Significance
Metal homeostasis is essential for living organisms. Metal transporters play key roles in metal uptake and compartmentalization. The tight regulation of metal homeostasis thus depends on accurate targeting of these metal transporters. Although the main metal transporters in plants have been identified, the mechanisms involved in their trafficking are still poorly understood. This study reveals that AtPH1, a pleckstrin homology (PH) domain containing protein binding to phosphatidylinositol 3-phosphate (PI3P), controls the subcellular localization of the iron and manganese transporter AtNRAMP1. Our results further indicate that, in addition to proteins containing the FYVE and PHOX domains, proteins containing the PH domain can decode the PI3P signal in endosomal function.
Keywords: metal transport, NRAMP, vacuole, late endosome, phosphatidylinositol 3-phosphate
Abstract
“Too much of a good thing” perfectly describes the dilemma that living organisms face with metals. The tight control of metal homeostasis in cells depends on the trafficking of metal transporters between membranes of different compartments. However, the mechanisms regulating the location of transport proteins are still largely unknown. Developing Arabidopsis thaliana seedlings require the natural resistance-associated macrophage proteins (NRAMP3 and NRAMP4) transporters to remobilize iron from seed vacuolar stores and thereby acquire photosynthetic competence. Here, we report that mutations in the pleckstrin homology (PH) domain-containing protein AtPH1 rescue the iron-deficient phenotype of nramp3nramp4. Our results indicate that AtPH1 binds phosphatidylinositol 3-phosphate (PI3P) in vivo and acts in the late endosome compartment. We further show that loss of AtPH1 function leads to the mislocalization of the metal uptake transporter NRAMP1 to the vacuole, providing a rationale for the reversion of nramp3nramp4 phenotypes. This work identifies a PH domain protein as a regulator of plant metal transporter localization, providing evidence that PH domain proteins may be effectors of PI3P for protein sorting.
The ability of iron (Fe) to switch between different oxidation states in cells is used in all biological kingdoms to catalyze essential biochemical reactions. Fe is found in the form of the Fe-S cluster, Heme, or Oxo di-iron as a cofactor in the catalytic center of a plethora of proteins involved in oxygen binding, electron transport chains, or DNA metabolism (1). As a consequence, Fe deficiency leads to severe symptoms in living organisms including anemia in humans or chlorosis and growth inhibition in plants. On the other hand, the excess of Fe is deleterious for cells because it may lead to the production of highly reactive oxygen species through the Fenton reaction. Therefore all organisms need to regulate Fe uptake and storage tightly according to their requirements. Fe homeostasis has been studied extensively in plants because of its strong impact on photosynthesis and therefore on biomass production and also because Fe biofortification of food crops is an important challenge to counteract Fe deficiency affecting billions of human beings (2, 3).
In dicotyledonous plants, the ferric iron (Fe3+) available as soluble chelates in soils is reduced to ferrous iron (Fe2+) at the root surface by a plasma membrane-bound ferric-chelate reductase known as “AtFRO2” in Arabidopsis thaliana (4). Fe2+ then is transported into root cells by the high-affinity iron-regulated transporter 1 (IRT1) from the ZIP/IRT family that is expressed in root epidermal cells in response to Fe starvation (5, 6). The natural resistance-associated macrophage protein1 from A. thaliana, AtNRAMP1, a homolog of the human DMT1 transporter, is able to transport Fe and manganese (Mn) when expressed in yeast (7, 8). In Arabidopsis, AtNRAMP1 was shown to mediate high-affinity Mn uptake (9) as well low-affinity Fe uptake (10).
In photosynthetic cells, most of the cellular Fe is found in chloroplasts, within components of the photosynthetic electron transport chain, or bound to ferritin (2, 11). The vacuole is also an important site for Fe compartmentalization. In the A. thaliana embryo, Fe is stored in the vacuole of endodermal cells under the action of the VIT1 transporter of the CCC1 family (12–14). The Fe pool stored in the vacuole of endodermal cells is remobilized during germination by two homologous vacuolar metal transporters, AtNRAMP3 and AtNRAMP4. The nramp3nramp4 double mutant is unable to develop into photosynthetic seedlings when germinated under Fe-limiting conditions because seed Fe is trapped in the vacuole of endodermal cells (13, 15). The nramp3nramp4 mutant is also hypersensitive to Cd and Zn for root growth, likely as a consequence of a globally unbalanced metal homeostasis (16–18).
Genetic analyses in yeast revealed that the regulation of metal homeostasis depends mainly on metal transporters and on the vacuolar protein-sorting pathway regulating the cellular trafficking of these transporters (19–21). The main Mn transporter in yeast, Smf1p, a homolog of AtNRAMP1, localizes on both the plasma membrane and endosomes under Mn-sufficient conditions. The endosomal fraction of Smf1p is targeted to the vacuole for degradation by the Trp/Bsd2p/Rsp5 ubiquitin-dependent pathway (22–26). However, under Mn-deficient conditions, the fraction of Smf1p present at the plasma membrane is increased, likely to stimulate Mn uptake. Recent studies revealed that the homeostasis of metals and minerals is regulated by the trafficking of transporters in plants as well. The Fe transporter IRT1 cycles between the trans-Golgi network (TGN)/early endosome (EE) compartment and the plasma membrane of root epidermal cells, but a fraction is targeted to the vacuole for degradation through the late endosome (LE)/multivesicular bodies (MVB) compartment (27). This pathway depends on the ubiquitination of IRT1 and its direct binding to FYVE1/FREE1 acting as noncanonical ESCRT-0 in the endosomal-sorting complex required for transport (ESCRT) machinery in plants (28, 29). Similarly, the trafficking of the boron transporter BOR1 and the phosphate transporter PHT1 between the plasma membrane and the vacuole is regulated by ubiquitin-dependent pathways, although the proteins involved in the trafficking of these transporters are not yet known (30–32).
To identify previously unknown elements regulating Fe homeostasis in the model plant A. thaliana, we looked for revertants of the nramp3nramp4 double mutant. Here we report that loss-of-function mutations in the pleckstrin homology (PH) domain containing protein AtPH1 alleviate the phenotypes of nramp3nramp4. We further show that the localization of AtPH1 in the LE/MVB compartment depends on binding to phosphatidylinositol 3-phosphate (PI3P) and that AtPH1 is involved in the regulation of AtNRAMP1 localization. Our results therefore reveal the involvement of AtPH1 in metal homeostasis in plants as well as the unsuspected role of PH domain-containing proteins as effectors of PI3P in protein sorting.
Results
The suppressor1 Mutation Mitigates nramp3nramp4 Growth Defect Under Fe-Deficient Conditions.
To identify genes involved in the regulation of metal homeostasis in plants, we screened for revertants of the Arabidopsis nramp3nramp4 mutant, which is hypersensitive to Fe starvation and to the presence of cadmium (Cd) at the early developmental stage (13, 15, 16). The nns1 (nramp3nramp4suppressor1) mutant was selected from a nramp3-1nramp4-1 ethyl methanesulfonate (EMS)-mutagenized M2 population (Ws accession) on the basis of a significant reduction of Cd sensitivity for root growth, used as a proxy for Fe deficiency (Fig. 1 A and B). We then directly tested whether the suppressor1 mutation also alleviates the hypersensitivity to Fe starvation of nramp3nramp4 seedlings (Fig. 1 C and D). After 7 d of growth on an Fe-limited medium, the nns1 root length (17.1 ± 5.3 mm) was intermediate between that of wild-type (31.8 ± 4.6 mm) and nramp3nramp4 (4.2 ± 2.6 mm) plants. This result indicated that the suppressor1 mutation partially reverts the Fe-deficient phenotype of nramp3nramp4.
Fig. 1.
nns1 partially reverts nramp3nramp4 phenotypes. (A) Wild-type (Ws), nramp3-1nramp4-1, and nns1 plants were grown vertically for 9 d on ABIS agar medium containing 30 µM CdCl2 (+Cd). (B) Root length of plants growing on ABIS medium (black bars) or supplemented with 30 µM CdCl2 (white bars). Data are shown as mean ± SD; n = 27–34 roots. (C) Wild-type (Ws), nramp3-1nramp4-1, and nns1 plants were grown vertically for 7 d on ABIS agar medium without Fe (−Fe). (D) Root length of plants growing on ABIS medium without Fe (white bars) or supplemented with 50 µM Fe-HBED (black bars). Data are shown as mean ± SD; n = 21–31. (Scale bars in A and C: 10 mm.)
The suppressor1 Mutation Affects the PH Domain-Containing Protein AtPH1.
To analyze the heredity of the suppressor 1 mutation, the nns1 mutant was backcrossed with the parental nramp3-1nramp4-1 mutant, and the F2 progeny was tested for the reversion of the nramp3nramp4 phenotype. The progeny showed a Mendelian 3:1 segregation ratio for the reversion of the nramp3nramp4 phenotype, suggesting that suppressor1 was a single-locus recessive mutation. To identify the suppressor1 mutation, we then generated an F2 recombinant population by outcrossing the nns1 mutant with the nramp3-2nramp4-2 mutant (Col accession). Using F2 plants showing the nns1-like phenotype, we mapped the suppressor1 mutation on chromosome 2 between the simple sequence-length polymorphism (SSLP) markers MSAT2-4 and MSAT2-11 using the polymorphism between Ws and Col accessions. We selected 19 F2 plants that had recombined between these two markers and sequenced the DNA of this subpopulation as a pool using next-generation sequencing technology. The analysis of the assembly of the sequencing reads to the A. thaliana Col genome further restricted the localization of the suppressor1 mutation to a region enriched in Ws-linked SNPs encompassing 580 kb on chromosome 2 (nucleotides 12,272,151–12,852,172). To identify the suppressor1 mutation, we searched for SNPs between the nns1-like population and the nramp3-1nramp4-1 mutant in this region containing 209 annotated genes. We identified a G-to-A transition covered by 18 reads in the ORF of At2g29700.1 (Fig. 2A). This transition results in the nonconservative substitution of Arg46 to His (R46H) in the predicted protein (Fig. 2 B and C). The At2g29700 gene encodes a PH domain-containing protein, originally called “AtPH1,” the biological function of which was still unknown (33, 34). This protein was later named “PH2” (35), but we will use the original name, AtPH1, in agreement with sequence databases.
