERp44 preferentially recognizes negatively charged regions in its client proteins. To gain insight into the mode of interaction between ERp44 and client proteins, we investigated which cysteine(s) of Ero1α engage(s) in covalent linkages with Cys29 of ERp44, using a set of Ero1α mutants (Fig. S3). Mixtures of ERp44 and wild-type (WT) Ero1α demonstrated that the two proteins form at least three types of mixed disulfide complexes (CI, CII, and CIII in Fig. 5A, lane 1). The CIII band disappeared upon mutation of Cys208/241Ser (WTSS) (lane 2), suggesting that CIII contains a mixed disulfide between Cys29 of ERp44 and Cys208 or Cys241 of Ero1α. A CI band was only observed for the hyperactive Ero1α (HA) mutant, its variant with Cys208/241Ser mutations (HASS), and the inactive (IA) Ero1α with Cys208/241Ser mutations (IASS) (lanes 3, 4, and 6, respectively). These three mutants all had Cys94 in common, which implies that Cys94 is the primary Ero1α cysteine involved in formation of the CI complex. In agreement with this, a cysteine-less mutant in which Cys94, Cys99, Cys104, and Cys131 were all mutated to Ala did not form a CI complex (lane 7). Whereas WTSS did display a CII band (lane 2), IASS did not (lane 6). These observations suggest that CII involves Cys99 or Cys104 of Ero1α. Collectively, these results led us to conclude that Cys29 forms mixed disulfide bonds with either one of the regulatory loop cysteines of Ero1α (23). Cys208 or Cys241 of Ero1α engages in complex formation with ERp44 more frequently when Cys94 is unavailable (see the bands marked with an asterisk in lanes 5 and 7).
Fig. S3.
Schematic representations of Ero1α variants used in this study. Cysteine residues are shown as red circles. Mutations of cysteine to alanine or serine are represented by yellow and green circles, respectively. In all constructs, Cys166 was mutated to alanine to avoid formation of aberrant disulfide-linked oligomers of Ero1α (black circle).
Fig. 5.
Modes of interaction between ERp44 and Ero1α. (A) Disulfide-bond formation of ERp44 with WT and cysteine mutants of Ero1α. ERp44 was mixed with each of the indicated Ero1α mutants at an equal molar ratio (5 µM). The mixture was treated with 1 mM NEM and then subjected to nonreducing SDS/PAGE. Asterisks indicate the complexes involving Cys208 or Cys241 of Ero1α. Lane 1, wild type; lane 2, WTSS (C208S/C241S); lane 3, HA (C104A/C131A); lane 4, HASS (C104A/C131A/C208S/C241S); lane 5, IA (C94A/C104A); lane 6, IASS (C94A/C104A/C208S/C241S); lane 7, Cysless (C94A/C99A/C104A/C131A); lane 8, CyslessSS (C94A/C99A/C104A/C131A/C208S/C241S). (B) ITC raw data (Upper) and binding isotherm data (Lower) for titration of Ero1α HA into ERp44 at pH 7.2 (Left) or 6.2 (Right). DP indicates a measured differential power between the reference and sample cells. Error bars represent errors estimated by a program NITPIC (49) for each integrated heat. (C) Proposed working model for the pH-dependent regulation of ERp44. Green dots represent cysteine residues that could be involved in the covalent complex. Dashed lines represent hydrogen bonds formed between the C tail and the main body of ERp44. CI, CII, and CIII denote the three types of mixed disulfide complexes between ERp44 and Ero1α suggested by experiments shown in A.
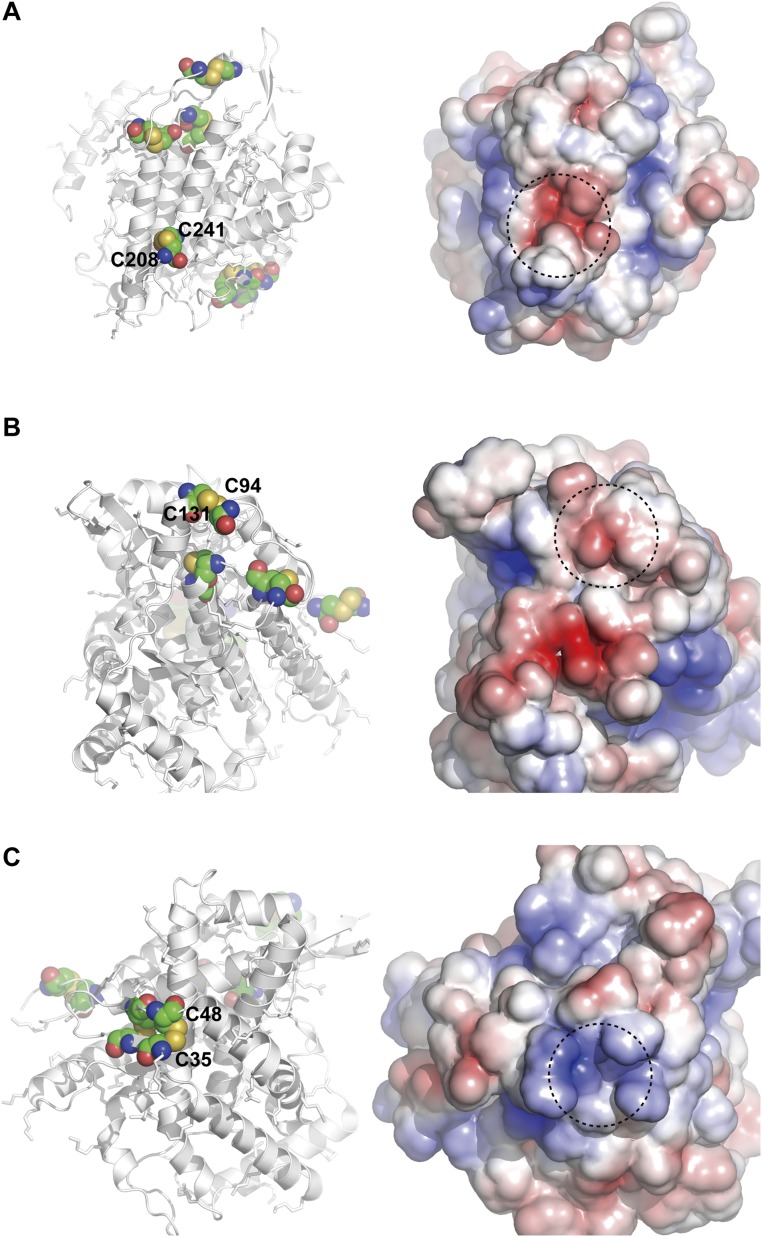
