Significance
Faithful and efficient genome duplication is essential for the propagation of life. Aberrant DNA replication can lead to genomic instability and cancer. In eukaryotes, the replication machinery is composed of the DNA-unwinding enzyme Cdc45–MCM–GINS (CMG) and dedicated DNA synthesis factors. Three different polymerases act sequentially on the leading-strand template to establish DNA replication. We describe the architecture and dynamics of the main leading-strand polymerase bound to the CMG helicase, and we propose a mechanism for the establishment of efficient leading-strand synthesis. Our findings provide important insights into how the eukaryotic replication machinery functions to ensure that genome integrity is maintained during replication.
Keywords: DNA replication, CMG helicase, DNA polymerase, single-particle electron microscopy
Abstract
The replisome unwinds and synthesizes DNA for genome duplication. In eukaryotes, the Cdc45–MCM–GINS (CMG) helicase and the leading-strand polymerase, Pol epsilon, form a stable assembly. The mechanism for coupling DNA unwinding with synthesis is starting to be elucidated, however the architecture and dynamics of the replication fork remain only partially understood, preventing a molecular understanding of chromosome replication. To address this issue, we conducted a systematic single-particle EM study on multiple permutations of the reconstituted CMG–Pol epsilon assembly. Pol epsilon contains two flexibly tethered lobes. The noncatalytic lobe is anchored to the motor of the helicase, whereas the polymerization domain extends toward the side of the helicase. We observe two alternate configurations of the DNA synthesis domain in the CMG-bound Pol epsilon. We propose that this conformational switch might control DNA template engagement and release, modulating replisome progression.
DNA replication is catalyzed by the replisome, a molecular machine that coordinates DNA unwinding and synthesis (1). These two functions must be tightly coordinated to prevent the rise of genome instability, which is a major cause of cancer. DNA unwinding by a replicative helicase involves single-strand translocation of a hexameric motor, whereas DNA synthesis requires template priming by a primase and extension by dedicated replicative DNA polymerases (2). In eukaryotes, the helicase function is performed by the Cdc45–MCM–GINS (CMG) complex (3, 4) and the primase function is played by Pol alpha (5), whereas DNA synthesis is catalyzed by two specialized DNA polymerases, Pol epsilon and delta. According to the consensus view, Pol epsilon synthesizes the leading and Pol delta the lagging strand (6–11). However, recent studies indicate that the division of labor between replicative polymerases might be more promiscuous than originally thought (12, 13). In in vitro-reconstituted DNA replication reactions, Pol delta can support leading-strand duplication (11, 14), but switching from Pol delta to epsilon is necessary for efficient establishment of leading-strand synthesis (14). The mechanism of substrate handoff between the two polymerases is currently unknown.
Recent breakthroughs in structural biology begin to provide an architectural framework to understand the interaction between helicase and polymerases at the replication fork. For example, studies on the CMG helicase and its subcomplexes have established that the MCM is a six-member ring with an N-terminal domain that serves as a processivity collar (15) and a C-terminal ATPase motor domain that provides the DNA unwinding function (16–21). High-resolution cryo-EM analysis has shown that the ATPase motor translocates on the leading-strand template (22), in agreement with work on Xenopus embryo extracts (23). The GINS and Cdc45 components of the CMG bind to the side and stabilize the N-terminal domain of the MCM ring (closing a dynamic Mcm5-2 gate), allowing for the motor to move on DNA (16, 20, 22).
Although previous work from us and others established that Pol alpha maps in proximity to the N-terminal face of the MCM ring (24, 25), recent data on the reconstituted yeast replisome indicate that Pol epsilon stably anchors onto the ATPase face of the helicase (24). Therefore, MCM not only functions as the motor that catalyzes fork progression but also is a central nexus around which the replication machinery is organized.
Pol epsilon is a heterotetramer consisting of two main modules with distinct functions. The first module is the Pol2 N-terminal catalytic domain, which is dispensable for viability (26, 27). The second module is the noncatalytic portion of the assembly comprising the essential Pol2 C-terminal domain (a catalytically defunct polymerase), Dpb2 (a defunct exonuclease), and the Dpb3 and Dpb4 ancillary factors (28–33). Coordinated action of the CMG and Pol epsilon supports leading-strand synthesis (14, 34–36), and emerging evidence indicates that the noncatalytic module of Pol epsilon plays a separate role in replication, being essential for CMG formation in cells and perhaps by stimulating the DNA unwinding function of the CMG helicase (14, 28, 33, 37, 38).
To understand how Pol epsilon and the CMG work together to duplicate the leading strand, we have reconstituted the intact CMG–Pol epsilon assembly for electron microscopy analysis. We describe here the complete structure of the helicase-leading strand polymerase complex. Using a combination of subunit dropout, domain deletion, and MBP fusion mutants, we can orient Pol epsilon with respect to the CMG helicase and define unexpected architectural features in the eukaryotic replisome. We uncover a conformational change of the DNA synthesis domain of Pol epsilon that (i) suggests a structural mechanism for the polymerase switch important during the establishment of leading-strand synthesis and (ii) provides a first insight into how replisome processivity might be regulated.
Results
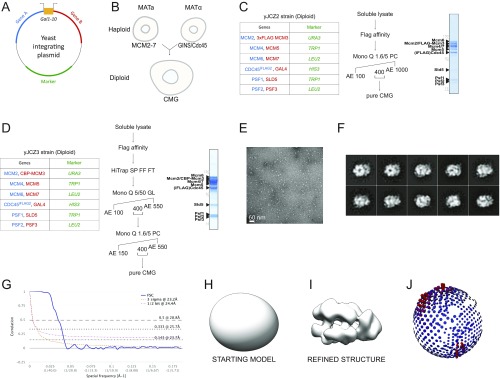
Pure, Active, and Homogeneous Yeast CMG from a Diploid Overexpression Strain.
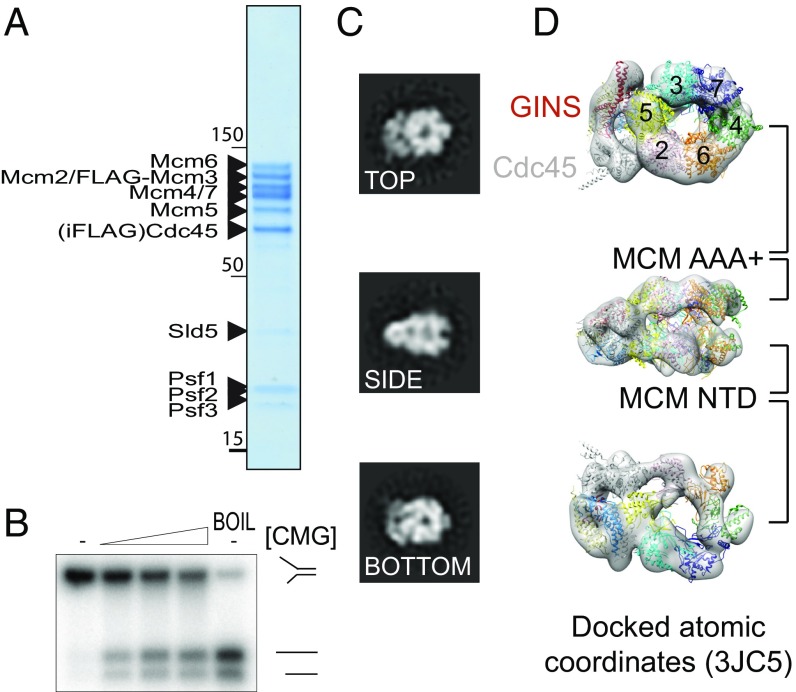
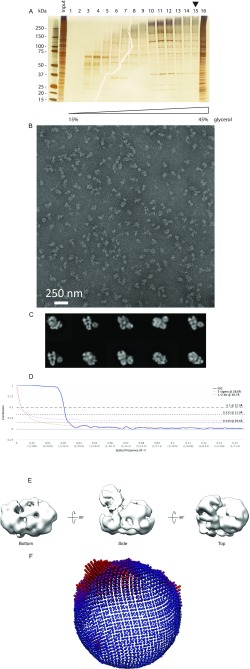
Catalytically active CMG has been previously purified from (i) Drosophila melanogaster embryo extracts (3), (ii) baculovirus-infected insect cells overexpressing fly or human proteins (4, 34), and (iii) a haploid Saccharomyces cerevisiae strain overexpressing yeast proteins (11). Some of these methods can be tedious, and preparation yields are often variable. Purification of a reconstituted, stoichiometric helicase–polymerase assembly, however, requires a reproducible approach to isolate large quantities of the CMG complex. To this end, we have devised a reliable strategy to produce yeast CMG, built on the established integration of plasmids bearing two codon-optimized genes under the control of a bidirectional galactose-inducible promoter (39) (Fig. S1A). Coexpressing as many as 11 different genes with this system is challenging due to the limited choice of selection markers required for plasmid integration. To circumvent this problem, we have mated a Mat a Mcm2–7 overexpression strain with a Mat α GINS–Cdc45 strain (Fig. S1B), yielding a diploid strain that produces yeast CMG. A similar method was recently used to produce the 15-member Ino80 complex (40). Purification strategies involved FLAG affinity [to capture FLAG-Mcm3/Cdc45internal FLAG, strain yJCZ2 (Fig. S1C) or alternatively Cdc45internal FLAG, strain yJCZ3 (Fig. S1D)], followed by anion exchange steps. This approach reproducibly yields Coomassie-stainable amounts of CMG helicase (Fig. 1A), in the low micromolar concentration range, which is necessary for reconstitution and further purification of a polymerase cocomplex (Materials and Methods). Importantly, we can show that our yeast CMG is as vigorous a helicase as the baculovirus-expressed Drosophila CMG (4, 22) (Fig. 1B). Likewise, 2D (Fig. 1C) and 3D (Fig. 1D) EM image analysis demonstrates that our yeast CMG structure is virtually identical to the published Drosophila (16) and yeast EM volumes (24) (Fig. S1). Docking the atomic coordinates of the yeast CMG [Protein Data Bank (PDB) ID code 3JC5 (20)] into our 3D EM map allows the unambiguous identification of each of the 11 helicase subunits in our structure (Fig. 1D).