Fig. 2.
The suppressor1 mutation affects AtPH1. (A) Alignment of reads from the nns1 mutant to the Col-0 reference genome (TAIR10) at the AtPH1/At2g27900 locus. Paired reads are in blue; broken pairs are in red and green. The translation of the At2g27900.1 gene model is given under the nucleotide sequence. The asterisk denotes the mutation identified in nns1 converting Arg46 to His. (B) Amino acid sequence of AtPH1/At2g27900.1. The PH domain is highlighted in gray, and the PPBM is highlighted in red. (C) Alignment of the AtPH1 PH domain N terminus showing conservation to other PH domain from Oryza sativa (XP_015638090.1), Physcomitrella patens (XP_001766896.1), Dictyostelium discoideum (XP_643886.1), A. thaliana DRP2A (NP_172500.1), Homo sapiens PKB/AKT1 (NP_001014431.1) and DAPP1 (NP_055210.1), and Saccharomyces cerevisiae ScOSH2 (NP_010265.1). The background color reflects sequence conservation. (D) Wild-type (Ws), nramp3-1nramp4-1, nns1, and homozygous T3 nns1 lines constitutively expressing AtPH1-GFP were grown vertically for 10 d on ABIS medium without iron (−Fe). (E) Wild-type (Col), nramp3-2nramp4-2, and nramp3-2nramp4-2atph1-2 plants were grown vertically for 8 d on ABIS medium without iron (−Fe) or for 10 d on medium containing 30 µM CdCl2 (+Cd). (Scale bars: 10 mm.)
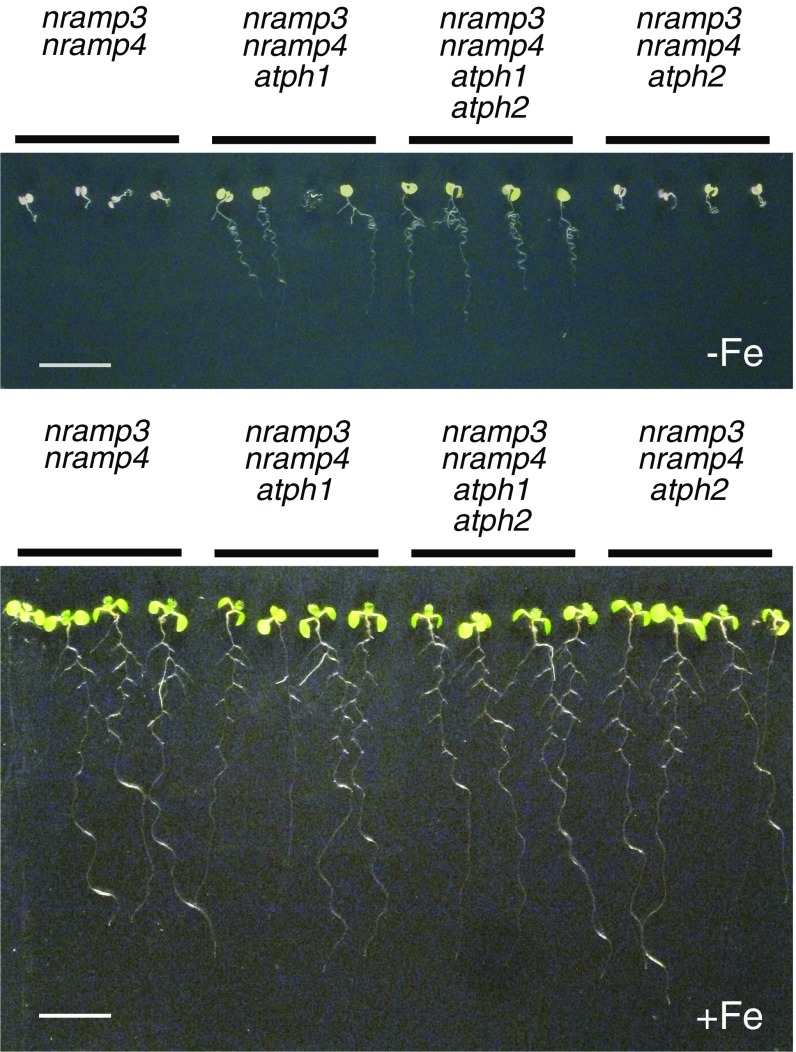
To demonstrate that the point mutation identified in AtPH1, hereafter called “atph1-1,” is responsible for the reversion of nramp3nramp4 phenotypes, we expressed AtPH1 fused to GFP at the C terminus (AtPH1-GFP) under the control of the 2x35S promoter in nns1. Transgenic nns1 plants expressing AtPH1-GFP grew normally on a medium supplemented with Fe (Fig. S1) but showed a reduction in root growth similar to that of nramp3-1nramp4-1 when grown on an Fe-limited medium (Fig. 2D), indicating that the expression of AtPH1-GFP complements the suppressor1 mutation. In addition, we isolated a T-DNA (transfer DNA) insertion mutant of AtPH1 in the Col accession, which was called “atph1-2” (Fig. S2). Neither atph1-1 (Ws) nor atph1-2 (Col) single mutants displayed a visible alteration of development or a change in sensitivity to Fe deficiency (Fig. S3). Like atph1-1, the atph1-2 mutation partially reverted the phenotype of nramp3-2nramp4-2 double mutant (Col) in the absence of Fe or in the presence of Cd (Fig. 2E), thus recapitulating the phenotype of nns1. All these data demonstrate that the loss of function of AtPH1 is responsible for the partial reversion of the nramp3nramp4 phenotypes. The fact that atph1 only partially reverts nramp3nramp4 phenotypes may be caused by the expression of an AtPH1 homolog playing a redundant function. To address this possibility, we isolated a knockout mutation in the At5g05710 gene, hereafter called “AtPH2,” which is the closest homolog to AtPH1 (Fig. S2). However, the analysis of the effect of the atph2-1 mutation in the nramp3nramp4 and nramp3nramp4atph1 mutant backgrounds indicated that AtPH2 does not play a redundant function with AtPH1 for the reversion of the nramp3nramp4 Fe-deficiency phenotype in roots (Fig. S4).
Fig. S1.
The expression of AtPH1 under the control of the 2x35S promoter does not affect root growth under Fe-sufficient conditions. Wild-type (Ws), nramp3-1nramp4-1, nns1, and homozygous T3 nns1 lines constitutively expressing AtPH1-GFP under the control of the 2x35S promoter were grown vertically for 10 d on ABIS-agar medium containing 50 µM Fe-HBED (+Fe). This experiment was performed side-by-side with the experiment presented in Fig. 2D. (Scale bar: 10 mm.)
Fig. S2.
Molecular characterization of the atph1 and atph2 mutants. (A and B) Structure of the AtPH1/At2g29700.1 (A) andAtPH2/At5g05710.1 (B) ORFs showing the localization of the mutations isolated in this study. The localization of the primers used for RT-PCR experiments is shown also. (C) The presence of AtPH1 and AtPH2 transcripts in different genetic backgrounds was analyzed by RT-PCR using AtPH1-for3/AtPH1-rev1 and AtPH2-for1/AtPH2-rev1 primers, respectively. The PCR products were analyzed on a 1.5% agarose gel with the GeneRuler 1 Kb Plus DNA ladder (Thermo Fisher Scientific).
Fig. S3.
atph1 mutants do not show developmental and Fe-deficiency phenotypes. (A and B) atph1-1 (A) and atph1-2 (B) mutants were sown on ABIS-agar medium without Fe (−Fe) together with nramp3-1nramp4-1, nramp3-2nramp4-2, Ws, and Col respectively. Plants were grown vertically for 11 d at 21 °C with 16 h of light. (Scale bars: 10 mm.)
Fig. S4.
The atph2 mutation does not revert the nramp3nramp4 Fe-deficiency root-length phenotype. The atph2-1 mutant (Col) was crossed with nramp3-2nramp4-2 and nramp3-2nramp4-2atph1-2 mutants (Col). Seeds of nramp3-2nramp4-2, nramp3-2nramp4-2atph1-2, nramp3-2nramp4-2atph1-2atph2-1, and nramp3-2nramp4-2atph2-1 mutants were sown on ABIS-agar medium without Fe (−Fe) or supplemented with 100 µM Fe-HBED (+Fe). Plants were grown vertically for 11 and 8 d, respectively, at 21 °C with 16 h of light. (Scale bars: 10 mm.)
AtPH1 Localization Depends on PI3P Binding in Vivo.