It is noteworthy that both Cys94–Cys131 and Cys208–Cys241 in Ero1α are located in loop regions rich in negative charges (Fig. S4 A and B), although the flexible segment around Cys99 is not resolved in the crystal structure of Ero1α (23). In contrast, the regions around the N-terminal cysteines (Cys35, Cys37, Cys46, and Cys48) of Ero1α, which were not involved in mixed disulfide formation with ERp44, are positively charged (Fig. S4C). These observations support our notion that the positively charged surface around Cys29 of ERp44 interacts preferentially with the negatively charged loop regions of Ero1α, leading to the efficient formation of the ERp44–Ero1α covalent complex.
Fig. S4.
Highlighted view of cysteine residues in an inactive Ero1α mutant with the Cys94–Cys131 disulfide bond. (Left) Solvent-exposed cysteine residues are represented by spheres. (Right) Electrostatic potential of Ero1α, in which positively and negatively charged regions are shown in blue and red, respectively. Dotted circles highlight the regions around the indicated cysteine residues.
The pH-Dependent Enthalpy-Driven Interactions Between ERp44 and Ero1α.
To further probe the electrostatic interaction between ERp44 and Ero1α, we investigated the thermodynamics of the binding of ERp44 to Ero1α by isothermal titration calorimetry (ITC). In the ITC experiments, the Ero1α hyperactive mutant was used as a titrant because this mutant bound strongly to ERp44, mainly through the single interaction mode (Fig. 5A, lane 3). At pH 7.2, ERp44 interacted weakly with Ero1α ΗΑ, with a Kd of 11 µM (Fig. 5B). At pH 6.2, however, the affinity of ERp44 for Ero1α ΗΑ was significantly increased (Kd 1.3 µM), as previously shown by surface plasmon resonance measurements (19). The ITC analyses uncovered that the interaction between ERp44 and Ero1α HA was an exothermic reaction at both pH 7.2 and 6.2. The large negative enthalpy (ΔH) values generated by titration of ERp44 with Ero1α indicate that electrostatic interactions and hydrogen bonds are the primary driving forces, consistent with the distribution of opposite charges on the surfaces of ERp44 and Ero1α as addressed above.
Discussion
Previous studies have indicated that the protonation state of Cys29 is a key determinant of the opened/closed configuration of the C tail (19). However, the observation that a number of hydrogen bonds and van der Waals contacts are formed between the C tail and the a and b′ domains (Fig. 4 D and E) suggested that additional factors are involved in the pH-dependent regulation of ERp44. The higher-resolution crystal structures of ERp44 at pH 6.5 and 7.2 described herein reveal that the protonation and orientation of the histidine bridging the a and b domains (His157) and of the conserved histidines constituting the His cluster (His299, His328, His332, and His333) (Figs. 2 A and B and 3 A and B) are also key to the pH-dependent conformational changes of ERp44. MD simulations demonstrate that acidic conditions induce the unwinding of the α16-helix, resulting in the disruption of several hydrogen bonds around the C tail. This enhances the dynamics of the C tail and facilitates the exposure of Cys29 to bulk solvent (Fig. 4G). Previous results showed that mutating these histidine residues abolished the pH sensitivity in living cells (22), which could be explained by the inhibited α16-helix unwinding.