Fig. S1.
Making homogeneous CMG from a diploid yeast strain and single-particle EM analysis. (A) Schematic representation of the integration plasmid containing two codon-optimized genes under the control of a bidirectional galactose-inducible promoter. (B) A diploid CMG overexpression strain was obtained by mating a Mat a Mcm2–7 overexpression strain and a Mat α GINS/Cdc45 strain. (C) Gene pairs and selection markers, purification protocol, and Coomassie-stained gel of the yeast CMG obtained from strain yJCZ2. (D) Gene pairs and selection markers, purification protocol, and Coomassie-stained gel of the yeast CMG obtained from strain yJCZ3. (E) Representative micrograph. (F) 2D class averages. (G) Fourier shell correlation indicates a resolution of 23.7 Å according to the 0.143 criterion. (H) Starting model for 3D refinement. (I) Refined 3D structure. (J) Angular distribution.
Fig. 1.
Catalytically active yeast CMG. (A) Coomassie-stained gel of the yeast CMG obtained from strain yJCZ2. (B) A DNA unwinding assay shows that the overexpressed yeast CMG is catalytically active. CMG quantities used in the reactions are (from left to right) 0, 159, 397, and 793 fmol. (C) 2D image analysis showing that our CMG preparations contain stable CMG particles, suitable for 3D reconstruction. (D) 3D reconstruction with atomic docking of the 11 different CMG subunits (PDB ID code 3JC5) showing that the purified CMG is very similar to the published yeast and Drosophila CMG complexes.
Pol Epsilon Architecture.
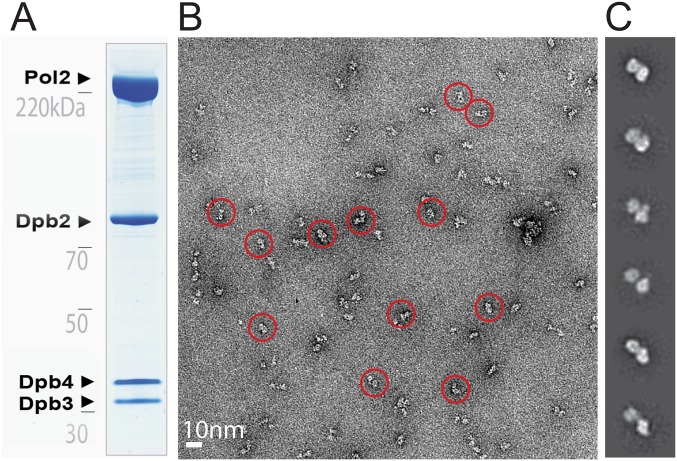
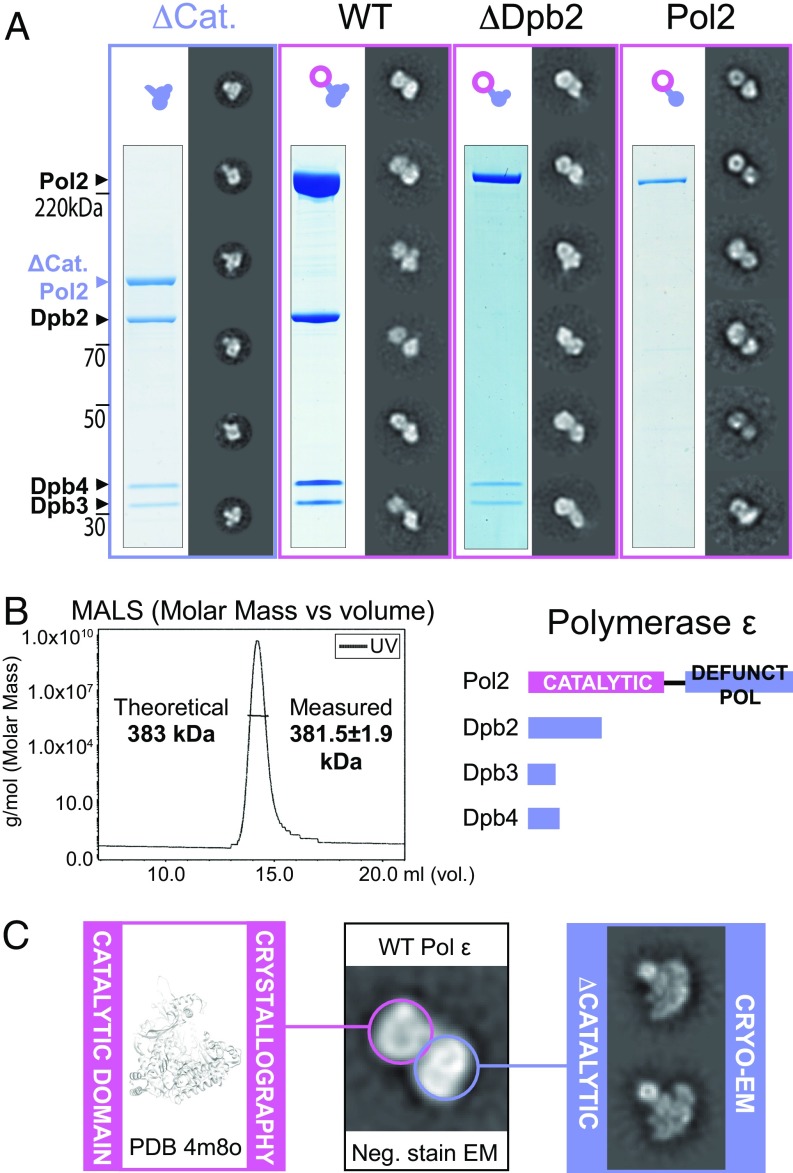
Wild-type yeast polymerase epsilon has been characterized in an early cryo-EM single-particle study (41), however the high-resolution X-ray structure of the catalytic domain (27) cannot be easily integrated with the lower resolution EM data. In an attempt to reconcile the two studies, we have decided to image yeast Pol epsilon by negative stain EM. To this end, we have characterized the active polymerase preparation used in the reconstituted yeast DNA replication system (38) (Fig. S2). We observed that wild-type Pol epsilon is composed of two lobes that appear connected by an isthmus of electron density (Fig. 2A and Movie S1). To establish whether the two lobes represent a dimeric form of the protein complex or rather a monomer, we have analyzed the same preparation by size-exclusion chromatography with multiangle light scattering. According to our measurements, the absolute molecular mass of Pol epsilon is 381.5 ± 1.9 kDa, in striking agreement with a predicted molecular mass of 383 kDa for a monomeric, CBP-tagged Pol epsilon complex (Fig. 2B). A maltose-binding protein (MBP) fused to one of four Pol epsilon subunits produces a polymerase particle decorated with one, not two, bright density feature proximal to the polymerase particle, in further support of the notion that Pol epsilon forms a single, not a double, heterotetramer. We conclude that Pol epsilon is formed by two spatially separated lobes, probably connected via a linker. To establish the identity of the two lobes, we have characterized a Pol epsilon complex lacking the Dpb2 subunit, to find that this assembly still exists as a bilobed entity (Fig. 2A, Fig. S3, and Movie S2). We then characterized the isolated Pol2 subunit of Pol epsilon and discovered that this polypeptide alone also contains a bilobed structure (Fig. 2A, Fig. S4, and Movie S3). We tentatively assign one lobe to the N-terminal catalytic domain and the second lobe to a C-terminal, catalytically defunct polymerase repeat. Our results on Pol2 differ from an earlier EM study on the same isolated subunit, which had been described as a singly lobed entity (41). To validate our results, we analyzed a deletion mutant of Pol epsilon that contains all subunits but lacks the N-terminal catalytic domain of Pol2 (“Δcat”; Fig. S5). As predicted, Δcat forms one singly lobed structure (Fig. 2A and Movie S4), and cryo-EM 2D analysis confirms that this is a large and compact protein assembly (Fig. 2C and Fig. S6), in contrast to earlier work that describes the noncatalytic portion of Pol epsilon as a poorly structured entity (41).
Fig. S2.