The PH domain of AtPH1 contains a putative phosphatidylinositol-3,4,5-trisphosphate-binding motif (PPBM). However, AtPH1 was shown to bind PI3P specifically in vitro (33). The R46H substitution identified in nns1 affects an invariant residue of the PPBM motif (Fig. 2C and Fig. S5), which is expected to interact directly with the 3-phosphate group of the inositol ring (36–39). Mutations of the corresponding amino acid in the human proteins PKB/AKT and BTK were shown to affect binding to phosphatidylinositol-3,4,5-trisphosphate and cause X-linked agammaglobulinemia disease, respectively (39, 40). Accordingly, the R46H mutation abrogated AtPH1 binding to PI3P in vitro (Fig. S5). In root cells, AtPH1-GFP was predominantly detected in small, punctate structures in the cytoplasm and on the vacuolar membrane (Fig. 3A). In contrast, AtPH1 carrying the R46H mutation (AtPH1R46H-GFP) was uniformly distributed in the cytosol, suggesting that the cellular localization of AtPH1 depends on binding to PI3P (Fig. 3A). Accordingly, AtPH1-GFP colocalized with the PI3P marker 2xFYVE-mCherry (Fig. 3B), and treatment of roots with the PI3K inhibitor wortmannin led to uniform redistribution of AtPH1-GFP in the cytosol (Fig. 3C). Together, these data indicate that AtPH1 cellular localization depends on binding to PI3P. The phenotype of the nns1 mutant further suggests that binding to PI3P is essential for AtPH1 function.
Fig. S5.
The R46H mutation affects the binding of AtPH1 to PI3P in vitro. (A) The structure of the AtPH1 PH domain was modeled using the structure of the PH domain of DAPP1 with inositol 1,3,4,5-tetrakisphosphate (IP4) as template (99.9% confidence). The position of the Arg46 lateral chain, corresponding to DAPP1 Arg184, is displayed (ProQ2 score = 0.00). (B) A close-up view of the area outlined in white in A. AtPH1 Arg46 is predicted to establish a hydrogen bond with the three-phosphate group of the IP4 inositol ring. (C) GST-AtPH1 and GST-AtPH1R46H proteins were purified from E. coli extracts. (D) GST-AtPH1 and GST-AtPH1R46H lipid overlay assay using a membrane spotted with serial dilutions of phospholipids (from 100–1.56 pmol per spot). A short (Upper) and a long (Lower) exposure are presented.
Fig. 3.
AtPH1 localization depends on binding to PI3P. (A) Confocal images of the root elongation zone of nns1 plants expressing AtPH1-GFP or AtPH1R46H-GFP under the control of the 2x35S promoter. The boxed area in the left panel is enlarged at right. Cell walls stained by PI are in magenta. The white arrow indicates the vacuolar membrane detaching from the cell periphery. (B) nns1 plants expressing AtPH1-GFP were crossed with plants expressing the PI3P marker 2xFYVE-mCherry. Colocalization of both fluorescent markers was analyzed by confocal microscopy on roots of F1 plants. On the merged picture the overlap of GFP (green) and mCherry (magenta) channels appears in white. Cell contours are represented by dashed lines. Pearson (rp) and Spearman (rs) correlation coefficients as well as M1 (GFP) and M2 (mCherry) Manders overlap coefficients above threshold were calculated. (C) The roots of nns1 plants expressing AtPH1-GFP were treated by 30 µM wortmannin or 0.1% DMSO for 3 h. (Scale bars: 10 µm.)
AtPH1 Localizes in the LE/MVB Compartment.
PI3P accumulates in the membranes delimiting the LE/MVB compartment and the vacuole of plant cells (28, 41). To characterize the localization of AtPH1 further, we outcrossed the nns1 line complemented by AtPH1-GFP with transgenic lines expressing cellular markers fused to monomeric RFP (mRFP) and analyzed F1 plants by confocal imaging. AtPH1 colocalized with the LE/MVB markers ARA7 and ARA6 but only very weakly with the TGN/EE marker SYP43 or the trans-Golgi marker ST (Fig. 4 and Fig. S6A). These data indicate that AtPH1 mainly localizes in the LE/MVB in plant cells. Using electron transmission microscopy, we observed a minor reduction of the diameter of LE/MVB vesicles but no difference in the number of intraluminal vesicles (ILV) in the nns1 mutant compared with the wild type (Fig. S7). These results indicate that AtPH1 does not play a major role in the biogenesis of LE/MVB and ILVs.
Fig. 4.
AtPH1 colocalizes with a marker of the LE/MVB. nns1 plants expressing AtPH1-GFP were crossed with plants expressing the trans-Golgi marker ST-mRFP, the TGN/EE marker mRFP-SYP43, or the LE/MVB marker mRFP-ARA7. Colocalization of fluorescent markers was analyzed by confocal microscopy on root epidermal cells of F1 plants. On merged pictures the overlap of GFP (green) and mRFP (magenta) channels appears white. Cell contours are represented by dashed lines. The regions within the white outline are enlarged in the lower panels (magnification: 3×). Pearson (rp) and Spearman (rs) correlation coefficients as well as M1 (GFP) and M2 (mRFP) Manders overlap coefficients above threshold were calculated. (Scale bars: 10 µm.)
Fig. S6.
Colocalization of AtPH1 with ARA6. Confocal images of the root elongation zone of transgenic F1 plants expressing AtPH1-GFP (A) or NRAMP1-GFP (B) and the LE/MVB marker mRFP-ARA6. Areas outlined in white on the left are enlarged on the right (magnification: 3×). On these merged images, the overlap of the GFP (green) and mRFP (magenta) channels appears in white. Dashed lines represent cell contours. Pearson (rp) and Spearman (rs) correlation coefficients as well as M1 (GFP) and M2 (mRFP) Manders overlap coefficients above threshold were calculated. (Scale bars: 10 µm.)
Fig. S7.
The nns1 mutant is not visibly affected in the biogenesis of ILVs. (A and B) Transmission electronic microscopy images of root cells from wild-type (Ws) (A) and the nns1 mutant (B). GA, Golgi apparatus; MVB, multivesicular bodies; V, vacuole. (Scale bars: 200 nm.) (C) The diameter of MVBs and the number of ILVs per MVB were quantified in Ws and nns1 strains. Results are shown as mean ± SD (Ws: 82 MVBs, 43 sections, two roots; nns1: 96 MVBs, 37 sections, two roots). Results were analyzed using a nonparametric Mann–Whitney test.
AtNRAMP1 Is Involved in the Reversion of nramp3nramp4 by atph1.
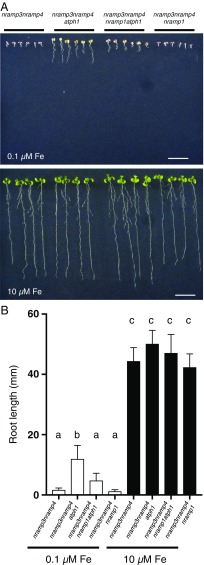
Because AtPH1 localizes in the LE/MVB, we hypothesized that atph1 might affect the trafficking of a metal influx transporter, leading to its localization on the vacuolar membrane where it could compensate for the loss of AtNRAMP3 and AtNRAMP4 activity. The metal transporter AtNRAMP1 acting at the plasma membrane for the uptake of Mn and Fe from the external medium to the cytoplasm (9, 10) was considered as a candidate for the reversion of nramp3nramp4 by atph1. We generated the nramp3nramp4atph1nramp1 quadruple mutant to test the effect of AtNRAMP1 loss of function on the nramp3nramp4atph1 phenotype. When grown on an Fe-limited medium, the root length of nramp3nramp4atph1nramp1 was intermediate between that of nramp3nramp4 and nramp3nramp4atph1 (Fig. 5). Under Fe-replete conditions, all genotypes displayed similar root growth. Therefore, our genetic analysis indicates that AtNRAMP1 is required for the reversion of nramp3nramp4 by atph1. However, because nramp1 does not fully suppress the effect of atph1, we cannot exclude the possibility that other mechanisms may be involved.
Fig. 5.
NRAMP1 loss of function partially reverts the effect of atph1. (A) The multiple mutants nramp3-2nramp4-2, nramp3-2nramp4-2atph1-2, nramp3-2nramp4-2nramp1-1atph1-2, and nramp3-2nramp4-2nramp1-1 (Col) were grown vertically for 10 d on Hoagland-agar medium containing a limited amount of Fe (0.1 µM Fe-HBED) or sufficient Fe (10 µM Fe-HBED). Representative plants from four independent culture plates were realigned for the pictures. (Scale bars: 10 mm.) (B) Quantification of root lengths shown in A. Results are shown as mean value ± SD; n = 21–27, four independent plates; letters indicate significant differences according to a Kruskal–Wallis test corrected by a Dunn’s multiple comparison test; P < 0.01.
atph1 Affects the Localization of AtNRAMP1.
To study the localization of AtNRAMP1 in planta, we first expressed NRAMP1-GFP under the control of the endogenous NRAMP1 promoter in the nramp1-1 mutant. However, the fluorescent GFP signal detected in roots was too weak to analyze NRAMP1-GFP localization in these lines by confocal microscopy. We therefore expressed NRAMP1-GFP under the control of the UBIQUITIN10 promoter in the nramp1-1 mutant. In four independent stable transgenic lines tested, the expression of AtNRAMP1-GFP complemented the Mn-deficient phenotype of nramp1-1, indicating that NRAMP1-GFP is functional (Fig. S8). We observed that NRAMP1-GFP localized mainly in cytoplasmic granular structures in epidermal cells and also frequently on the plasma membrane, notably in young epidermal cells (Fig. 6A). Colocalization analyses indicated that NRAMP1-GFP is distributed across the trans-Golgi, TGN/EE, and LE/MVB compartments (Fig. 6B and Fig. S6B).