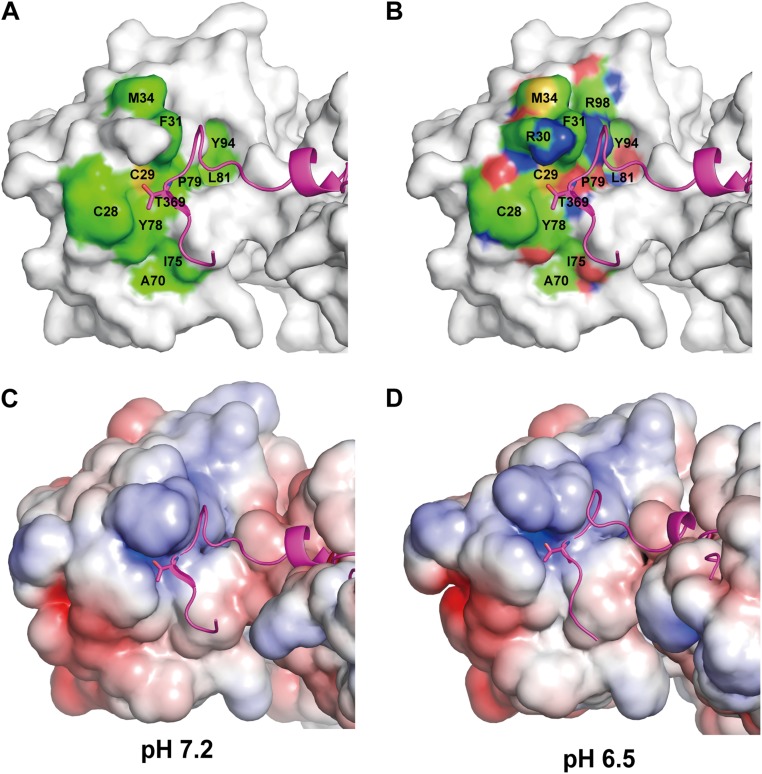
Our systematic mutation assays demonstrated that Cys29 located at the positively charged surface of ERp44 forms the mixed disulfide with cysteines located within the negatively charged loop regions of Ero1α. These results suggest that electrostatic interactions play an essential role in client recognition in ERp44. However, previous studies suggested that the surface around Cys29 in the ERp44 a domain mediates “hydrophobic” interaction with client proteins (18, 19). This discrepancy is likely due to inappropriate representation of the molecular surfaces (Fig. S5A). Reanalysis of the chemical properties of the ERp44 surfaces in light of the present higher-resolution crystal structures shows that the substrate-binding site around Cys29 is less hydrophobic (Fig. S5B) but rather positively charged (Fig. S5 C and D). This structural feature therefore appears suited to interact with the negatively charged surface of Ero1α.
Fig. S5.
Reanalysis of the molecular surface of the client-binding site in the a domain. The C-terminal tail is represented by a ribbon diagram colored magenta. (A) The atoms of the indicated hydrophobic residues are colored in green. The thiol group of Cys29 is shown in yellow. (B) The carbon, nitrogen, and oxygen atoms of the indicated hydrophobic residues are colored green, blue, and red, respectively. (C and D) Electrostatic surface potential at pH 7.2 (C) or pH 6.5 (D). Positively and negatively charged regions are denoted in blue and red, respectively.
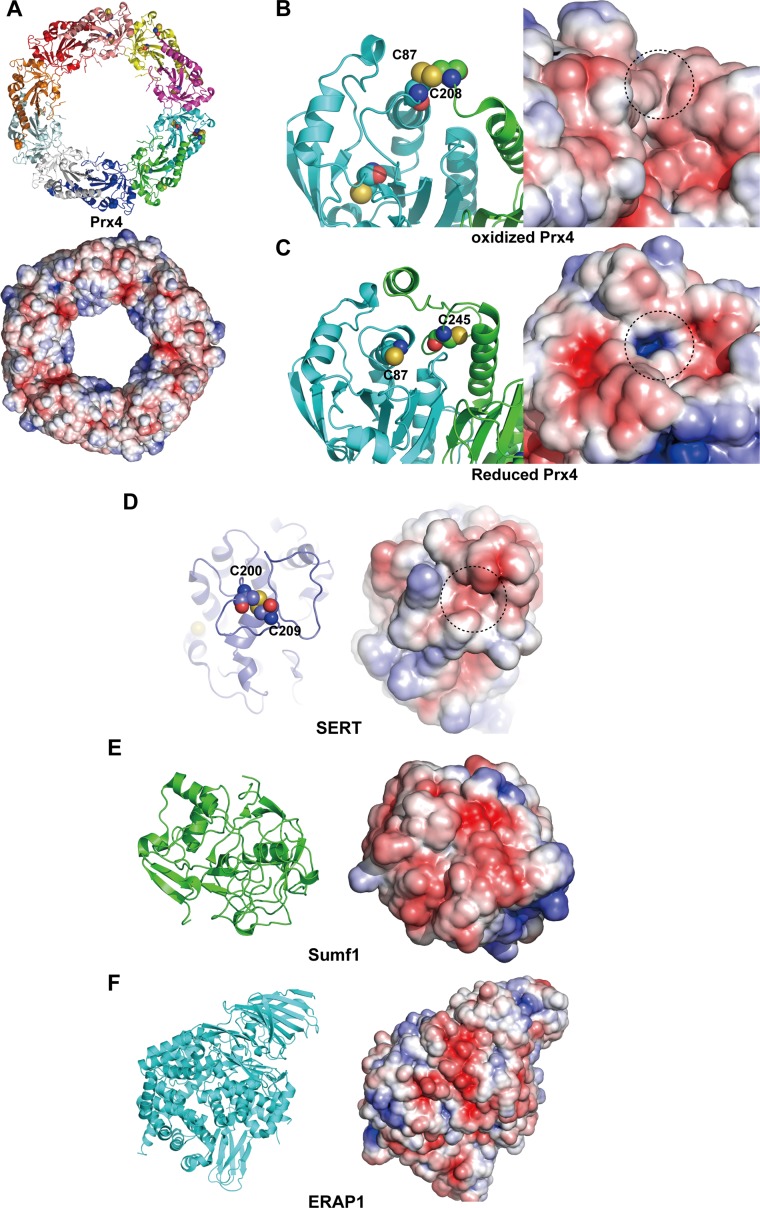
Importantly, other ERp44 clients also share similar surface properties with Ero1α. Prx4 contains three solvent-exposed cysteines (Cys51, Cys124, and Cys245) (24–26). In the oxidized form of Prx4, the peroxidatic (Cys124) cysteine of one protomer forms a disulfide bond with the resolving (Cys245) cysteine of another protomer, whereas Cys51 is not visible in the electron density in the reported crystal structures (24, 25). Notably, the molecular surface around the Cys124–Cys245 disulfide bond in Prx4 is negatively charged, as are the surfaces surrounding the ERp44-targeting cysteines in Ero1α (Fig. S6 A and B). The crystal structure of SERT also reveals that the target disulfide bond (Cys200–Cys209) is surrounded by negatively charged loops (Fig. S6D) (27). Similarly, other client proteins, Sumf1 and ERAP1, have negatively charged regions, although their putative ERp44-binding sites are not resolved in the reported crystal structures (28–31) (Fig. S6 E and F). Altogether, a negatively charged flexible loop is likely a common structural feature in ERp44 clients.