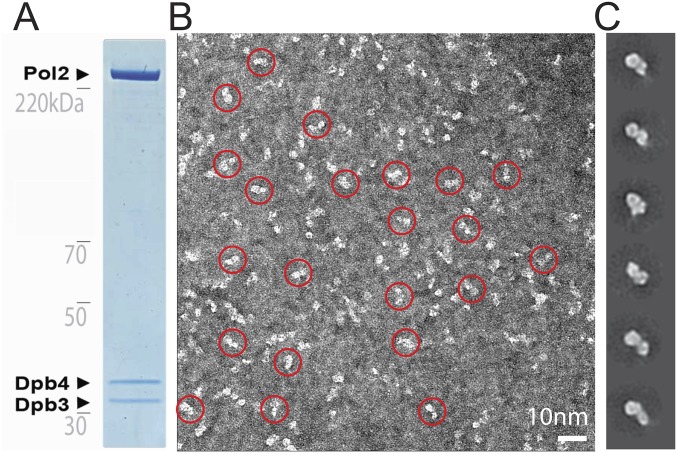
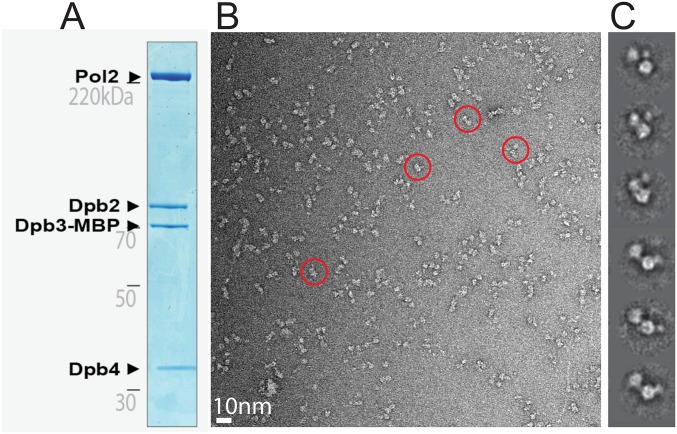
Single-particle EM analysis of the wild-type yeast Pol epsilon. (A) Coomassie-stained gel of the purified Pol epsilon assembly. (B) Representative electron micrograph with a few circled particles, containing the characteristic bilobed structure. (C) 2D class averages.
Fig. 2.
Architecture of the Pol epsilon complex. (A) From the left: A tetrameric complex lacking the catalytic domain of Pol2 (Δcat) forms a singly lobed assembly. The WT complex forms a bilobed assembly. A Dpb2 subunit dropout trimeric complex forms a bilobed assembly. Surprisingly, the isolated Pol2 is also bilobed. (B, Left) Size-exclusion chromatography with multiangle light scattering shows that the wild-type Pol epsilon complex is a single homotetramer in solution and not a dimer of tetramers. (Right) A stick diagram representing the Pol epsilon tetramer. (C) The Pol epsilon assembly is formed of two lobes. One lobe comprises the N-terminal catalytic domain of Pol2 (for which a crystal structure is available; PDB ID code 4m8o), and the second lobe comprises the noncatalytic portion of the complex (for which we determined a 2D cryo-EM structure).
Fig. S3.
Single-particle EM analysis of the ΔDpb2 yeast Pol epsilon. (A) Coomassie-stained gel of the purified Pol epsilon assembly. (B) Representative electron micrograph with a few circled particles, containing the characteristic bilobed structure. (C) 2D class averages.
Fig. S4.
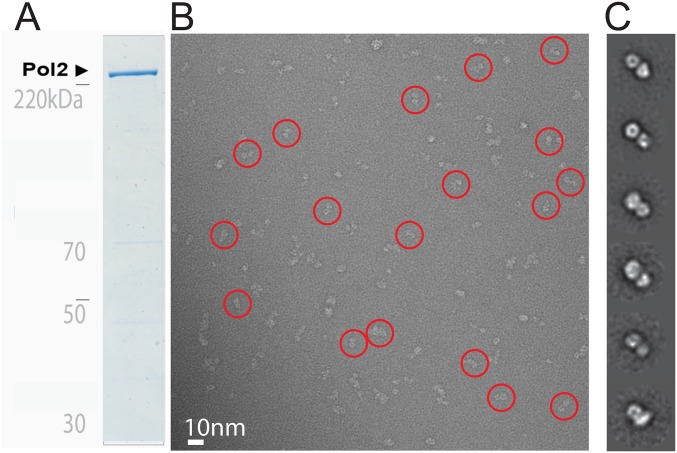
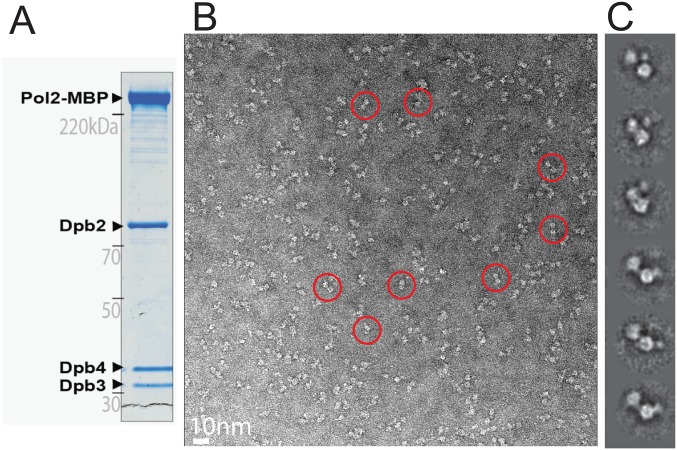
Single-particle EM analysis of the Pol2 subunit of the yeast Pol epsilon. (A) Coomassie-stained gel of the purified Pol2 subunit. (B) Representative electron micrograph with a few circled particles, containing the characteristic bilobed structure. (C) 2D class averages.
Fig. S5.
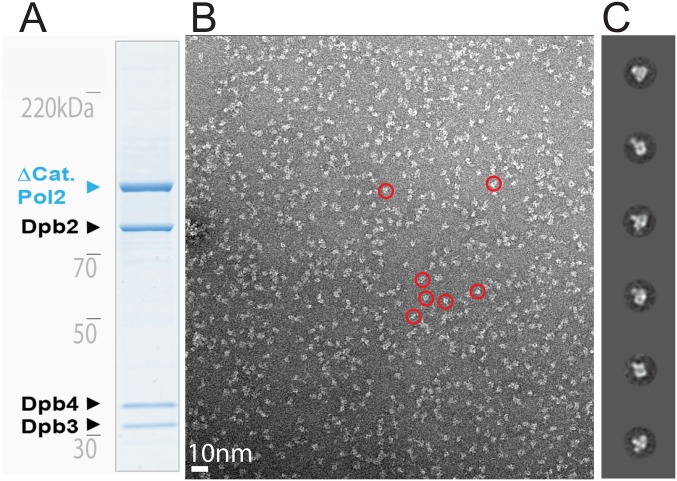
Single-particle EM analysis of the Δcat yeast Pol epsilon. (A) Coomassie-stained gel of the purified Pol epsilon assembly. (B) Representative electron micrograph with a few circled particles, containing a singly lobed structure. (C) 2D class averages.
Fig. S6.
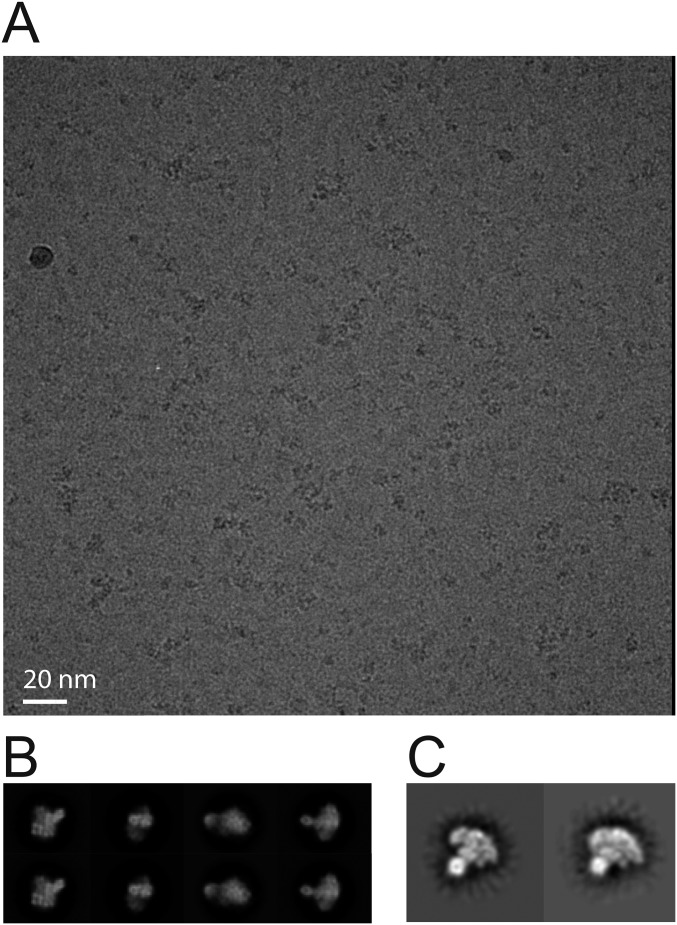
Single-particle cryo-EM analysis of the Δcat yeast Pol epsilon. (A) Representative sum of the aligned movie frames. (B) 2D class averages. (C) Notable anchor-shaped class averages visualized after imposing a 30–5 Å band-pass filter.
In summary, we conducted negative stain and cryo-EM studies on wild-type, subunit dropout, and domain deletion mutants. We find that Pol epsilon contains a bilobed structure, where the catalytic domain of Pol2 constitutes one lobe and the noncatalytic modules in the assembly form the second lobe (Fig. 2C). Our findings support the notion that functionally separated modules in Pol epsilon are indeed spatially separated.
CMG–Pol Epsilon Reconstitution.