Fig. S8.
The expression of NRAMP1-GFP complements the Mn deficiency phenotype of nramp1. The nramp1-1 mutant (Col) was transformed with pMUBI83-NRAMP1. Among four independent lines showing complementation of nramp1-1 phenotype, the homozygous single-locus line #K is shown (two homozygous T3 progenies). Plants were grown vertically for 9 d on Hoagland-agar medium made with EDTA-washed agar containing 20 µM Fe-HBED and supplemented (+Mn) or not (−Mn) with 5 µM MnSO4. (Scale bars: 10 mm.)
Fig. 6.
NRAMP1 is present in the endosomal pathway and on the plasma membrane. Root epidermal cells of nramp1 plants complemented by NRAMP1-GFP were imaged by confocal microscopy. (A) The vacuolar plane of elongating cells (Upper) and cortical plane of dividing cells (Lower) showing that NRAMP1-GFP was present mostly in cytoplasmic vesicles but also was detected on the plasma membrane (white arrow), more clearly in younger cells. Transmitted-light (differential interference contrast, DIC) images are shown also. (B, Upper) NRAMP1-GFP plants were crossed with plants expressing the trans-Golgi marker ST-mRFP, the TGN/EE marker mRFP-SYP43, and the LE/MVB marker mRFP-ARA7. Colocalization of fluorescent markers was analyzed in root epidermal cells of F1 plants. On the merged pictures the overlap of GFP (green) and mRFP (magenta) channels appears white. Cell contours are represented by dashed lines. (Lower) Enlarged view of the region marked by the white outline in the upper row (magnification: 3×). Pearson (rp) and Spearman (rs) correlation coefficients as well as M1 (GFP) and M2 (mRFP) Manders overlap coefficients above threshold were calculated. (Scale bars: 10 µm.)
The localization of NRAMP1-GFP was not visibly affected in the nramp3-1nramp4-1 mutant (Fig. 7A). However, in the nns1 (nramp3-1nramp4-1atph1-1) mutant, we observed that a significant fraction of NRAMP1-GFP colocalized with the vacuolar membrane marker TagRFP-SYP22 (Fig. 7 B and C). When nramp3-1nramp4-1 and nns1 plants expressing NRAMP1-GFP were placed in the dark, a condition reducing vacuolar proteolysis of GFP (42, 43), we observed the appearance of a GFP signal in the lumen of vacuoles, indicating that a fraction of NRAMP1-GFP is targeted in the vacuole for degradation in both genetic backgrounds (Fig. 7D). Treatment of seedlings with Concanamycin A, an inhibitor of vacuolar ATPase (V-ATPase), which also led to a reduction of vacuolar protease activity (27, 43), confirmed these observations (Fig. S9). Together, these results suggest that NRAMP1 traffics through the endosomal system en route to its degradation in the vacuolar compartment and that the atph1 mutation leads to the mislocalization of a fraction of NRAMP1 on the vacuolar membrane.
Fig. 7.
NRAMP1 localizes on the vacuole in the nns1 mutant. (A and B) NRAMP1-GFP and TagRFP-SYP22 were expressed in nramp3-1nramp4-1 (A) and nramp3-1nramp4-1atph1-1 (nns1) (B) mutants. Root epidermal cells in the elongation zone were imaged on a spinning disk confocal microscope for GPF and tagRFP. (Scale bars: 10 µm.) (C) NRAMP1 localization on the vacuole was quantified in single-plane images corresponding to individual cells of nramp3nramp4 (n = 48) and nramp3nramp4atph1 (n = 42) as a normalized fraction of NRAMP1-GFP signal colocalizing with the TagRFP-SYP22 signal. Note that because of the close proximity of cytoplasmic NRAMP1 structures with the vacuole, the value of the vacuolar membrane localization index is never null. In the graph, boxes include the two central quartiles, separated by the median. The whiskers extend to the 5th and 95th percentiles, and outliers are represented by a dot. P < 0.0001; Mann–Whitney test. (D) Root epidermal cells of nramp3nramp4 and nns1 mutants expressing NRAMP1-GFP were imaged across vacuolar planes using spinning disk confocal imaging. GPF fluorescence and transmitted-light (DIC) images are displayed. The asterisks denote the position of vacuoles. Plants were placed in darkness for 14 h (Dark) before imaging to reduce GFP degradation in the vacuoles. The contrast of the images was adjusted for the visualization of GFP in vacuoles. (Scale bar: 10 µm.)
Fig. S9.
Effect of Concanamycin A on NRAMP1-GFP localization. Root epidermal cells of nramp3nramp4 and nns1 mutants expressing NRAMP1-GFP were imaged across vacuolar planes by spinning disk confocal microscopy. GPF fluorescence and transmitted light images (DIC) are displayed. Plants were treated with 2 µM Concanamycin A (ConcA) for 3 h before imaging or with DMSO (0.1%) as control. The arrows indicate the position of vacuolar structures in which GFP signal accumulates upon ConcA treatment. (Scale bar: 10 µm.)
Discussion
The aim of this work was to identify elements involved in the regulation of metal homeostasis in plants using a forward genetic screen to isolate revertants of the nramp3nramp4 mutant. This screen identified a loss-of-function mutation in the gene coding for the PH domain-containing protein AtPH1 (Fig. 2), whose cellular localization and biological function were unknown. We further showed that the atph1 mutation reverts the defects of the nramp3nramp4 mutant by an original mechanism of suppression: atph1 leads to mislocalization of at least one metal transporter, AtNRAMP1, to the vacuolar membrane, where it substitutes for AtNRAMP3 and AtNRAMP4 function. More generally, our results uncover the involvement of AtPH1 in the regulation of plant metal homeostasis and highlight the role of a PH domain protein specifically binding PI3P in intracellular protein sorting.
AtPH1 Loss of Function Partially Reverts the nramp3nramp4 Fe-Deficiency Phenotype.
Mutations affecting AtPH1 partially revert the Fe-deficient phenotype of the nramp3nramp4 mutant (Fig. 1). Genetic evidence further suggests that the metal transporter NRAMP1 is required for the reversion of nramp3nramp4 phenotypes by atph1 (Fig. 5). NRAMP1 was previously shown to localize on the plasma membrane of A. thaliana root hair cells (9). Although we also observed NRAMP1 at the plasma membrane of root epidermal cells, the main pool of NRAMP1 is localized in the endosomal system (Fig. 6), suggesting that NRAMP1 localization depends on culture conditions and cell types. Indeed, we observed that NRAMP1 is mistargeted to the vacuolar membrane in the presence of the atph1 mutation (Fig. 7). Therefore, we propose that the fraction of NRAMP1 present on the vacuolar membrane in the nns1 mutant can compensate for the lack of NRAMP3 and NRAMP4 activities in the remobilization of Fe from vacuolar stores (Fig. 8). Because atph1 mutations partially rescue nramp3nramp4 phenotypes (Figs. 1 and 2E), it could be argued that the effect of the atph1 mutation is partially masked by genes playing redundant functions. The A. thaliana genome encodes 50 PH domain-containing proteins (35). Our genetic analysis, however, indicates that AtPH1 and its closest homolog, AtPH2/At5g05710.1, do not have redundant functions in the reversion of the nramp3nramp4 Fe-deficiency phenotype in roots (Fig. S4). This result does not exclude the possibility that these two proteins might have redundant functions in other pathways or different tissues, nor can we completely exclude the possibility that more distantly related PH domain proteins might play redundant roles with AtPH1. The partial reversion of nramp3nramp4 could be the consequence of insufficient expression or activity of NRAMP1 and possibly other Fe transporters that are targeted on the vacuolar membrane in root cells in atph1. Although it was previously shown that NRAMP1 is expressed in vascular tissues, as are NRAMP3 and NRAMP4 (9), this transporter has a lower affinity for Fe than NRAMP3 and NRAMP4 (8, 10), thus providing a rationale for the partial reversion of nramp3nramp4. Interestingly, the identification of AtNRAMP1, which is the high-affinity Mn transporter in Arabidopsis, as a target of AtPH1 opens the possibility that this PI3P-binding protein might have a role in the regulation of Mn homeostasis.
Fig. 8.
Possible hypotheses to explain the role of AtPH1 in the reversion of nramp3nramp4 through the regulation of NRAMP1 trafficking. (A) In wild-type plants, NRAMP1 could cycle between the plasma membrane and TGN/EE. A fraction of NRAMP1 is directed to the vacuole, where it is degraded. AtPH1 that localizes in LE/MVB and the vacuolar membrane could be involved in the targeting of NRAMP1 in ILVs (1), in the recycling of NRAMP1 from the LE/MVB compartment to the plasma membrane (2), or in the degradation of NRAMP1 on the vacuolar membrane (3). (B) The absence of functional AtPH1 in the nns1 mutant (nramp3nramp4atph1) leads to the accumulation of NRAMP1 on the membrane of the vacuole, thus complementing the activity of NRAMP3 and NRAMP4 in the remobilization of Fe from the vacuole.
Possible Modes of Action of AtPH1.