Fig. S6.
Surface properties of other ERp44 client proteins. (A) Overall structure of Prx4 in ribbon representation (Upper) and surface representation with electrostatic surface potential (Lower). (B and C) Close-up view of the active-site cysteine residues of the oxidized (B) and reduced (C) forms of Prx4. (Left) The indicated cysteine residues are represented by spheres. (Right) Electrostatic surface potential of oxidized Prx4, in which positively and negatively charged regions are shown in blue and red, respectively. The dotted circles highlight regions around the indicated cysteine residues. (D, Left) Close-up view of the Cys200–Cys209 disulfide bond of SERT, in which the indicated cysteine residues are represented by spheres. (D, Right) Electrostatic surface potential of SERT, in which positively and negatively charged regions are shown in blue and red, respectively. The dotted circle highlights a region around the indicated cysteine residues. (E and F) Overall structures of Sumf1 (E) and ERAP1 (F) in ribbon representation (Left) and surface representation with electrostatic surface potential (Right).
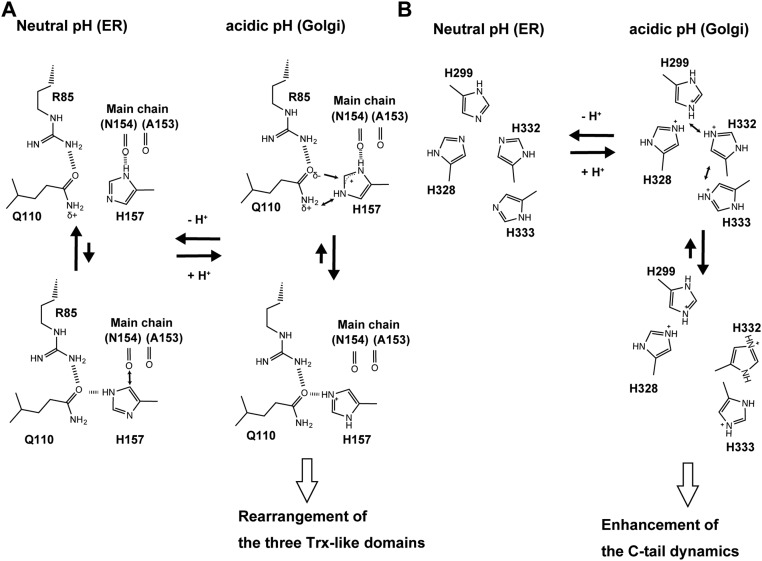
Based on these findings, we can draw a molecular model of the pH-dependent protein quality control cycle exerted by ERp44 (Fig. 5C). In the ER (pH ∼7.2), the Nδ of His157 is presumably protonated and makes hydrogen bonds with the carbonyl oxygens of Ala153 and Asn154 (Fig. 2B and Fig. S7A). The histidine-rich region in the C tail assumes the α16-helix, interacting with the subsequent extended loop (Fig. 3A). At this stage, the client-binding site around Cys29 is masked by the C tail. In the more acidic Golgi, however, both the Nδ and Nε of His157 are likely to be protonated, and the imidazole ring is flipped. This flipped conformation seems to be induced or stabilized by the hydrogen bond between the Nε of His157 and the Oε of Gln110 (Fig. 2B and Fig. S7A), leading to the significant rotation of the a and b′ domains and an enlargement of the positively charged surface area around the client-binding site (Fig. 2 D and E). Concomitantly, electrostatic repulsion is likely to occur between the protonated His299, His328, His332, and His333 in the His cluster, which unwinds the α16-helix and disrupts the hydrogen bonds around the C tail (Fig. 3B and Fig. S7B). As a result, the C-tail dynamics is enhanced and the positively charged surface involving Cys29 is more exposed, facilitating electrostatic interactions, hydrogen bonds, and mixed disulfide formation between ERp44 and its client proteins.