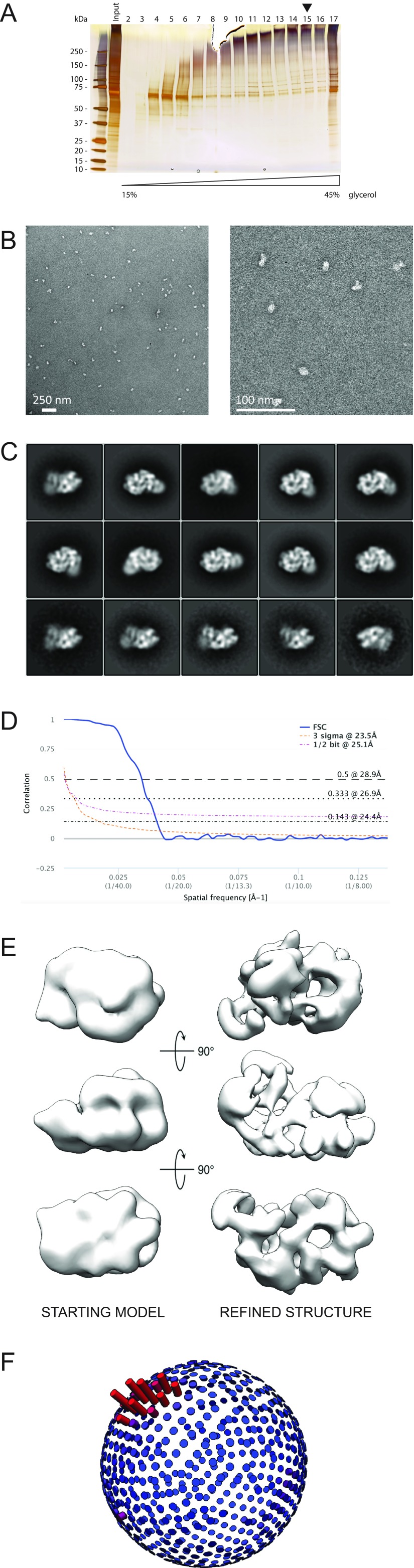
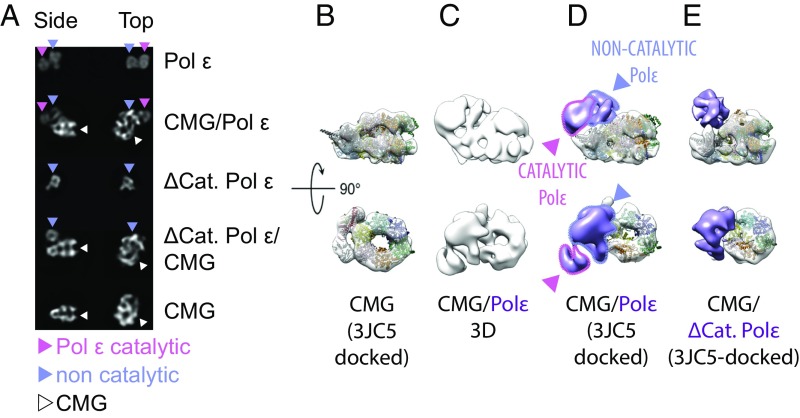
To establish how leading/lagging-strand segregation is achieved at the eukaryotic replication fork, an exhaustive description of the CMG–Pol epsilon complex is needed (42). Recent structural work on the eukaryotic replisome shows that Pol epsilon is anchored to the ATPase side of the helicase ring (24). To experimentally locate the catalytic domain of Pol epsilon in the helicase–polymerase complex, we have developed a protocol to reconstitute the CMG–Pol epsilon assembly, followed by mild cross-linking (XL) and purification over a glycerol gradient (Materials and Methods). Our preparation yielded homogeneous, monodisperse, stabilized particles that are suitable for EM analysis (Fig. S7). As discernible in 2D averages of side views, CMG bound to wild-type Pol epsilon appears decorated with a bilobed feature. One lobe is proximal to the ATPase tier of the MCM motor, whereas the second lobe is more peripheral. Our observation agrees with the notion that the isolated, monomeric Pol epsilon is a bilobed entity, indicating a CMG:Pol epsilon stoichiometry of 1:1 (Fig. 3A). To establish the orientation of Pol epsilon bound to the CMG, we repeated the helicase–polymerase reconstitution experiment using the Pol epsilon Δcat mutant (Fig. S8). As expected from our characterization of the isolated deletion mutant, the CMG appears decorated with one lone polymerase lobe (Fig. 3A). The 3D reconstruction (Fig. S7 D–F) of the full wild-type assembly yields a recognizable CMG structure, bound to polymerase density that departs from the ATPase tier and extends toward the outer perimeter of the helicase structure, contacting the peripheral helicase component Cdc45 (Fig. 3 B–D). Comparing the 3D structure of the full assembly with the 2D averages and the 3D structure of the reconstituted CMG–Δcat Pol epsilon, the catalytic domain of Pol epsilon can be unambiguously located at a 90° offset to the CMG helicase ring pore and not proximal to the ATPase tier (Fig. 3 and Fig. S8 C–F).
Fig. S7.
Single-particle EM analysis of the CMG–Pol epsilon complex. (A) Silver-stained SDS/PAGE gel of the CMG–Pol epsilon preparation. The arrowhead indicates the lane analyzed by EM. (B) Representative micrograph (Left) and a magnified view thereof (Right). (C) 2D class averages. (D) Fourier shell correlation indicates a resolution of 24.4 Å according to the 0.143 criterion. (E) The yeast CMG was used as a starting model for 3D classification and refinement. The resulting structure is a CMG bound to a bilobed Pol epsilon assembly. (F) Angular distribution.
Fig. 3.
CMG–Pol epsilon reconstitution and 3D structure. (A) The isolated Pol epsilon has a bilobed structure (top row). In complex with Pol epsilon, the CMG is decorated with a bilobed feature (second row). The isolated Δcat Pol epsilon is a singly-lobed entity (third row). In complex with Δcat Pol epsilon, the CMG is decorated with a singly lobed feature (fourth row). Characteristic side and top views of the CMG (bottom row). (B) Yeast CMG reconstruction with docked atomic coordinates (PDB ID code 3JC5). (C) 3D structure of the CMG–Pol epsilon complex. (D) 3D structure of CMG–Pol epsilon complex with docked atomic coordinates of the CMG and assigned catalytic domain and noncatalytic portion of Pol epsilon. The catalytic domain departs radially from the core particle. Density corresponding to the polymerase is highlighted in purple. (E) 3D structure of the CMG–Δcat Pol epsilon complex color-coded as in C, with docked atomic coordinates.
Fig. S8.
Single-particle EM analysis of the CMG–Δcat Pol epsilon complex. (A) Silver-stained SDS/PAGE gel of the CMG–Δcat Pol epsilon preparation. The arrowhead indicates the lane analyzed by EM. (B) Representative micrograph. (C) 2D class averages. (D) Fourier shell correlation indicates a resolution of 29.6 Å according to the 0.143 criterion. (E) The yeast CMG in complex with wild-type Pol epsilon was used as a starting model for 3D classification and refinement. The resulting structure is a CMG bound to a singly lobed Pol epsilon assembly. (F) Angular distribution.
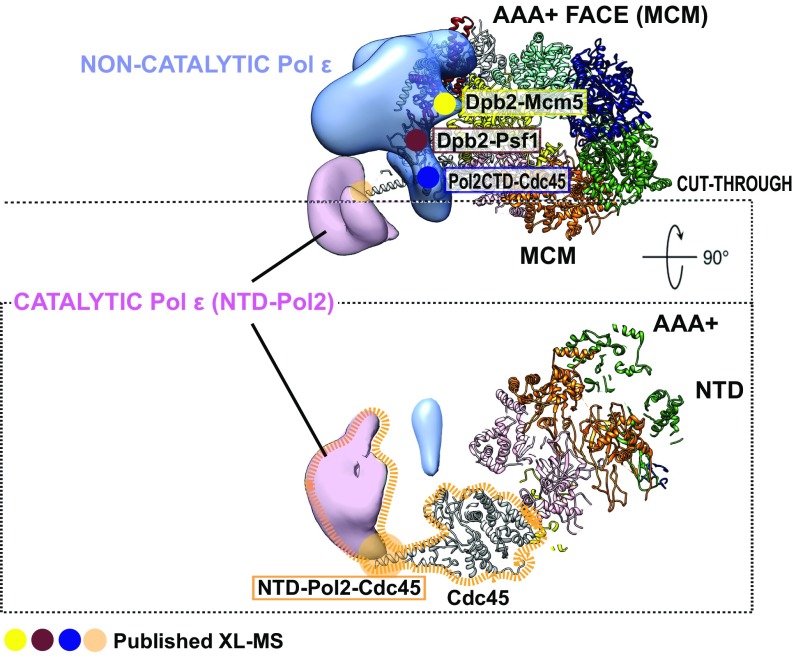
We wondered whether short exposure to mild XL might have introduced artefacts in our preparations. Three lines of evidence argue against this notion. First, with the exception of the recovered density of the catalytic domain, our structure of the CMG–Pol epsilon appears similar to the previously published structure (with particular emphasis on the Pol epsilon anchor; Movie S5) (42). Second, the bilobed feature in the cross-linked CMG–Pol epsilon is highly reminiscent of the non–cross-linked, isolated wild-type Pol epsilon assembly (Fig. 3A). Third, our structure agrees with the published XL–mass spectrometry (XL-MS) characterization of the CMG–Pol epsilon architecture (24). According to the XL-MS study, one major CMG contact made by the Pol epsilon catalytic domain is with Cdc45 helix α6 (20, 24, 43). This Cdc45 element projects radially from the CMG core and intimately contacts the catalytic domain of Pol epsilon in our structure (Fig. 4). Other detected contacts between the noncatalytic portion of Pol epsilon and the CMG (including Pol2-CTD·Cdc45, Dpb2·Psf1, and Dpb2·Mcm5) are summarized in Fig. 4. Altogether, our data indicate that Pol epsilon can exist in an extended bilobed configuration when CMG-bound, with the catalytic domain of Pol epsilon departing radially from the globular core of the complex. The previous structure reported by O’Donnell, Li and colleagues might have captured a distinct state of the polymerase, where the catalytic module is markedly flexible (hence invisible in the averaged 3D structure) or alternatively very tightly compacted against the noncatalytic polymerase-anchor domain (24).