Our results indicate that the mutation of AtPH1 leads to an accumulation of NRAMP1 on the membrane of the vacuole, thus compensating for the lack of vacuolar Fe export activity in the nramp3nramp4 mutant background (Fig. 8). Several hypotheses may account for the mislocalization of NRAMP1. First, the accumulation of NRAMP1 on the vacuolar membrane could result from its stabilization on the outer membrane of LE/MVB, which subsequently fuses with the vacuole (Fig. 8). A similar mislocalization of the auxin transporter PIN2 on the vacuolar membrane was observed in the fyve1/free1-knockout mutant that does not produce ILVs (29). The PI3P-binding protein FYVE1/FREE1 plays an essential role as the ESCRT-0 component of the plant ESCRT machinery that regulates the trafficking and the degradation in the vacuole of several ubiquitinated proteins, including PIN2, IRT1, and the ABA receptor PYL4 (28, 29, 44). As a consequence, the fyve1/free1-knockout mutant is strongly affected in LE/MVB functions and in the biogenesis of vacuoles, resulting in growth arrest at a very early stage of seedling development (28, 29, 45). In contrast, the atph1-knockout mutant is not lethal, does not display a visible developmental phenotype, and is not affected in the biogenesis of ILVs (Figs. S1 and S6). These observations suggest that AtPH1 might play a more specific role than FYVE1/FREE1 in LE/MVB, possibly targeting a limited number of cargo proteins, including NRAMP1, to ILVs for their degradation in the vacuole. Further experiments will be required to confirm the role of AtPH1 in this pathway and consequently to determine if AtPH1 acts independently or in concert with FYVE1/FREE1. Alternatively, the accumulation of NRAMP1 on the outer membrane of LE/MVB could be the consequence of a defect in the recycling of NRAMP1 from LE/MVB back to the plasma membrane in the nns1 mutant (Fig. 8). Although still debated in the literature (46), the existence of this recycling pathway in plants is supported by the function of the plant-specific GTPase of the Rab5 family, ARA6/AtRABF1 (47–49), that colocalizes with AtPH1 (Fig. S6A). Finally, the mislocalization of NRAMP1 in the absence of functional AtPH1 may result from other processes, e.g., a defect of NRAMP1 degradation on the vacuole by invagination of the vacuolar membrane. This process, called “microphagy,” has been proposed to regulate the turnover of the vacuolar potassium channel TPK1 (50, 51). The identification of AtPH1-interacting proteins and the analysis of the trafficking of other cargo proteins in the atph1 mutant should help in discriminating between the different hypotheses regarding the mode of action of AtPH1.
AtPH1 as an Effector of PI3P in the Endosomal Pathway.
Independently of its mode of action, our results demonstrate that the function of AtPH1 depends on PI3P binding in vivo. Using a phospholipid overlay assay, it was previously shown that AtPH1 binds specifically PI3P in vitro (33). We confirmed these data (Fig. S5) and, more importantly, showed that the localization of AtPH1 in the LE/MVB compartment of root cells depends on its ability to bind PI3P in vivo (Figs. 3 and 4). The loss-of-function mutation identified in the nns1 mutant affects the highly conserved residue Arg46 in the PH domain of AtPH1, which is predicted to bind the 3-phosphate group of the inositol ring of phosphoinositides (36–39). Accordingly, the R46H mutation abrogates both AtPH1 binding to PI3P in vitro (Fig. S5) and its localization in the LE/MVB compartment (Fig. 3A). These results strongly suggest that the direct binding of AtPH1 to PI3P is necessary for AtPH1 function in the endosomal pathway. More generally, this work provides genetic evidence supporting a role for a PH domain protein directly binding PI3P in endosomal functions. Indeed, the involvement of PH domain proteins in endosomal functions has largely been overlooked compared with proteins containing FYVE or PHOX domains, because the binding specificity and affinity of PH domains for PI3P are usually lower than those of FYVE and PHOX domains (52–54). AtPH1 has no obvious orthologs outside the plant kingdom, but several PH domain-containing proteins specifically binding PI3P in vitro have been identified from yeast and human genomes (33, 55–57). Future genetic and functional studies might uncover a role for some of these proteins in the fine-tuning of endosomal trafficking.
Materials and Methods
A. thaliana Genotypes.
The A. thaliana mutants nramp3-1nramp4-1 (Ws accession), nramp3-2nramp4-2 (Col accession), and nramp1-1 (Col) were described previously (9, 13, 16). The atph1-2 allele (Col) isolated in this study corresponds to the GABI-Kat line GK-310H12 (58). The single atph1-1 mutant was isolated after outcrossing the nns1 mutant with the wild-type (Ws accession) strain. The multiple mutants nramp3-2nramp4-2atph1-2 (Col), nramp1-1nramp3-2nramp4-2 (Col), and nramp1-1nramp3-2nramp4-2atph1-2 (Col) were obtained by outcrossing available mutants. Homozygous mutants of the expected genotypes were selected by PCR using gene- and T-DNA–specific primers or a derived cleaved amplified polymorphic sequence (dCAPS) approach (SI Materials and Methods and Table S1).
Table S1.
Primers used in this study
| Target | Primer | Sequence | Mutant analysis |
| NRAMP3/At2g23150 | NRAMP3-for1 | GAAATACCGAGTCCAAGAAGC | To amplify NRAMP3 from Ws DNA. Use NRAMP3-for1 and LB102 for nramp3-1. |
| NRAMP3-rev1 | TCTCGTCTCTGGGACCTAACT | ||
| NRAMP3-for2 | TGTCCCAGAAAAACAAACAAAAC | To amplify NRAMP3 from Col DNA. Use NRAMP3-for2 and LBb1 for nramp3-2. | |
| NRAMP3-rev2 | TCCTGATTCCAACAAAAAGACC | ||
| NRAMP4/At5g67330 | NRAMP4-for1 | TTTGGTCAGACGAAACCCAGT | To amplify NRAMP4 from Ws DNA. Use NRAMP4-for1 and JL202 for nramp4-1. |
| NRAMP4-rev1 | TAAAACGACTTGGCAAACACC | ||
| NRAMP4-for2 | AGTCGTCACTTTAGTCGCAGC | To amplify NRAMP4 from Col DNA. Use NRAMP4-rev2 and LBa1 for nramp4-2. | |
| NRAMP4-rev2 | TAAAACGACTTGGCAAACACC | ||
| NRAMP1/ At1g80830 | NRAMP1-for1 | CCCATGATTAAACCACCAATG | To amplify NRAMP1 from Col DNA. Use NRAMP1-for1 and LBb1 for nramp1-1. |
| NRAMP1-rev1 | TCTCGCACTTCAAAAATACGG | ||
| AtPH1/At2g29700 | At2g29700-fwd | ACGCAGAGAGCCAGCATAAT | To amplify AtPH1 from Col DNA. Use At2g29700-rev2 and GABI-LB for atph1-2. |
| At2g29700-rev2 | TCCTCCTTCAGAAAACCCCTA | ||
| AtPH1-dCrev1 | CCTCGTTTGAGAACGAACCAT | Cut the At2g29700-fwd/AtPH1-dCrev1 PCR product with TaqI for atph1-1. | |
| AtPH1-for3 | GTCGTGGTGAGATTTTAG | To amplify AtPH1 transcript by RT-PCR. | |
| AtPH1-rev1 | CCACTCTTCTTTCTCCTTCTC | ||
| AtPH2/At5g05710 | At5g05710-fwd1 | TTTTGGGGCAAAAGATAAACA | To amplify AtPH2 from Col DNA. Use At5g05710-rev1 and LBa1 for atph2-1. |
| At5g05710-rev1 | GTCTTTGAACCAGAAGCGTTG | ||
| AtPH2-for1 | CTGTGGCGAGCAGTAATC | To amplify AtPH2 transcript by RT-PCR. | |
| AtPH2-rev1 | CCAATCTTCTTTCTCCTTCTCC | ||
| T-DNA | LB102 | GATGCACTCGAAATCAGCCAATTTTAGAC | |
| JL202 | CATTTTATAATAACGCTGCGGACATCTAC | ||
| LBa1 | TGGTTCACGTAGTGGGCCATCG | ||
| LBb1 | GCGTGGACCGCTTGCTGCAACT | ||
| GABI-LB | TATTGACCATCATACTCATTGC |
Arabidopsis nramp3nramp4 Suppressor Screening.
Seeds of the Arabidopsis thaliana nramp3-1nramp4-1 (Ws accession) were mutagenized by EMS as previously described (15). Approximately 36,000 seeds corresponding to the progeny of 1,800 mutagenized M1 seeds were sown on ABIS-Agar plates supplemented with 20 μM CdCl2 and were grown vertically for 14 d at 21 °C with 16 h light/d. Plantlets showing longer roots than nramp3-1nramp4-1 were selected as nramp3nramp4 suppressors (nns). The nns1 mutant was backcrossed twice with nramp3-1nramp4-1 to perform physiological studies.
Culture of Plants for Root-Growth Assays.
Plants were grown vertically at 21 °C with 16 h of light for 5–9 d on ABIS or Hoagland-Agar medium (16, 59). When indicated, the medium was supplemented with Fe-HBED [N,N′-di(2-hydroxybenzyl) ethylenediamine-N,N′diacetic acid monochloride hydrate] (Strem Chemicals) and CdCl2 (Sigma-Aldrich). For the Mn-depleted condition, MnCl2 was omitted from the Hoagland medium, and metal-free agar was prepared as previously described (60).
Identification of the nns1 Mutation.