Fig. S7.
(A) Proposed model of the protonation states of His157 and the protonation-induced rearrangement of hydrogen bonds between His157 and its surrounding residues. (B) Plausible charge repulsion between the protonated histidine residues (His299, His328, His332, and His333) in the His cluster at acidic pH.
During the evaluation process of this paper, Yang et al. reported the crystal structure of the ERp44–Prx4 complex (32), which overall supports our proposed model of ERp44 binding to the client proteins. The observation that ERp44 prefers the oxidized form of Prx4 to its reduced form (32) can be explained by the different electrostatic surface properties between these two redox states. The Cys87–Cys208 disulfide bond of Prx4 in the oxidized state is surrounded by negatively charged regions, whereas there is a positively charged region near the solvent-exposed Cys87 in the reduced state (Fig. S6 B and C). Thus, electrostatic interactions are likely critical for ERp44 to approach the target disulfide bond in Prx4. The crystal structure of the ERp44–Prx4 complex (32) reveals that a Cys29 (ERp44)–Cys208 (Prx4) intermolecular disulfide bond and a number of hydrogen bonds form at the interface between the ERp44 a domain and the Prx4 homodimer. In this context, the flexible C-terminal region of Prx4 assumes a β-strand, which forms hydrogen bonds with the β4-strand of ERp44 (32). Taken together, these observations suggest the following general mechanism of ERp44–client complex formation. In the weakly acidic Golgi, the enlarged positively charged surface of ERp44 binds clients’ negatively charged loops, increasing C-tail opening. The ERp44–client complex is further stabilized by hydrogen bonds and intermolecular disulfide linkages between Cys29 of ERp44 and specific cysteines of client proteins.
The partial release of the C tail induced by low pH may explain why ERp44 localizes primarily in distal ESP compartments (ERGIC and cis-Golgi) unless abundant clients are expressed (10). The feasible benefit from client binding to ERp44 is the even greater exposure of the C-terminal RDEL motif, ensuring recognition by KDELR. In agreement with this notion, the overexpression of a client protein significantly inhibits the secretion of ERp44 mutants with impaired pH sensitivity that lack the conserved histidines (22).
As exemplified by ERp44, the ESP is equipped with several pH-dependent regulatory systems involving protonation of histidine residues. The lectin activity of ERGIC53, which functions as a transport receptor for some secretory glycoproteins, is controlled by a conserved histidine switch (33). The ER-resident collagen-specific chaperone HSP47 binds its folded clients in the ER and releases them in the Golgi. The bind-and-release cycle of HSP47 is regulated by protonation of a tandem histidine pair (34). A histidine switch also controls the pH-dependent interaction of receptor-associated proteins with LDL receptor family members in the ESP (35). In these cases, protonation of the histidine residues increases their mutual electrostatic repulsion and thereby induces partial unfolding of the client-binding sites, resulting in client release. Similarly, in ERp44, histidine protonation induces domain movements and local helix unwinding, and these modulate the affinity for client proteins. Thus, pH-dependent histidine protonation likely controls both anterograde and retrograde transport in the ESP.
In summary, our results reveal how histidine protonation determines the pH-dependent regulation of ERp44. It is reasonable that other folding assistants residing in the ESP use this strategy to regulate their localization and functionality. In all likelihood, they also sense the redox states of client proteins and the surrounding redox environment. Our in vitro analysis showed that ERp44 forms covalent complexes with the hyperactive state of Ero1α more efficiently than with its inactive state (Fig. 5A). Likewise, ERp44 binds oxidized Prx4 better than reduced Prx4 (32). ERp44 may thus selectively transport active oxidoreductases back to the ER to promote oxidative protein folding in this organelle. It will be interesting to further explore the hypothesis that ERp44 selectively binds to clients in a particular redox state to maintain the redox status and protein homeostasis in the ESP.