Fig. 4.
Integration of the 3D EM structure of the CMG–Pol epsilon with published XL-MS data. The noncatalytic portion of Pol epsilon sits on top of the ATPase tier of the MCM. Dpb2 contacts Mcm5 (yellow) and Psf1 (brown), and Pol2-CTD contacts Cdc45 (blue). The catalytic domain of Pol2 contacts the tip of helix α6 in Cdc45 (orange).
CMG–Pol Epsilon Dynamics.
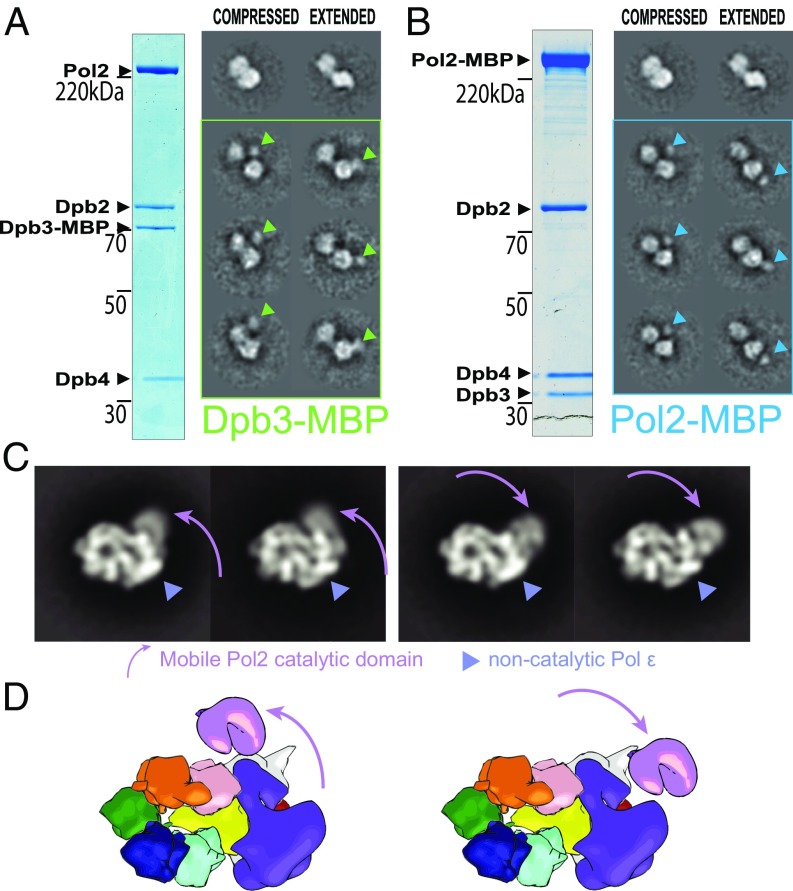
We have so far established that Pol epsilon is a bilobed entity, with two globular domains that appear to be connected by a linker (Fig. 2). Using a protocol that involved reconstitution, XL, glycerol gradient purification, and imaging, we have described the intact helicase–polymerase complex, containing two recognizable globular domains for Pol epsilon (Fig. 3). Taken together, these data point to an inherent flexible nature of Pol epsilon, which might play an important role in replisome dynamics, in particular during leading-strand replication establishment (Discussion). To further characterize the different conformational states of Pol epsilon, we revisited the 2D image analysis of the isolated wild-type enzyme. By aligning all particles to one lone Pol epsilon domain, it appears that the second domain exists in two alternate states, either compact or extended (Movie S6). To verify that what we observe is a real conformational switch and not distinct 2D views of the same 3D object, we have repeated the analysis using a Pol epsilon derivative that contains a C-terminal MBP fusion of the Dpb3 subunit (Fig. S9). Our 2D analysis indicates that the MBP maps either in close proximity to the interface between the two lobes (equator) or alternatively at the tip of one lobe (south pole; Fig. 5A and Movie S7). This result supports the notion that the Dpb3-containing noncatalytic portion of Pol epsilon rotates with respect to the catalytic domain. We cannot rule out, however, from this experiment alone, a second scenario whereby Dpb3 has two alternate binding sites on Pol epsilon, resulting in two possible locations for the MBP tag. To exclude this second hypothesis, we repeated the MBP fusion experiment, tagging the C-terminal domain of Pol2, which is the only subunit that spans the two lobes of Pol epsilon (Fig. S10). As in the previous experiment, the MBP density can be observed either at the equator or at the south pole of the assembly (Fig. 5B and Movie S7). Collectively, our data establish that Pol epsilon undergoes a large-scale movement with one globular domain rotating with respect to the other. In conclusion, the isolated Pol epsilon undergoes a conformational change reminiscent of a switchblade knife being ejected from its handle.
Fig. S9.
Single-particle EM analysis of the yeast CMG–Pol epsilon complex containing an MBP fused to the C terminus of Dpb3. (A) Coomassie-stained gel of the purified Pol epsilon assembly. (B) Representative electron micrograph with a few circled particles, containing the characteristic bilobed structure. (C) 2D class averages.
Fig. 5.
CMG–Pol epsilon dynamics. (A) The isolated Pol epsilon complex with a fused MBP tag at the C terminus of Dpb3 contains two configurations: compressed (with the MBP tag mapping at the interface between lobes, equator) or extended (with the MBP tag mapping at the tip of one lobe, south pole). (B) The isolated Pol epsilon complex with a fused MBP tag at the C terminus of Pol2 also contains two configurations: compressed (MBP tag at the equator) or extended (MBP-tag at the south pole). (C) The CMG–Pol epsilon contains a flexible catalytic domain of Pol2. This can be found in proximity to the MCM ring (Left) or departing radially from the core particle (Right). (D) Cartoon representation depicting the flexibility of the CMG–Pol epsilon.
Fig. S10.
Single-particle EM analysis of the yeast CMG–Pol epsilon complex containing an MBP fused to the C terminus of Pol2. (A) Coomassie-stained gel of the purified Pol epsilon assembly. (B) Representative electron micrograph with a few circled particles, containing the characteristic bilobed structure. (C) 2D class averages.
Integrating the rotating Pol epsilon complex into the helicase–polymerase superassembly could provide new important insights into replisome dynamics. We have therefore extended the analysis on polymerase flexibility to the CMG–Pol epsilon complex. We found that the catalytic domain of Pol epsilon moves with respect to the core particle and exists in two alternate states (Fig. 5 C and D and Movie S8), either projecting outward from Cdc45 or bent inwards and in closer proximity to the helicase ring (two conformations captured in a recent XL/MS analysis) (24). The inherent flexibility of Pol epsilon hence persists in the helicase–polymerase assembly. We postulate that the two Pol epsilon configurations reflect distinct functional states, and DNA engagement might select for one of these two forms.
Discussion
We have reconstituted and imaged a 15-member assembly of the eukaryotic replisome, comprising the CMG helicase and the leading-strand polymerase, Pol epsilon. The DNA synthesis domain in this complex is highly dynamic, providing important insights into the function of the DNA replication machinery.
According to the commonly accepted model of the replication fork, lagging-strand synthesis occurs discontinuously, with cycles of priming and extension of Okazaki fragments, catalyzed by Pol alpha and delta, respectively. Conversely, the leading-strand template is copied continuously by Pol epsilon (44). Lagging-strand synthesis is highly dynamic, and Pol alpha and delta might be recycled as the replication fork progresses. In agreement with this notion, no stable association of Pol delta with the replisome core has been reported to date, and loss of the (Ctf4-mediated) association between Pol alpha and the CMG helicase does not seem to affect fork progression rates (14, 43, 45, 46). A dynamic interplay between different polymerases might not, however, be restricted to the duplication of the lagging strand (12, 13). Recent findings, in fact, suggest that two subsequent polymerase switching events—(i) Pol alpha to Pol delta and (ii) Pol delta to Pol epsilon—might both be required to establish leading-strand duplication (14). This highly choreographed process could revolve around a stable, however dynamic, CMG and Pol epsilon assembly.