The nns1 mutant was outcrossed with the nramp3-2nramp4-2 mutant (Col accession). The initial mapping of the suppressor1 mutation in the F2 population was performed using the MSAT collection of SSLP markers (www7.inra.fr/vast/msat.php) (61). The genomic DNA was purified from selected F2 plants showing an nns1-like phenotype and from the parental nramp3-1nramp4-1 mutant. The two pools of genomic DNA were sequenced using Illumina 100-bp paired-end reads technology (Fasteris SA). We obtained 70.4 × 106 reads for the nns1-like pool and 88.2 × 106 reads for nramp3-1nramp4-1. The reads were mapped to the A. thaliana Columbia-0 reference genome (TAIR10) using CLCbio Genomic Workbench 5.5.1 (QIAGEN). The genomic region linked to the nns1 phenotype was mapped by analysis of SNP distribution between Ws and Col accessions using the variant detection tool. To identify the suppressor1 EMS mutation, we restricted the SNP analysis to the genomic region linked to the nns1 phenotype, selected SNPs corresponding to G-to-A and C-to-T transitions, filtered out SNPs present in parental nramp3-1nramp4-1, and focused on SNPs affecting the coding sequence of annotated genes.
Cloning of AtPH1 and NRAMP1 Coding Sequences.
The AtPH1 (AT2G29700.1) and NRAMP1 (AT1G80830.1) coding sequences without a stop codon were amplified by PCR using high-fidelity Phusion polymerase (Thermo Scientific) from genomic DNA and cDNA respectively. AtPH1R46H was obtained using nns1 genomic DNA as template. Coding sequences were recombined using Gateway cloning (Invitrogen) into the pDONR207 vector and then in the expression vectors pMDC83 (62) and pMUBI83 (63) to generate pMDC83-AtPH1, pMDC83-AtPH1R46H, and pMUBI83-NRAMP1. All constructs were verified by Sanger sequencing (GATC Biotech).
Expression of AtPH1 and NRAMP1 in Transgenic Plants.
A. thaliana nns1 (nramp3-1nramp4-1atph1-1) plants were transformed with pMDC83-AtPH1 and pMDC83-AtPH1R46H by floral dipping using the agrobacterium AGL0 strain to express AtPH1-GFP and AtPH1R46H-GFP fusion proteins, respectively. The nramp1-1, nramp3-1nramp4-1, and nns1 mutants were transformed with pMUBI83-NRAMP1 to express NRAMP1-GFP fusion protein.
Transgenic lines were selected in vitro on half-strength Murashige and Skoog (MS) agar medium supplemented with 50 μg/mL Carbenicillin and 15 μg/mL Hygromycin B. Homozygous single-locus-insertion transgenic T3 lines were selected for further experiments.
Confocal Imaging and Colocalization Analyses.
For colocalization analyses, the homozygous Arabidopsis transgenic lines nns1/pMDC83-AtPH1 and nramp1-1/pMUBI83-NRAMP1 were outcrossed with lines expressing mRFP-SYP43, ARA6-mRFP, and mRFP-ARA7 under their endogenous promoters, ST-mRFP under the control of the 35S promoter (47, 64, 65), and 2xFYVEHRS-mCherry under the control of the UBI10 promoter (28, 41). F1 plants were used for colocalization analysis. Homozygous nramp3-1nramp4-1 and nns1 lines expressing NRAMP1-GFP (pMUBI83-NRAMP1) were transformed with pBGW/pSYP22:tagRFP-SYP22 (66), and primary transformants were selected on half-strength MS agar medium supplemented with 15 μg/mL Basta. T2 lines expressing both fluorescent markers were selected for further experiments.
Seedlings were grown vertically for 5–9 d on Hoagland Agar medium containing 1% sucrose and 50 µM Fe-HBED before imaging. When indicated, seedlings were treated for 3 h with 30 µM wortmannin (Sigma) or 0.1% DMSO as control and with 1% (wt/vol) propidium iodide (PI) for 5 min. Seedlings were mounted in culture medium, and confocal images of root epidermal cells in the division and elongation zone were obtained on either a Leica SP8 microscope to acquire GFP (λex= 490 nm, λem= 500–550 nm) and mCherry/mRFP/PI (λex= 590 nm, λem= 600–650 nm) fluorescence simultaneously or on a Nipkow spinning disk confocal system by high-speed (300 ms) sequential acquisition of the two channels (GFP and TagRFP) using a Hamamatsu ORCA-Flash 4.0_LT or a Photometrics EMCCD Evolve camera (67).
Image processing (cropping, contrast adjustment, and background subtraction) and analysis were performed with ImageJ (Wayne Rasband; National Institutes of Health). Colocalization coefficients were computed with the JACoP plugin [rp, Pearson correlation coefficient; M1 and M2, Manders overlap coefficients above threshold (68)] or the Coloc2 plugin (rs, Spearman correlation coefficient) of the FiJi distribution of ImageJ (69). To evaluate the differential colocalization of NRAMP1-GFP with tagRFP-SYP22, an automated procedure was used. First, the fraction of total GFP fluorescence above threshold (established by the Huang method of ImageJ) present on SYP22 regions (segmented by thresholding with the Isodata method) was computed. These values then were normalized to the extent (fraction of total cytoplasmic surface) of vacuolar membrane segmented in each image, to obtain a vacuolar membrane localization index. Such an index corresponds to a Manders overlap coefficient normalized to the relative extent of vacuolar membrane (SYP22 signal) in each image. The data shown in Fig. 7C represent the pool of two independent experiments, performed on different dates, which gave virtually identical results. In each experiment, 20–27 epidermal cells from the root elongation zones of four to seven seedlings were analyzed individually for each genotype. The quantification was conducted on single-plane images corresponding to the vacuolar plane.
Statistical Analyses.
Statistical analyses were performed using GraphPad Prism version 7.00 (GraphPad Software, www.graphpad.com).
Accession Numbers.
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/NCBI databases under the following accession numbers: AtPH1 (AT2G29700), AtPH2 (AT5G05710), NRAMP1 (AT1G80830), NRAMP3 (AT2G23150), and NRAMP4 (AT5G67330).
SI Materials and Methods
Mutant Genotyping.
The A. thaliana mutants were genotyped by PCR using gene- and T-DNA–specific primers according to Table S1. Genomic DNA was amplified by 40 cycles of PCR (94 °C for 30 s; 60 °C for 30–45 s; 72 °C for 30–75 s) using standard conditions. To detect atph1-1, the PCR fragment was digested with TaqI, and DNA fragments were analyzed on a 3% agarose gel. The homozygous AtPH2 (At5g05710) T-DNA insertion line isolated in this study, atph2-1 (Col), corresponds to the SALK line 069686.
To amplify the AtPH1 and AtPH2 transcripts, total RNA was extracted from 10-d-old plantlets grown in vitro using the Qiagen RNeasy kit with on-column DNase treatment. cDNA was synthesized using SuperScript IV (Invitrogen) with an anchored oligo(dT)20 primer according to the manufacturer’s recommendation. Transcripts were amplified by 30 cycles of PCR (94 °C for 20 s; 55 °C for 30 s; 72 °C for 30 s) using standard conditions with the primers described in Table S1.
Protein Structure Modeling.
The model of the AtPH1 structure was obtained using the Phyre2 server (70). The structure of HsDAPP1 with inositol 1,3,4,5–3 tetrakisphosphate (Protein Data Bank ID code 1FAO) was selected as template. The resulting model for the PH domain of AtPH1 (residues 24–128) was obtained with 99.9% confidence.
Lipid-Binding Assay.
To produce GST fusion proteins, AtPH1 and AtPH1R46H ORFs were recombined from pDON207-AtPH1stop and pDON207-AtPH1R46Hstop to pDEST15 to produce pDEST15-AtPH1 and pDEST15-AtPH1R46H, respectively. These constructs were transformed into Escherichia coli Rosetta2(DE3) pLysS, and a 200-mL culture was grown at 37 °C to OD600nm 0.3 in 2YT (16 g bacto tryptone, 10 g bacto yeast extract, and 5 g NaCl per liter) medium containing antibiotics. Expression of GST fusion proteins then was induced by 0.5 µM isopropyl β-d-1-thiogalactopyranoside (IPTG) for 4 h at 25 °C. Cells were collected by centrifugation, suspended in 4 mL ice-cold TBS buffer (pH 7.5) containing 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 0.1% 2-Mercaptoethanol, 1 mM PMSF, and protease inhibitor mixture (11873580001; Roche) and were lysed by sonication. The lysates were centrifuged at 3,500 × g for 20 min at 4 °C, and the cleared supernatant was incubated with 300 µL 50% slurry glutathione-Sepharose (Pharmacia) for 120 min at 4 °C on a rotary wheel. Beads were collected by centrifugation at 300 × g for 1 min and were washed four times with 5 mL TBS buffer (pH 7.5) containing 1 mM DTT. Fusion proteins were eluted from the resin with 250 µL 50 mM Tris⋅HCl (pH 8), 150 mM NaCl, 20 mM reduced glutathione, and 1 mM DTT for 60 min at 4 °C. The elution was repeated once. The amount of eluted recombinant protein was measured by Bradford assay, and the quality of the protein purification was checked by SDS/PAGE.
The protein–lipid overlay assay was performed with GST-AtPH1 and GST-AtPH1R46H using PIP Arrays (P-6100; Echelon Biosciences) essentially as previously described (71), with the following modifications. The PIP array membrane was blocked with TBS buffer (pH 7.5) containing 0.1% Tween 20 (TBST) and 1% (wt/vol) nonfat dry milk for 60 min at room temperature. The PIP array membrane then was incubated overnight at 4 °C under gentle stirring in the same solution containing 10 nM of recombinant protein. All subsequent steps were performed at room temperature with gentle shaking. The membrane was washed five times for 10 min in TBST at room temperature and then was incubated with goat anti-GST antibody (1/1,000 dilution) (27-4577-01; GE Healthcare) for 120 min in TBST containing 1% (wt/vol) nonfat dry milk. The membrane was washed as before and was incubated with HRP-peroxidase rabbit anti-goat IgG antibody (1/10,000 dilution) (401515; Calbiochem) as for the primary antibody. The membrane was washed as before and then was rinsed in TBS. HRP activity was revealed using the Amersham ECL chemiluminescence method according to the manufacturer’s recommendations and was measured using a ChemiDoc Touch Imaging System (Bio-Rad).