Materials and Methods
Purification and Crystallization of ERp44 at Neutral and Weakly Acidic pHs.
Expression and purification of recombinant WT ERp44 and a ΔRDEL mutant were performed as described previously (19). All crystallizations were performed by the sitting-drop vapor-diffusion method at 277 K. Crystals of ERp44 ΔRDEL at neutral pH were obtained in a few days by mixing 1.2 to 1.5 µL of protein solution (30 mg/mL protein in 20 mM Tris⋅HCl, pH 7.5 and 150 mM NaCl) with 1.2 to 1.5 µL of precipitant solution containing 0.1 M Tris⋅HCl (pH 7.6) and 19 to 21% (wt/vol) polyethylene glycol (PEG) 8000 (final pH of 7.2). Crystals of ERp44 ΔRDEL at weakly acidic pH were grown at 277 K for a week by mixing 0.7 µL of the protein solution (20 mg/mL protein in 20 mM Bis-Tris⋅HCl, pH 6.5 and 150 mM NaCl) with 0.7 µL of precipitant solution containing 0.1 M Bis-Tris⋅HCl (pH 6.1) and 0.6 to 0.7 mM Na/K tartrate (final pH of 6.5). Crystals of full-length ERp44 were also obtained under the same conditions with the same space group, showing that deletion of the RDEL motif hardly affected the overall structure, although the diffraction qualities of these crystals were poor (3- to 4-Å resolution), compared with ERp44 ΔRDEL crystals.
Before data collection, a cryoprotectant solution containing 0.1 M Tris⋅HCl (pH 7.6), 22% (wt/vol) PEG 8000, and from 5 to 15% (vol/vol) PEG 400 (for neutral pH) or 0.1 M Bis-Tris⋅HCl (pH 6.1), 0.68 mM K/Na tartrate, and 3.5 M Na formate (for weakly acidic pH) was added to the drop in a stepwise manner, and then crystals were flash-cooled in a nitrogen stream at 100 K.
Data Collection and Structure Determination.
The X-ray diffraction data were collected on beamline BL44XU at SPring-8 (proposal nos. 2013A6804, 2013B6804, 2014A6904, 2014B6904, 2015A6558, and 2015B6558) and beamline BL-1A at the Photon Factory. The diffraction images were processed with the HKL2000 package (36) or XDS (37). The structures of ERp44 at neutral and weakly acidic pHs were determined by the molecular replacement method using MOLREP (38), with the previously determined ERp44 structure (Protein Data Bank ID code 2R2J) as a search model. The initial electron density map was improved with RESOLVE (39). Manual model building was performed with Coot (40), refined with PHENIX (41), and validated with MolProbity (42). Electrostatic surface potentials were calculated using PDB2PQR (43) and APBS (44). Protonation states of the E and G forms were assigned at pH 7.2 and 6.5, respectively, using PROPKA (45). Structural figures were prepared with PyMOL (www.pymol.org).
MD Simulation.
MD simulations were performed with GROMACS 4.6.7 (46) using the AMBER03 force field (47). The disordered extended loop in the structure of the G form was manually modeled using the equivalent structure in the E form. The RDEL motif was manually fused to the C terminus of the crystal structures of the E and G forms determined in the present work. Hydrogen atoms were added to the protein model using the pdb2gmx module in GROMACS. In the simulation of the E form, both protonated and deprotonated forms of Cys29 were modeled. The structures thus modified were placed in cubic boxes filled with TIP3P water molecules (46). Systems were electrically neutralized by replacing water molecules with an appropriate number of chloride ions. After energy minimization, NVT (isothermal-isochoric: constant number of particles, volume, and temperature) and NPT (isothermal-isobaric: constant number of particles, pressure, and temperature) equilibrations (46) were performed, each for 100 ps at 300 K. Production MD simulations were carried out for 30 ns with integration steps of 2 fs. The number of hydrogen bonds between the C tail (residues 350 to 378) and the Trx domains (residues 1 to 340) was determined using the g_hbond module in GROMACS with default parameters.
Disulfide-Bond Formation Between ERp44 and Ero1α.
WT and mutants of Ero1α were prepared as described previously (23, 48). In all constructs, Cys166 was mutated to alanine to avoid formation of aberrant disulfide-linked oligomers of Ero1α. ERp44 (5 µM) and WT or mutant Ero1α (5 µM) were mixed and incubated on ice for 30 min in 20 mM Bis-Tris⋅HCl (pH 6.2) and 150 mM NaCl. Mixtures were then treated with 1 mM N-ethylmaleimide (NEM) for 10 min on ice and separated by nonreducing SDS/PAGE (8%).
Isothermal Titration Calorimetry Analysis.
ITC experiments were carried out using a MicroCal iTC200 calorimeter (Malvern) in 20 mM Bis-Tris⋅HCl (pH 6.2 or 7.2) and 150 mM NaCl. After an initial injection (0.4 µL), a total of 24 injections of 1.5 µL Erο1α HA (420 µM) were titrated into ERp44 (30 µM) at 180-s intervals with a stirring speed of 750 rpm at 283 K. Integration of the injection peaks was performed with NITPIC (49), and global analysis was performed with SEDPHAT (50) using a 1:1 binding model. All experiments were performed at least twice.