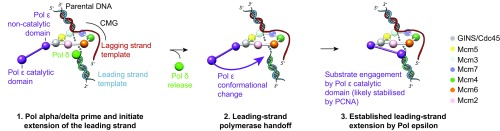
An overview of our current knowledge on Pol epsilon processivity is useful to understand how the CMG helicase and Pol epsilon work together at the replication fork. In all characterized replication systems, processivity factors help tether the replicative polymerase to newly duplicated DNA, often by topologically enclosing the double helix (1, 47, 48). However, the requirement for a dedicated, canonical processivity factor by Pol epsilon has been a matter of debate. Crystallographic analysis on the isolated catalytic domain of Pol2 has revealed a unique insertion that allows the polymerase to encircle the nascent duplex DNA, providing what appears to be an in-built processivity collar (27). Furthermore, in work reported by Langston, O’Donnell, and colleagues, the CMG helicase and Pol epsilon could be purified from yeast cells as a stable protein complex. Because the MCM motor component of the CMG helicase is a ring that encircles DNA, the authors suggested that the helicase itself might act as the main processivity factor that links Pol epsilon to the replication fork (35). These findings are in line with early observations that the DNA sliding clamp PCNA (the processivity factor in the eukaryotic replication fork) only mildly stimulates processivity in a minimal replisome system (11, 14, 49). DNA tethering by the helicase alone, however, is not sufficient to ensure optimal Pol epsilon processivity. In fact, recent reconstitution studies of a more complete replisome that includes the Mrc1, Tof1, and Csm3 fork stabilization factors indicate that cellular rates of fork progression can only be achieved when Pol epsilon is PCNA-associated (14). Therefore, one would predict that the DNA synthesis domain of Pol epsilon is functionally separated and only a PCNA link can lead to stable association between the Pol2 catalytic domain and the DNA template (whereas the helicase-anchor module would play an important role in the correct assembly of the CMG helicase and possibly stimulate DNA unwinding) (14). Using negative stain EM, O’Donnell, Li, and colleagues have recently analyzed a helicase–polymerase supercomplex produced by mixing DNA-coincubated yeast CMG with Pol epsilon. The derived structure appears highly compact, and the proposed assignment placed the catalytic domain of Pol2 next to the ATPase ring of the CMG helicase. The Pol epsilon density, however, appears not to account for the expected molecular mass of Pol epsilon, and the authors entertained the possibility that the helicase–polymerase assembly might be incomplete (24). Using an overexpression strain to produce the CMG and a multistep protocol that combines salt-dialysis reconstitution, protein XL, and purification over a glycerol gradient for electron microscopy sample preparation, we have now obtained a complete structure of Pol epsilon bound to the CMG helicase and experimentally located the DNA synthesis domain in the superassembly. Our results indicate that the ATPase-anchor module does not contain the catalytic domain, but rather the polymerase domain extends radially from the side of the CMG helicase and is free to switch between two positions around the equator of the protein assembly. We speculate that the dynamic nature of the DNA-synthesis module in Pol epsilon might relate to DNA binding and release, with only one form being competent for substrate binding. In turn, PCNA engagement by Pol epsilon might stabilize the interaction between the DNA synthesis domain of Pol2 and the leading strand, for fully processive leading-strand synthesis. We note that DNA binding and release by the catalytic domain of Pol epsilon might facilitate substrate handoff between Pol delta and epsilon, while Pol epsilon remains physically tethered to the moving fork via the CMG helicase (14). Such a handoff mechanism has been invoked during the establishment of leading-strand synthesis in the early stages of replisome progression and in reestablishing coupled leading-strand synthesis after DNA damage repair (Fig. 6). This model will be tested in the future, using higher resolution cryo-EM on a CMG–Pol epsilon complex, imaged in the act of processively duplicating the leading strand.
Fig. 6.
A polymerase-switch mechanism for the establishment of leading-strand synthesis, facilitated by a reconfiguration of the Pol epsilon catalytic domain.
Materials and Methods
Full details of experimental procedures are described in SI Materials and Methods.
Protein Purification.
Yeast strains for the expression of either CMG or Pol epsilon were first induced with galactose, then harvested and lysed with a freezer mill. CMG extracts were first purified by affinity column followed by successive anion-exchange steps. Pol epsilon extracts were first purified by affinity column followed by Heparin and size-exclusion steps.
CMGE Reconstitution.
Purified CMG and Pol epsilon were dialyzed together in the reconstitution buffer, cross-linked with 0.05% glutaraldehyde, and then separated by glycerol gradient sedimentation. Fractions were collected and used for EM preparation.
EM Sample Preparation and Data Collection.
For negative staining, protein sample was applied to glow-discharged, carbon-coated copper grids; stained with successive drops of 2% uranyl formate solution; and then blotted dry. Grids were either imaged on a Tecnai G2 Spirit transmission electron microscope (FEI) at 120 kV (for all Pol epsilon constructs and CMG–Δcat) or using a JEM-2100 LaB6 electron microscope (JEOL) at 120 kV (for CMG and CMG–Pol epsilon). For cryopreparation of Δcat Pol epsilon, purified sample was applied to glow-discharged C-flat grids, blotted, and plunge-freezed. Data were collected on a Tecnai F30 Polara electron microscope at 300 kV with a K2 Summit direct electron detector (Gatan, Inc.).
Single-Particle Image Processing.
Particles were picked semiautomatically in EMAN2 (50). Contrast transfer function parameters were estimated using CTFFIND4 (51). All further processing was performed in RELION (52).
Note Added in Proof.
After submission of this manuscript, a paper describing new CMG–DNA structures was published by O'Donnell, Li, and colleagues (53). This study supports the notion that the ATPase motor of the CMG is a single-stranded DNA translocase.
SI Materials and Methods
Yeast Expression Strains.
All strains are based on W303. The diploid strains yJCZ2 and yJCZ3 expressing the 11 yeast CMG subunits are each the mating product of two intermediate strains.
yJCZ2 has the following genotype: yJF39 (39): MATa pep4::KanMx4 bar1::Hph-NT1 trp1-1::TRP1pJF3 (MCM5-GAL1-10-MCM4) leu2-3::LEU2pJF4 (MCM7-GAL1-10-MCM6) ura3-1::URA3pJF6 (MCM2-GAL1-10–3xFLAG-MCM3) CDT1::CDT1-TAPTCP(HIS3)/yJCZ1: MATα pep4::KanMx4 bar1::Hph-NT1 his3-11::HIS3pJY13 (CDC45iFLAG2-GAL1-10-GAL4) (38) trp1-1::TRP1pJCZ1 (PSF1-GAL1-10-SLD5) leu2-3::LEU2pJCZ2 (PSF2-GAL1-10-PSF3) ade2-1::pJCZ3 (ADE2).
yJCZ3 has the following genotype: yAM22: MATa pep4::KanMx4 bar1::Hph-NT1 trp1-1::TRP1pJF3 (MCM5-GAL1-10-MCM4) leu2-3::LEU2pJF4 (MCM7-GAL1-10-MCM6) ura3-1::URA3pRS306/MCM2-GAL1-10-CBP-TEV-MCM3/yJCZ1: MATα pep4::KanMx4 bar1::Hph-NT1 his3-11::HIS3pJY13 (CDC45iFLAG2-GAL1-10-GAL4) (38) trp1-1::TRP1pJCZ1 (PSF1-GAL1-10-SLD5) leu2-3::LEU2pJCZ2 (PSF2-GAL1-10-PSF3) ade2-1::pJCZ3 (ADE2).
The yAJ2 strain expressing full-length Pol epsilon has been previously described (38) and has the following genotype: MATa pep4::KanMx4 bar1::Hph-NT1 trp1-1::TRP1pRS304/Pol2-GAL1-10-DPB4-TEV-CBP ura3-1::URA3pRS306/DPB2-GAL1-10-DPB3. yAJ25, yAJ17, yAJ19, and AJ43 have identical genotypes to yAJ2 with the following exceptions: yAJ25 contains Pol2 Δ1–1262aa, and yAJ17 and yAJ19 carry Pol2-MBP and Dpb3-MBP, respectively. To generate the yAJ43 strain lacking Dpb2, the pRS306 vector containing Dpb2 and Dpb3 was modified to remove Dpb2.
Protein Purification.
CMG.
Strains were induced with 2% galactose for 3 h at 30 °C, frozen dropwise in liquid nitrogen, and ground into powder in a cryogenic grinding mill (SPEX SamplePrep 6870). For CMG expressed from strain yJCZ2, cell powder was resuspended in lysis buffer (20 mM Hepes pH 7.6, 10% glycerol, 1 mM EDTA, 4 mM MgCl2, 0.1 M KCl, 1 mM ATP, 2 mM DTT) supplemented with complete protease inhibitors mixture (Roche). Following centrifugation, the cleared lysate was incubated with anti-Flag M2 affinity resin (Sigma Aldrich) at 4 °C, washed with wash buffer (25 mM Tris, pH 8.0, 10% glycerol, 1 mM MgCl2, 20 mM KOAc, 0.1 mM KCl, 1 mM ATP, 1 mM DTT), and eluted with wash buffer supplemented with 1 mg/mL Flag peptide. Peak fractions were pooled and loaded onto Mono Q 1.6/5 PC (GE Healthcare). The column was washed with the same wash buffer and eluted over 0.1 M to 1 M KCl gradient in the same buffer. CMG expressed from strain yJCZ3 was purified as previously described (22). Briefly, cell powder was resuspended in buffer C (25 mM Hepes, pH 7.6, 0.02% Tween-20, 10% glycerol, 1 mM EDTA, 1 mM EGTA) supplemented with 15 mM KCl, 2 mM MgCl2, 2 mM β-mercaptoethanol, and complete protease inhibitors mixture. KCl was then added to 100 mM, and the lysate cleared by centrifugation. The clear lysate was incubated with anti-Flag M2 affinity resin at 4 °C, washed with C-100 buffer (buffer C with 100 mM KCl and 1 mM DTT), and eluted with C-100 buffer supplemented with 0.5 mg/mL Flag peptide and complete protease inhibitors mixture. Peak fractions were pooled and loaded onto a HiTrap SP FF (GE Healthcare). The flow-through was collected and loaded onto a Mono Q 5/50 GL (GE Healthcare), washed with C-100 buffer, and eluted over 100 mM to 550 mM KCl gradient in C-100 buffer supplemented with 1 mM DTT. Peak fractions were pooled, diluted to 150 mM KCl in buffer C, loaded onto Mono Q 1.6/5 PC in buffer D (25 mM Hepes, pH 7.6, 1 mM EDTA, 1 mM EGTA, 1 mM DTT) supplemented with 150 mM KCl, and eluted over 150 mM to 550 mM KCl gradient in buffer D.