Transmission Electronic Microscopy and Analyses.
A. thaliana Ws and nns1 mutant root tips were transferred into 200-µm cupules (16706897; Leica) containing 1-hexadecen and were frozen with a high-pressure freezing device (EMPACT2; Leica). Freeze substitution was carried out in a freeze-substitution unit (AFS2; Leica) in acetone supplemented with 2% osmium tetroxide. Samples were treated as follows: −90 °C for 37 h, then warmed to −60 °C over 15 h, −60 °C for 14 h, warmed to −30 °C over 15 h, −30 °C for 10 h, warmed to 0 °C for 1 h. Samples then were rinsed three times in anhydrous acetone and gradually were embedded in epoxy resin at room temperature (Low Viscosity Premix Kit Medium; Agar Scientific). For polymerization, samples were placed in flat plastic molds and polymerized for 17 h at 60 °C. Ultrathin sections of root tips (80 nm) were obtained with an ultramicrotome EM UC6 (Leica Microsystems) and were collected on formvar carbon-coated copper grids (Agar Scientific) and stained with 2% uranyl acetate for 20 min and with lead citrate for 10 min. Observations were made with a JEOL JEM-1400 transmission electron microscope operating at 120 kV. Images were acquired using a postcolumn high-resolution (11 megapixels) high-speed camera (SC1000 Orius; Gatan) and were processed with Digital Micrograph (Gatan).
For image analysis, epidermis and cortex cells were considered. MVBs were defined as a vesicular cytoplasmic structure with a diameter <500 nm, delimited by a single membrane, with a clear lumen and containing at least one ILV. MVB diameters were computed from the measured surface, applying the formula for circles (area = πr2). The number of ILVs was estimated by implementing an automated procedure with ImageJ based on the detection of maxima of intensity after Gaussian blurring of manually selected MVBs.
Acknowledgments
We thank Takashi Ueda, Emi Ito, and Tomohiro Uemura for kindly providing mRFP marker lines and the pBGW/pSYP22:tagRFP-SYP22 construct and Gregory Vert for critical reading of the manuscript. This work was supported by Grants ANR-11-BSV6-0004 (to S.T.) and ANR-2011-ISV6-001-01 (to C.C.). A.A. was the recipient of a Postdoctoral Fellowship from the Spanish Ministry of Science and Innovation. This work has benefited from the core facilities of Imagerie-Gif (www.i2bc.paris-saclay.fr/spip.php?rubrique184), a member of Infrastructures en Biologie Santé et Agronomie (IBiSA) (www.ibisa.net), supported by France BioImaging Grant ANR-10INBS-04-01 and the Saclay Plant Science Labex Grant ANR-11-IDEX-0003-02.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1702975114/-/DCSupplemental.
References
- 1.Kaplan J, Ward DM. The essential nature of iron usage and regulation. Curr Biol. 2013;23:R642–R646. doi: 10.1016/j.cub.2013.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Briat JF, Dubos C, Gaymard F. Iron nutrition, biomass production, and plant product quality. Trends Plant Sci. 2015;20:33–40. doi: 10.1016/j.tplants.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 3.Murgia I, Arosio P, Tarantino D, Soave C. Biofortification for combating ‘hidden hunger’ for iron. Trends Plant Sci. 2012;17:47–55. doi: 10.1016/j.tplants.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Robinson NJ, Procter CM, Connolly EL, Guerinot ML. A ferric-chelate reductase for iron uptake from soils. Nature. 1999;397:694–697. doi: 10.1038/17800. [DOI] [PubMed] [Google Scholar]
- 5.Eide D, Broderius M, Fett J, Guerinot ML. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA. 1996;93:5624–5628. doi: 10.1073/pnas.93.11.5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vert G, et al. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell. 2002;14:1223–1233. doi: 10.1105/tpc.001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curie C, Alonso JM, Le Jean M, Ecker JR, Briat JF. Involvement of NRAMP1 from Arabidopsis thaliana in iron transport. Biochem J. 2000;347:749–755. [PMC free article] [PubMed] [Google Scholar]
- 8.Thomine S, Wang R, Ward JM, Crawford NM, Schroeder JI. Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proc Natl Acad Sci USA. 2000;97:4991–4996. doi: 10.1073/pnas.97.9.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cailliatte R, Schikora A, Briat JF, Mari S, Curie C. High-affinity manganese uptake by the metal transporter NRAMP1 is essential for Arabidopsis growth in low manganese conditions. Plant Cell. 2010;22:904–917. doi: 10.1105/tpc.109.073023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castaings L, Caquot A, Loubet S, Curie C. The high-affinity metal Transporters NRAMP1 and IRT1 Team up to Take up Iron under Sufficient Metal Provision. Sci Rep. 2016;6:37222. doi: 10.1038/srep37222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Briat JF, Duc C, Ravet K, Gaymard F. Ferritins and iron storage in plants. Biochim Biophys Acta. 2010;1800:806–814. doi: 10.1016/j.bbagen.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Kim SA, et al. Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science. 2006;314:1295–1298. doi: 10.1126/science.1132563. [DOI] [PubMed] [Google Scholar]
- 13.Lanquar V, et al. Mobilization of vacuolar iron by AtNRAMP3 and AtNRAMP4 is essential for seed germination on low iron. EMBO J. 2005;24:4041–4051. doi: 10.1038/sj.emboj.7600864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roschzttardtz H, Conéjéro G, Curie C, Mari S. Identification of the endodermal vacuole as the iron storage compartment in the Arabidopsis embryo. Plant Physiol. 2009;151:1329–1338. doi: 10.1104/pp.109.144444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mary V, et al. Bypassing Iron Storage in Endodermal Vacuoles Rescues the Iron Mobilization Defect in the natural resistance associated-macrophage protein3natural resistance associated-macrophage protein4 Double Mutant. Plant Physiol. 2015;169:748–759. doi: 10.1104/pp.15.00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Molins H, et al. Mutants impaired in vacuolar metal mobilization identify chloroplasts as a target for cadmium hypersensitivity in Arabidopsis thaliana. Plant Cell Environ. 2013;36:804–817. doi: 10.1111/pce.12016. [DOI] [PubMed] [Google Scholar]
- 17.Oomen RJ, et al. Functional characterization of NRAMP3 and NRAMP4 from the metal hyperaccumulator Thlaspi caerulescens. New Phytol. 2009;181:637–650. doi: 10.1111/j.1469-8137.2008.02694.x. [DOI] [PubMed] [Google Scholar]
- 18.Pottier M, et al. Identification of mutations allowing Natural Resistance Associated Macrophage Proteins (NRAMP) to discriminate against cadmium. Plant J. 2015;83:625–637. doi: 10.1111/tpj.12914. [DOI] [PubMed] [Google Scholar]
- 19.Arita A, et al. A genome-wide deletion mutant screen identifies pathways affected by nickel sulfate in Saccharomyces cerevisiae. BMC Genomics. 2009;10:524. doi: 10.1186/1471-2164-10-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eide DJ, et al. Characterization of the yeast ionome: a genome-wide analysis of nutrient mineral and trace element homeostasis in Saccharomyces cerevisiae. Genome Biol. 2005;6:R77. doi: 10.1186/gb-2005-6-9-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruotolo R, Marchini G, Ottonello S. Membrane transporters and protein traffic networks differentially affecting metal tolerance: a genomic phenotyping study in yeast. Genome Biol. 2008;9:R67. doi: 10.1186/gb-2008-9-4-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hettema EH, Valdez-Taubas J, Pelham HR. Bsd2 binds the ubiquitin ligase Rsp5 and mediates the ubiquitination of transmembrane proteins. EMBO J. 2004;23:1279–1288. doi: 10.1038/sj.emboj.7600137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen LT, et al. Down-regulation of a manganese transporter in the face of metal toxicity. Mol Biol Cell. 2009;20:2810–2819. doi: 10.1091/mbc.E08-10-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu XF, Culotta VC. Post-translation control of Nramp metal transport in yeast. Role of metal ions and the BSD2 gene. J Biol Chem. 1999;274:4863–4868. doi: 10.1074/jbc.274.8.4863. [DOI] [PubMed] [Google Scholar]