Supplementary Material
Acknowledgments
We thank S. Ogawa and M. Okumura for preparation of Ero1 variants, Y. Amagai for constructive discussions, and the beamline scientists of SPring-8 and the Photon Factory for their help with X-ray data collection. This work was supported by Grants-in-Aid for Scientific Research (15H04335 and 26116005; to K.I.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, the Platform Project for Supporting Drug Discovery and Life Science Research (Platform for Drug Discovery, Informatics, and Structural Life Science) from the Japan Agency for Medical Research and Development (AMED) (to K.I.), and Telethon (GGP15059), Ministero della Salute (PE-2011-02352286), Associazione Italiana per la Ricerca sul Cancro (IG18824), and Fondazione Cariplo (to R.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 5GU6 (E form) and 5GU7 (G form)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1621426114/-/DCSupplemental.
References
- 1.Barlowe CK, Miller EA. Secretory protein biogenesis and traffic in the early secretory pathway. Genetics. 2013;193:383–410. doi: 10.1534/genetics.112.142810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Görlach A, Klappa P, Kietzmann T. The endoplasmic reticulum: Folding, calcium homeostasis, signaling, and redox control. Antioxid Redox Signal. 2006;8:1391–1418. doi: 10.1089/ars.2006.8.1391. [DOI] [PubMed] [Google Scholar]
- 3.Okumura M, Kadokura H, Inaba K. Structures and functions of protein disulfide isomerase family members involved in proteostasis in the endoplasmic reticulum. Free Radic Biol Med. 2015;83:314–322. doi: 10.1016/j.freeradbiomed.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 4.Ellgaard L, Helenius A. Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- 5.Anelli T, Sitia R. Protein quality control in the early secretory pathway. EMBO J. 2008;27:315–327. doi: 10.1038/sj.emboj.7601974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anelli T, Sannino S, Sitia R. Proteostasis and “redoxtasis” in the secretory pathway: Tales of tails from ERp44 and immunoglobulins. Free Radic Biol Med. 2015;83:323–330. doi: 10.1016/j.freeradbiomed.2015.02.020. [DOI] [PubMed] [Google Scholar]
- 7.Anelli T, et al. Thiol-mediated protein retention in the endoplasmic reticulum: The role of ERp44. EMBO J. 2003;22:5015–5022. doi: 10.1093/emboj/cdg491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anelli T, et al. Sequential steps and checkpoints in the early exocytic compartment during secretory IgM biogenesis. EMBO J. 2007;26:4177–4188. doi: 10.1038/sj.emboj.7601844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiang L, Wang H, Farmer SR. Adiponectin secretion is regulated by SIRT1 and the endoplasmic reticulum oxidoreductase Ero1-L alpha. Mol Cell Biol. 2007;27:4698–4707. doi: 10.1128/MCB.02279-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang ZV, et al. Secretion of the adipocyte-specific secretory protein adiponectin critically depends on thiol-mediated protein retention. Mol Cell Biol. 2007;27:3716–3731. doi: 10.1128/MCB.00931-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hampe L, et al. Regulation and quality control of adiponectin assembly by endoplasmic reticulum chaperone ERp44. J Biol Chem. 2015;290:18111–18123. doi: 10.1074/jbc.M115.663088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freyaldenhoven S, et al. The role of ERp44 in maturation of serotonin transporter protein. J Biol Chem. 2012;287:17801–17811. doi: 10.1074/jbc.M112.345058. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Anelli T, et al. ERp44, a novel endoplasmic reticulum folding assistant of the thioredoxin family. EMBO J. 2002;21:835–844. doi: 10.1093/emboj/21.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otsu M, et al. Dynamic retention of Ero1alpha and Ero1beta in the endoplasmic reticulum by interactions with PDI and ERp44. Antioxid Redox Signal. 2006;8:274–282. doi: 10.1089/ars.2006.8.274. [DOI] [PubMed] [Google Scholar]
- 15.Kakihana T, et al. Dynamic regulation of Ero1α and peroxiredoxin 4 localization in the secretory pathway. J Biol Chem. 2013;288:29586–29594. doi: 10.1074/jbc.M113.467845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mariappan M, Radhakrishnan K, Dierks T, Schmidt B, von Figura K. ERp44 mediates a thiol-independent retention of formylglycine-generating enzyme in the endoplasmic reticulum. J Biol Chem. 2008;283:6375–6383. doi: 10.1074/jbc.M709171200. [DOI] [PubMed] [Google Scholar]
- 17.Hisatsune C, et al. ERp44 exerts redox-dependent control of blood pressure at the ER. Mol Cell. 2015;58:1015–1027. doi: 10.1016/j.molcel.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 18.Wang L, et al. Crystal structure of human ERp44 shows a dynamic functional modulation by its carboxy-terminal tail. EMBO Rep. 2008;9:642–647. doi: 10.1038/embor.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vavassori S, et al. A pH-regulated quality control cycle for surveillance of secretory protein assembly. Mol Cell. 2013;50:783–792. doi: 10.1016/j.molcel.2013.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casey JR, Grinstein S, Orlowski J. Sensors and regulators of intracellular pH. Nat Rev Mol Cell Biol. 2010;11:50–61. doi: 10.1038/nrm2820. [DOI] [PubMed] [Google Scholar]