Pol epsilon.
Cells were arrested in G1 phase with 100 ng/mL alpha factor for 3 h at 30 °C. Protein expression was then induced by adding galactose to 2%, and growth continued for 3 h at 30 °C. Cells were then frozen and lysed to powder as described for the CMG strains. Pol epsilon was purified as described previously (38). Briefly, cell powder was resuspended in buffer E (25 mM Hepes, pH 7.6, 10% glycerol, 1 mM DTT) supplemented with 500 mM KOAc and complete protease inhibitors mixture. The lysate was cleared by centrifugation, and CaCl2 was added to 2 mM. The extract was incubated with calmodulin affinity resin (GE Healthcare) at 4 °C, washed with buffer E supplemented with 400 mM KOAc and 2 mM CaCl2, and eluted with buffer E supplemented with 400 mM KOAc, 2 mM EGTA, and 2 mM EDTA. The peak fractions were pooled, loaded onto a Hi-Trap Heparin column (GE Healthcare), washed with buffer E supplemented with 450 mM KOAc, and eluted over 450 mM to 1 M KOAc gradient in buffer E. Peak fractions were pooled and loaded onto a Superdex 200 10/300 GL column (GE Healthcare) equilibrated in buffer E supplemented with 400 mM KOAc.
CMGE Reconstitution.
Purified CMG was dialyzed with molar excess amounts of either full-length Pol epsilon or Pol epsilon with Pol2 Δcat (eluted from CBP affinity resin) in 25 mM Hepes, pH 7.6, 10% glycerol, 0.15 M KOAc, 1 mM DTT, 1 mM ATP, and 10 mM MgOAc at 4 °C for 1 h. The reconstituted products were incubated in the presence of 0.05% glutaraldehyde for 30 min on ice. XL was then quenched by adding glycine to a final concentration of 80 mM. The cross-linked sample was separated over 5 mL 15–45% (vol/vol) glycerol gradient in the same reconstitution buffer at 50,000 rpm, 4 °C, for 16 h in a Beckman ultracentrifuge using a SW 55Ti rotor. Fractions (310 μL each) were collected from the top of the gradient.
CMG Helicase Assay.
Helicase assay was performed as previously described (22). In brief, purified CMG was incubated with 25 mM Hepes, pH 7.6, 50 mM NaCl, 0.5 mM ATP, 10 mM MgOAc, 1 mM DTT, 0.1 μg/μL BSA, 27 μM ATPγS, and [γ-32P]-ATP–labeled fork DNA substrate for 3 h at 30 °C. Unwinding was then induced by adding ATP to 7 mM for 5 min at 30 °C and stopped with stop buffer (150 mM Tris, pH 8.0, 3% SDS, 120 mM EDTA). The unwinding product was separated using 8% nondenaturing PAGE in 0.1% SDS 1 × TBE. For the fork DNA, the sequence of the leading strand is as follows:
5′-GGCAGGCAGGCAGGCAGGCAGGCAGGCAGGCAGGCAGGCAGGCAGGCAGGCAGGCAGGCAGGCAGGCAGGTTGGCGAATTCCCATTGCCTCAGCAGATATCACCTCAGCACGACGTTGTAAAACGAG-3′.
The sequence of the lagging strand is as follows:
5′-GGCTCGTTTTACAACGTCGTGCTGAGGTGATATCTGCTGAGGCAATGGGAATTCGCCAACCTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT-3′.
Negative Stain Grid Preparation.
Carbon was evaporated onto freshly cleaved mica using a Q150TE carbon coater (Quorum Technologies) and baked overnight at 50 °C, before floating onto 400-mesh copper grids (Agar Scientific). Oven-dried carbon grids were glow-discharged for 30 s at 45 mA, with a 100× glow discharger (Electron Microscopy Sciences). For all permutations of the isolated Pol epsilon complex and for the CMG, a 4 μL drop of purified protein assembly was applied onto the grid for 2 min before staining. For the CMG–Pol epsilon and the CMG–Δcat assemblies, freshly glow-discharged grids were laid on top of a 50 μL drop from fraction 15 of the glycerol gradient and incubated overnight at 4 °C in a custom-made humidity chamber. Grids were subsequently stained directly (isolated Pol epsilon and the CMG) or quickly rinsed with two 75 μL drops of deionized water before staining (CMG–Pol epsilon and CMG–Δcat Pol epsilon), with four subsequent 75 μL drops of a fresh 2% uranyl formate solution. The staining solution was then blotted dry, and the grids were stored at room temperature before imaging.
Negative Stain EM Data Collection.
All isolated Pol epsilon preparations and the CMG–Δcat assembly were imaged on a Tecnai G2 Spirit transmission electron microscope (FEI) operated at 120 kV (Electron Microscopy Science Technology Platform, The Francis Crick Institute). Images were recorded manually using a 2k × 2k GATAN Ultrascan 100 camera at a nominal magnification of 30,000× (3.45 Å per pixel at the specimen level) at around 35 electrons per Å2. One exception was ΔDpb2, for which a 42,000× nominal magnification was used (2.46 Å per pixel at the specimen level). Between 100 and 250 micrographs were collected for each isolated Pol epsilon data set, whereas for the CMG–Δcat sample, 3,010 micrographs were recorded. The isolated CMG preparation was imaged using a JEM-2100 LaB6 electron microscope (JEOL) operated at 120 kV. Images (148) were recorded manually at a nominal magnification of 40,000× on a Ultrascan 4k × 4k CCD camera (Gatan), resulting in a 2.73 Å pixel size at the specimen level. The CMG–Pol epsilon assembly was imaged at a nominal magnification of 30,000×, resulting in a 3.65 Å pixel size. For CMG–Pol epsilon, micrographs (3,490) were recorded using the JEOL Automated Data Acquisition System (JADAS) (53) to automatically collect low-dose images at around 35 electrons per Å2. All images were acquired using a −0.5–2.0-µm defocus range.
Cryogrid Preparation, Data Collection for Δcat Pol Epsilon, and Movie Alignment.
Four microliters of reconstituted Δcat Pol epsilon at a concentration of 200 nM were applied onto a freshly glow-discharged C-flat 1/1 grid. After a 30-s incubation in 80% humidity, the grid was double-side blotted for 2.5 s using a Vitrobot (FEI) and plunged into liquid ethane. Data were collected on a Tecnai F30 Polara electron microscope operated at 300 kV and equipped with a K2 Summit direct electron detector (Gatan, Inc.) and an energy filter in zero-loss mode (GIF Quantum, Gatan, Inc.). Twenty-five–frame movies (1,772) were manually collected, with a single frame duration of 0.4 ms. Movies were acquired using SerialEM, in single-electron counting mode with a total dose of 48 e−·Å−2 and a −4.5 to 2.5 defocus range, at a constant nominal magnification of 37,037×, yielding a 1.35-Å pixel size. To correct for beam-induced drift, whole frame alignment and averaging was performed using MotionCorr (cryoem.ucsf.edu/software/driftcorr.html) (54).
Single Particle Image Processing.
Particles were picked semiautomatically in EMAN2 (50). Contrast transfer function parameters were estimated using CTFFIND4 (51). All further processing was performed in RELION (52). Image analysis was limited to 2D classification for all isolated Pol epsilon complexes. The 3D structures of the CMG and CMG–Pol epsilon assemblies were determined using routine combinations of 3D classification and refinement. Gaussian noise was used as an initial model to determine the CMG structure. The CMG structure was used as the initial model to determine the CMG–Pol epsilon structure. The CMG–Pol epsilon structure was used as an initial model to determine the CMG–Δcat structure. Eman2 (50) and Imagic (55) were used for further handling of 2D class averages.
Supplementary Material
Acknowledgments
We thank Gideon Coster, Ann Early, and Lucy Drury for help with yeast strain; Ali Alidoust and Namita Patel for yeast cultures; Svend Kjaer for support with the multi-angle light scattering measurements; Raffaella Carzaniga and Lucy Collinson (The Francis Crick Institute) and Kirsty MacLellan-Gibson (National Institute for Biological Standards and Control) for technical support with EM; and Hasan Yardimci and Hazal Kose for generous help with helicase assay. This work was supported by The Francis Crick Institute [core funding from Cancer Research UK, the UK Medical Research Council, and the Wellcome Trust (WT); to J.F.X.D. and A.C.] and two PhD fellowships from the Boehringer Ingelheim Fonds (to A.J. and F.A.A.). J.F.X.D. is the recipient of European Research Council Grant 669424-CHROMOREP and Wellcome Senior Investigator Award 106252/Z/14/Z. A.K. is supported by the WT. The Division of Structural Biology–Particle Imaging Center Electron Microscopy Facility at the University of Oxford was founded by the WT JIF Award 060208/Z/00/Z and is supported by WT Equipment Grant 093305/Z/10/Z. The WT, Medical Research Council, and Biotechnology and Biological Sciences Research Council also support the National EM facility, which enabled provision of the K2 detector at Oxford.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The EM 3D maps have been deposited in the EMDataBank, www.emdatabank.org [EMD-3642 (CMGE), EMD-3643 (CMGE ΔCat.), and EMD-3644 (CMG)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700530114/-/DCSupplemental.