- 25.Portnoy ME, Liu XF, Culotta VC. Saccharomyces cerevisiae expresses three functionally distinct homologues of the nramp family of metal transporters. Mol Cell Biol. 2000;20:7893–7902. doi: 10.1128/mcb.20.21.7893-7902.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stimpson HE, Lewis MJ, Pelham HR. Transferrin receptor-like proteins control the degradation of a yeast metal transporter. EMBO J. 2006;25:662–672. doi: 10.1038/sj.emboj.7600984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barberon M, et al. Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc Natl Acad Sci USA. 2011;108:E450–E458. doi: 10.1073/pnas.1100659108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barberon M, et al. Polarization of IRON-REGULATED TRANSPORTER 1 (IRT1) to the plant-soil interface plays crucial role in metal homeostasis. Proc Natl Acad Sci USA. 2014;111:8293–8298. doi: 10.1073/pnas.1402262111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao C, et al. A unique plant ESCRT component, FREE1, regulates multivesicular body protein sorting and plant growth. Curr Biol. 2014;24:2556–2563. doi: 10.1016/j.cub.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 30.Kasai K, Takano J, Miwa K, Toyoda A, Fujiwara T. High boron-induced ubiquitination regulates vacuolar sorting of the BOR1 borate transporter in Arabidopsis thaliana. J Biol Chem. 2011;286:6175–6183. doi: 10.1074/jbc.M110.184929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin WY, Huang TK, Chiou TJ. Nitrogen limitation adaptation, a target of microRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. Plant Cell. 2013;25:4061–4074. doi: 10.1105/tpc.113.116012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takano J, et al. Polar localization and degradation of Arabidopsis boron transporters through distinct trafficking pathways. Proc Natl Acad Sci USA. 2010;107:5220–5225. doi: 10.1073/pnas.0910744107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dowler S, et al. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem J. 2000;351:19–31. doi: 10.1042/0264-6021:3510019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mikami K, Takahashi S, Katagiri T, Yamaguchi-Shinozaki K, Shinozaki K. Isolation of an Arabidopsis thaliana cDNA encoding a pleckstrin homology domain protein, a putative homologue of human pleckstrin. J Exp Bot. 1999;50:729–730. [Google Scholar]
- 35.van Leeuwen W, Okrész L, Bögre L, Munnik T. Learning the lipid language of plant signalling. Trends Plant Sci. 2004;9:378–384. doi: 10.1016/j.tplants.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 36.Baraldi E, et al. Structure of the PH domain from Bruton’s tyrosine kinase in complex with inositol 1,3,4,5-tetrakisphosphate. Structure. 1999;7:449–460. doi: 10.1016/s0969-2126(99)80057-4. [DOI] [PubMed] [Google Scholar]
- 37.Ferguson KM, et al. Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol Cell. 2000;6:373–384. doi: 10.1016/s1097-2765(00)00037-x. [DOI] [PubMed] [Google Scholar]
- 38.Lietzke SE, et al. Structural basis of 3-phosphoinositide recognition by pleckstrin homology domains. Mol Cell. 2000;6:385–394. doi: 10.1016/s1097-2765(00)00038-1. [DOI] [PubMed] [Google Scholar]
- 39.Thomas CC, Deak M, Alessi DR, van Aalten DM. High-resolution structure of the pleckstrin homology domain of protein kinase b/akt bound to phosphatidylinositol (3,4,5)-trisphosphate. Curr Biol. 2002;12:1256–1262. doi: 10.1016/s0960-9822(02)00972-7. [DOI] [PubMed] [Google Scholar]
- 40.Hyvönen M, Saraste M. Structure of the PH domain and Btk motif from Bruton’s tyrosine kinase: molecular explanations for X-linked agammaglobulinaemia. EMBO J. 1997;16:3396–3404. doi: 10.1093/emboj/16.12.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simon ML, et al. A multi-colour/multi-affinity marker set to visualize phosphoinositide dynamics in Arabidopsis. Plant J. 2014;77:322–337. doi: 10.1111/tpj.12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kleine-Vehn J, et al. Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc Natl Acad Sci USA. 2008;105:17812–17817. doi: 10.1073/pnas.0808073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamura K, et al. Why green fluorescent fusion proteins have not been observed in the vacuoles of higher plants. Plant J. 2003;35:545–555. doi: 10.1046/j.1365-313x.2003.01822.x. [DOI] [PubMed] [Google Scholar]
- 44.Belda-Palazon B, et al. FYVE1/FREE1 Interacts with the PYL4 ABA Receptor and Mediates its Delivery to the Vacuolar Degradation Pathway. Plant Cell. 2016;28:2291–2311. doi: 10.1105/tpc.16.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kolb C, et al. FYVE1 is essential for vacuole biogenesis and intracellular trafficking in Arabidopsis. Plant Physiol. 2015;167:1361–1373. doi: 10.1104/pp.114.253377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson DG. Endocytosis: Is There Really a Recycling from Late Endosomes? Mol Plant. 2015;8:1554–1556. doi: 10.1016/j.molp.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 47.Ebine K, et al. A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat Cell Biol. 2011;13:853–859. doi: 10.1038/ncb2270. [DOI] [PubMed] [Google Scholar]
- 48.Ebine K, et al. Endosomal trafficking pathway regulated by ARA6, a RAB5 GTPase unique to plants. Small GTPases. 2012;3:23–27. doi: 10.4161/sgtp.18299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nielsen ME, Feechan A, Böhlenius H, Ueda T, Thordal-Christensen H. Arabidopsis ARF-GTP exchange factor, GNOM, mediates transport required for innate immunity and focal accumulation of syntaxin PEN1. Proc Natl Acad Sci USA. 2012;109:11443–11448. doi: 10.1073/pnas.1117596109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maîtrejean M, Vitale A. How are tonoplast proteins degraded? Plant Signal Behav. 2011;6:1809–1812. doi: 10.4161/psb.6.11.17867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maîtrejean M, et al. Assembly and sorting of the tonoplast potassium channel AtTPK1 and its turnover by internalization into the vacuole. Plant Physiol. 2011;156:1783–1796. doi: 10.1104/pp.111.177816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moravcevic K, Oxley CL, Lemmon MA. Conditional peripheral membrane proteins: facing up to limited specificity. Structure. 2012;20:15–27. doi: 10.1016/j.str.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raiborg C, Schink KO, Stenmark H. Class III phosphatidylinositol 3-kinase and its catalytic product PtdIns3P in regulation of endocytic membrane traffic. FEBS J. 2013;280:2730–2742. doi: 10.1111/febs.12116. [DOI] [PubMed] [Google Scholar]
- 54.Schink KO, Raiborg C, Stenmark H. Phosphatidylinositol 3-phosphate, a lipid that regulates membrane dynamics, protein sorting and cell signalling. BioEssays. 2013;35:900–912. doi: 10.1002/bies.201300064. [DOI] [PubMed] [Google Scholar]
- 55.Catimel B, et al. The PI(3)P interactome from a colon cancer cell. J Proteomics. 2013;82:35–51. doi: 10.1016/j.jprot.2013.01.031. [DOI] [PubMed] [Google Scholar]
- 56.Vonkova I, et al. Lipid Cooperativity as a General Membrane-Recruitment Principle for PH Domains. Cell Reports. 2015;12:1519–1530. doi: 10.1016/j.celrep.2015.07.054. [DOI] [PubMed] [Google Scholar]
- 57.Yu JW, et al. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol Cell. 2004;13:677–688. doi: 10.1016/s1097-2765(04)00083-8. [DOI] [PubMed] [Google Scholar]
- 58.Kleinboelting N, Huep G, Kloetgen A, Viehoever P, Weisshaar B. GABI-Kat SimpleSearch: new features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res. 2012;40:D1211–D1215. doi: 10.1093/nar/gkr1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lanquar V, et al. Export of vacuolar manganese by AtNRAMP3 and AtNRAMP4 is required for optimal photosynthesis and growth under manganese deficiency. Plant Physiol. 2010;152:1986–1999. doi: 10.1104/pp.109.150946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quinn JM, Merchant S. Copper-responsive gene expression during adaptation to copper deficiency. Methods Enzymol. 1998;297:263–279. doi: 10.1016/s0076-6879(98)97020-3. [DOI] [PubMed] [Google Scholar]
- 61.Loudet O, Chaillou S, Camilleri C, Bouchez D, Daniel-Vedele F. Bay-0 x Shahdara recombinant inbred line population: a powerful tool for the genetic dissection of complex traits in Arabidopsis. Theor Appl Genet. 2002;104:1173–1184. doi: 10.1007/s00122-001-0825-9. [DOI] [PubMed] [Google Scholar]
- 62.Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Merlot S, et al. The metal transporter PgIREG1 from the hyperaccumulator Psychotria gabriellae is a candidate gene for nickel tolerance and accumulation. J Exp Bot. 2014;65:1551–1564. doi: 10.1093/jxb/eru025. [DOI] [PubMed] [Google Scholar]
- 64.Inada N, et al. Modulation of Plant RAB GTPase-Mediated Membrane Trafficking Pathway at the Interface Between Plants and Obligate Biotrophic Pathogens. Plant Cell Physiol. 2016;57:1854–1864. doi: 10.1093/pcp/pcw107. [DOI] [PubMed] [Google Scholar]
- 65.Uemura T, et al. Qa-SNAREs localized to the trans-Golgi network regulate multiple transport pathways and extracellular disease resistance in plants. Proc Natl Acad Sci USA. 2012;109:1784–1789. doi: 10.1073/pnas.1115146109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Uemura T, et al. Vacuolar/pre-vacuolar compartment Qa-SNAREs VAM3/SYP22 and PEP12/SYP21 have interchangeable functions in Arabidopsis. Plant J. 2010;64:864–873. doi: 10.1111/j.1365-313X.2010.04372.x. [DOI] [PubMed] [Google Scholar]
- 67.Le Bars R, Marion J, Le Borgne R, Satiat-Jeunemaitre B, Bianchi MW. ATG5 defines a phagophore domain connected to the endoplasmic reticulum during autophagosome formation in plants. Nat Commun. 2014;5:4121. doi: 10.1038/ncomms5121. [DOI] [PubMed] [Google Scholar]
- 68.Bolte S, Cordelières FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 69.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dowler S, Kular G, Alessi DR. Protein lipid overlay assay. Sci STKE. 2002;2002:pl6. doi: 10.1126/stke.2002.129.pl6. [DOI] [PubMed] [Google Scholar]