- 21.Paroutis P, Touret N, Grinstein S. The pH of the secretory pathway: Measurement, determinants, and regulation. Physiology. 2004;19:207–215. doi: 10.1152/physiol.00005.2004. [DOI] [PubMed] [Google Scholar]
- 22.Sannino S, et al. Progressive quality control of secretory proteins in the early secretory compartment by ERp44. J Cell Sci. 2014;127:4260–4269. doi: 10.1242/jcs.153239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inaba K, et al. Crystal structures of human Ero1α reveal the mechanisms of regulated and targeted oxidation of PDI. EMBO J. 2010;29:3330–3343. doi: 10.1038/emboj.2010.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao Z, Tavender TJ, Roszak AW, Cogdell RJ, Bulleid NJ. Crystal structure of reduced and of oxidized peroxiredoxin IV enzyme reveals a stable oxidized decamer and a non-disulfide-bonded intermediate in the catalytic cycle. J Biol Chem. 2011;286:42257–42266. doi: 10.1074/jbc.M111.298810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Wang L, Wang X, Sun F, Wang CC. Structural insights into the peroxidase activity and inactivation of human peroxiredoxin 4. Biochem J. 2012;441:113–118. doi: 10.1042/BJ20110380. [DOI] [PubMed] [Google Scholar]
- 26.Sato Y, et al. Synergistic cooperation of PDI family members in peroxiredoxin 4-driven oxidative protein folding. Sci Rep. 2013;3:2456. doi: 10.1038/srep02456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coleman JA, Green EM, Gouaux E. X-ray structures and mechanism of the human serotonin transporter. Nature. 2016;532:334–339. doi: 10.1038/nature17629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kochan G, et al. Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming. Proc Natl Acad Sci USA. 2011;108:7745–7750. doi: 10.1073/pnas.1101262108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen TT, et al. Structural basis for antigenic peptide precursor processing by the endoplasmic reticulum aminopeptidase ERAP1. Nat Struct Mol Biol. 2011;18:604–613. doi: 10.1038/nsmb.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dierks T, et al. Molecular basis for multiple sulfatase deficiency and mechanism for formylglycine generation of the human formylglycine-generating enzyme. Cell. 2005;121:541–552. doi: 10.1016/j.cell.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 31.Roeser D, et al. A general binding mechanism for all human sulfatases by the formylglycine-generating enzyme. Proc Natl Acad Sci USA. 2006;103:81–86. doi: 10.1073/pnas.0507592102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang K, et al. Crystal structure of the ERp44-peroxiredoxin 4 complex reveals the molecular mechanisms of thiol-mediated protein retention. Structure. 2016;24:1755–1765. doi: 10.1016/j.str.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 33.Appenzeller-Herzog C, Roche AC, Nufer O, Hauri HP. pH-induced conversion of the transport lectin ERGIC-53 triggers glycoprotein release. J Biol Chem. 2004;279:12943–12950. doi: 10.1074/jbc.M313245200. [DOI] [PubMed] [Google Scholar]
- 34.Oecal S, et al. The pH-dependent client release from the collagen-specific chaperone HSP47 is triggered by a tandem histidine pair. J Biol Chem. 2016;291:12612–12626. doi: 10.1074/jbc.M115.706069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee D, et al. RAP uses a histidine switch to regulate its interaction with LRP in the ER and Golgi. Mol Cell. 2006;22:423–430. doi: 10.1016/j.molcel.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 36.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 37.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 39.Terwilliger TC. Automated structure solution, density modification and model building. Acta Crystallogr D Biol Crystallogr. 2002;58:1937–1940. doi: 10.1107/s0907444902016438. [DOI] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Robertson AD, Jensen JH. Very fast empirical prediction and rationalization of protein pKa values. Proteins. 2005;61:704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 46.Pronk S, et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duan Y, et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 48.Ramming T, et al. Cysteines 208 and 241 in Ero1alpha are required for maximal catalytic turnover. Redox Biol. 2016;7:14–20. doi: 10.1016/j.redox.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keller S, et al. High-precision isothermal titration calorimetry with automated peak-shape analysis. Anal Chem. 2012;84:5066–5073. doi: 10.1021/ac3007522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao H, Piszczek G, Schuck P. SEDPHAT—A platform for global ITC analysis and global multi-method analysis of molecular interactions. Methods. 2015;76:137–148. doi: 10.1016/j.ymeth.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Afonine PV, et al. FEM: Feature-enhanced map. Acta Crystallogr D Biol Crystallogr. 2015;71:646–666. doi: 10.1107/S1399004714028132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.