References
- 1.Zhang D, O’Donnell M. The eukaryotic replication machine. Enzymes. 2016;39:191–229. doi: 10.1016/bs.enz.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Yao NY, O’Donnell ME. Evolution of replication machines. Crit Rev Biochem Mol Biol. 2016;51:135–149. doi: 10.3109/10409238.2015.1125845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci USA. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pellegrini L. The Pol α-primase complex. Subcell Biochem. 2012;62:157–169. doi: 10.1007/978-94-007-4572-8_9. [DOI] [PubMed] [Google Scholar]
- 6.Clausen AR, et al. Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation. Nat Struct Mol Biol. 2015;22:185–191. doi: 10.1038/nsmb.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pursell ZF, Isoz I, Lundström EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyabe I, Kunkel TA, Carr AM. The major roles of DNA polymerases epsilon and delta at the eukaryotic replication fork are evolutionarily conserved. PLoS Genet. 2011;7:e1002407. doi: 10.1371/journal.pgen.1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu C, et al. Strand-specific analysis shows protein binding at replication forks and PCNA unloading from lagging strands when forks stall. Mol Cell. 2014;56:551–563. doi: 10.1016/j.molcel.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Georgescu RE, et al. Mechanism of asymmetric polymerase assembly at the eukaryotic replication fork. Nat Struct Mol Biol. 2014;21:664–670. doi: 10.1038/nsmb.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson RE, Klassen R, Prakash L, Prakash S. A major role of DNA polymerase δ in replication of both the leading and lagging DNA strands. Mol Cell. 2015;59:163–175. doi: 10.1016/j.molcel.2015.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stillman B. Reconsidering DNA polymerases at the replication fork in eukaryotes. Mol Cell. 2015;59:139–141. doi: 10.1016/j.molcel.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeeles JT, Janska A, Early A, Diffley JF. How the eukaryotic replisome achieves rapid and efficient DNA replication. Mol Cell. 2017;65:105–116. doi: 10.1016/j.molcel.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barry ER, McGeoch AT, Kelman Z, Bell SD. Archaeal MCM has separable processivity, substrate choice and helicase domains. Nucleic Acids Res. 2007;35:988–998. doi: 10.1093/nar/gkl1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costa A, et al. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol. 2011;18:471–477. doi: 10.1038/nsmb.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pape T, et al. Hexameric ring structure of the full-length archaeal MCM protein complex. EMBO Rep. 2003;4:1079–1083. doi: 10.1038/sj.embor.7400010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fletcher RJ, et al. The structure and function of MCM from archaeal M. thermoautotrophicum. Nat Struct Biol. 2003;10:160–167. doi: 10.1038/nsb893. [DOI] [PubMed] [Google Scholar]
- 19.Li N, et al. Structure of the eukaryotic MCM complex at 3.8 Å. Nature. 2015;524:186–191. doi: 10.1038/nature14685. [DOI] [PubMed] [Google Scholar]
- 20.Yuan Z, et al. Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation. Nat Struct Mol Biol. 2016;23:217–224. doi: 10.1038/nsmb.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGeoch AT, Trakselis MA, Laskey RA, Bell SD. Organization of the archaeal MCM complex on DNA and implications for the helicase mechanism. Nat Struct Mol Biol. 2005;12:756–762. doi: 10.1038/nsmb974. [DOI] [PubMed] [Google Scholar]
- 22.Abid Ali F, et al. Cryo-EM structures of the eukaryotic replicative helicase bound to a translocation substrate. Nat Commun. 2016;7:10708. doi: 10.1038/ncomms10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu YV, et al. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell. 2011;146:931–941. doi: 10.1016/j.cell.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun J, et al. The architecture of a eukaryotic replisome. Nat Struct Mol Biol. 2015;22:976–982. doi: 10.1038/nsmb.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simon AC, et al. A Ctf4 trimer couples the CMG helicase to DNA polymerase α in the eukaryotic replisome. Nature. 2014;510:293–297. doi: 10.1038/nature13234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng W, D’Urso G. Schizosaccharomyces pombe cells lacking the amino-terminal catalytic domains of DNA polymerase epsilon are viable but require the DNA damage checkpoint control. Mol Cell Biol. 2001;21:4495–4504. doi: 10.1128/MCB.21.14.4495-4504.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hogg M, et al. Structural basis for processive DNA synthesis by yeast DNA polymerase ϵ. Nat Struct Mol Biol. 2014;21:49–55. doi: 10.1038/nsmb.2712. [DOI] [PubMed] [Google Scholar]
- 28.Johansson E, Macneill SA. The eukaryotic replicative DNA polymerases take shape. Trends Biochem Sci. 2010;35:339–347. doi: 10.1016/j.tibs.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 29.Tahirov TH, Makarova KS, Rogozin IB, Pavlov YI, Koonin EV. Evolution of DNA polymerases: An inactivated polymerase-exonuclease module in Pol epsilon and a chimeric origin of eukaryotic polymerases from two classes of archaeal ancestors. Biol Direct. 2009;4:11. doi: 10.1186/1745-6150-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Araki H, Hamatake RK, Johnston LH, Sugino A. DPB2, the gene encoding DNA polymerase II subunit B, is required for chromosome replication in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1991;88:4601–4605. doi: 10.1073/pnas.88.11.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dua R, Levy DL, Campbell JL. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae pol epsilon and its unexpected ability to support growth in the absence of the DNA polymerase domain. J Biol Chem. 1999;274:22283–22288. doi: 10.1074/jbc.274.32.22283. [DOI] [PubMed] [Google Scholar]
- 32.Isoz I, Persson U, Volkov K, Johansson E. The C-terminus of Dpb2 is required for interaction with Pol2 and for cell viability. Nucleic Acids Res. 2012;40:11545–11553. doi: 10.1093/nar/gks880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kesti T, Flick K, Keränen S, Syväoja JE, Wittenberg C. DNA polymerase epsilon catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol Cell. 1999;3:679–685. doi: 10.1016/s1097-2765(00)80361-5. [DOI] [PubMed] [Google Scholar]
- 34.Kang YH, Galal WC, Farina A, Tappin I, Hurwitz J. Properties of the human Cdc45/Mcm2-7/GINS helicase complex and its action with DNA polymerase epsilon in rolling circle DNA synthesis. Proc Natl Acad Sci USA. 2012;109:6042–6047. doi: 10.1073/pnas.1203734109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langston LD, et al. CMG helicase and DNA polymerase ε form a functional 15-subunit holoenzyme for eukaryotic leading-strand DNA replication. Proc Natl Acad Sci USA. 2014;111:15390–15395. doi: 10.1073/pnas.1418334111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Georgescu RE, et al. Reconstitution of a eukaryotic replisome reveals suppression mechanisms that define leading/lagging strand operation. eLife. 2015;4:e04988. doi: 10.7554/eLife.04988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hogg M, Johansson E. DNA polymerase ε. Subcell Biochem. 2012;62:237–257. doi: 10.1007/978-94-007-4572-8_13. [DOI] [PubMed] [Google Scholar]
- 38.Yeeles JT, Deegan TD, Janska A, Early A, Diffley JF. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature. 2015;519:431–435. doi: 10.1038/nature14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frigola J, Remus D, Mehanna A, Diffley JF. ATPase-dependent quality control of DNA replication origin licensing. Nature. 2013;495:339–343. doi: 10.1038/nature11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurat CF, Yeeles JT, Patel H, Early A, Diffley JF. Chromatin controls DNA replication origin selection, lagging-strand synthesis, and replication fork rates. Mol Cell. 2017;65:117–130. doi: 10.1016/j.molcel.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Asturias FJ, et al. Structure of Saccharomyces cerevisiae DNA polymerase epsilon by cryo-electron microscopy. Nat Struct Mol Biol. 2006;13:35–43. doi: 10.1038/nsmb1040. [DOI] [PubMed] [Google Scholar]
- 42.Sun J, Yuan Z, Georgescu R, Li H, O’Donnell M. The eukaryotic CMG helicase pumpjack and integration into the replisome. Nucleus. 2016;7:146–154. doi: 10.1080/19491034.2016.1174800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simon AC, Sannino V, Costanzo V, Pellegrini L. Structure of human Cdc45 and implications for CMG helicase function. Nat Commun. 2016;7:11638. doi: 10.1038/ncomms11638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burgers PM, Gordenin D, Kunkel TA. Who is leading the replication fork, Pol ε or Pol δ? Mol Cell. 2016;61:492–493. doi: 10.1016/j.molcel.2016.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pellegrini L, Costa A. New insights into the mechanism of DNA duplication by the eukaryotic replisome. Trends Biochem Sci. 2016;41:859–871. doi: 10.1016/j.tibs.2016.07.011. [DOI] [PubMed] [Google Scholar]
- 46.Gambus A, et al. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO J. 2009;28:2992–3004. doi: 10.1038/emboj.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee SJ, Richardson CC. Choreography of bacteriophage T7 DNA replication. Curr Opin Chem Biol. 2011;15:580–586. doi: 10.1016/j.cbpa.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lewis JS, Jergic S, Dixon NE. The E. coli DNA replication fork. Enzymes. 2016;39:31–88. doi: 10.1016/bs.enz.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 49.Chilkova O, et al. The eukaryotic leading and lagging strand DNA polymerases are loaded onto primer-ends via separate mechanisms but have comparable processivity in the presence of PCNA. Nucleic Acids Res. 2007;35:6588–6597. doi: 10.1093/nar/gkm741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 51.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scheres SH. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Georgescu R, et al. Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation. Proc Natl Acad Sci USA. 2017;114:E697–E706. doi: 10.1073/pnas.1620500114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.