Significance
Despite significant advances in the prediction of natural product biosynthetic gene clusters (BGCs) from microbial genomes, challenges remain for those belong to the lesser-known classes. Using a bioactivity-guided library screening approach, we have identified a BGC encoding the biosynthesis of anisomycin, an important pyrrolidine-containing protein synthesis inhibitor. The biosynthetic pathway is distinct from known bacterial alkaloid pathways and involves a class of natural product backbone biosynthesis genes encoding an α-keto acid-incorporating transketolase. A cryptic but crucial glycosylation, unexpected transaminations, and a multistep pyrrolidine-forming reaction catalyzed by a single enzyme are also required to complete the assembly of the core benzylpyrrolidine scaffold. These findings open up new avenues for genomics-guided natural product discovery and engineering of pyrrolidine antibiotics.
Keywords: pyrrolidine antibiotics, anisomycin biosynthetic pathway, protein synthesis inhibitor, cryptic glycosylation, Streptomyces
Abstract
The protein synthesis inhibitor anisomycin features a unique benzylpyrrolidine system and exhibits diverse biological and pharmacologic activities. Its biosynthetic origin has remained obscure for more than 60 y, however. Here we report the identification of the biosynthetic gene cluster (BGC) of anisomycin in Streptomyces hygrospinosus var. beijingensis by a bioactivity-guided high-throughput screening method. Using a combination of bioinformatic analysis, reverse genetics, chemical analysis, and in vitro biochemical assays, we have identified a core four-gene ensemble responsible for the synthesis of the pyrrolidine system in anisomycin: aniQ, encoding a aminotransferase that catalyzes an initial deamination and a later reamination steps; aniP, encoding a transketolase implicated to bring together an glycolysis intermediate with 4-hydroxyphenylpyruvic acid to form the anisomycin molecular backbone; aniO, encoding a glycosyltransferase that catalyzes a cryptic glycosylation crucial for downstream enzyme processing; and aniN, encoding a bifunctional dehydrogenase that mediates multistep pyrrolidine formation. The results reveal a BGC for pyrrolidine alkaloid biosynthesis that is distinct from known bacterial alkaloid pathways, and provide the signature sequences that will facilitate the discovery of BGCs encoding novel pyrrolidine alkaloids in bacterial genomes. The biosynthetic insights from this study further set the foundation for biosynthetic engineering of pyrrolidine antibiotics.
Anisomycin (1), a pyrrolidine antibiotic (Fig. 1), was first isolated from Streptomyces roseochromogenes and Streptomyces griseolus in 1954 (1). Later it was also found in Streptomyces hygrospinosus var. beijingensis (2). The structure and stereochemistry of 1 were solved by X-ray crystallographic analysis (3). 1 features a unique pyrrolidine structure with a trans-diol and exhibits diverse biological and pharmacologic activities. Owing to its reversible 60S ribosomal subunit binding, 1 can block peptide bond formation and shows potent selective activity against pathogenic protozoa and fungi (4). It has been successfully used in the clinics for the treatment of amoebic dysentery and trichomoniasis, and it is also one of the important effective components in Agricultural Antibiotic 120, which has been widely used for controlling crop diseases in China (2). 1 has been shown to disrupt traumatic memory consolidation, and thus it has emerged as a useful tool for studying the memory processes and as a potential psychiatric drug (5, 6). Moreover, 1 has been identified as a potential antitumor substance exhibiting potent cytotoxicity against human tumor cell lines in vitro (7, 8). Recently, 1 has been widely used as an activation agent in mitogen-activated protein kinase signaling pathways (9).
Fig. 1.
Identification of the biosynthetic gene cluster for anisomycin (1) and functional analysis by gene deletions. (A) Bioactivity-guided high-throughput library screening for localizing the ani cluster. (B) Organization of the ani gene cluster. Each gene was deleted in-frame. (C) Putative anisomycin biosynthetic route based on the accumulated intermediates in individual gene-deletion mutants.
The diverse biological activities and structural features of 1 have attracted the attention of many chemists and biologists. Chemical synthesis of 1 and its analogs have been achieved (10, 11); however, biosynthetic studies have been limited to preliminary isotope feeding experiments. A previous precursor feeding study showed that the aromatic ring and C2 of the pyrrolidine ring of 1 to be originated from tyrosine, whereas C4 and C5 were proposed to be derived from glycine or acetate (12). The methyl group of 1 was demonstrated to be originated from methionine, with deacetylanisomycin as an intermediate during 1 biosynthesis. Nevertheless, the origin of the nitrogen and the carbon (C3) for the biosynthesis of pyrrolidine has remained unknown (12).
Here we report the identification of the biosynthetic gene cluster (BGC) of 1 from S. hygrospinosus var. beijingensis using a bioactivity-guided library screening approach. The identified BGC does not fit into any of the known classes. Gene functional analysis, in vivo gene inactivation, in vitro biochemical assays, and detailed chemical analysis have identified nine genes responsible for the production of 1. The results reveal a previously unknown pathway for the biosynthesis of pyrrolidine that is strikingly different from the previously characterized nonribosomal peptide synthetase (NRPS) pathways (13), in which several unexpected enzymes and cryptic biosynthetic steps are involved in the biosynthesis of 1.
Results
Identification and Verification of the Anisomycin BGC.
The genome of S. hygrospinosus var. beijingensis was previously sequenced (genome size 9.09 Mbp) (14). Given the lack of knowledge regarding the biosynthesis of this class of compounds, the BGC for 1 cannot be confidently predicted from the genome. Thus, we used a bioactivity-guided high-throughput library screening approach to identify the 1 gene cluster (15) (Fig. 1A). First, we constructed an S. hygrospinosus var. beijingensis genomic library and transferred it into Streptomyces lividans TK24 (Fig. 1A and SI Appendix, Fig. S1). Because 1 has antifungal activity, the resulting S. lividans exconjugants were screened by a high-throughput anti-yeast assay. Four exconjugants exhibited strong inhibition against Saccharomyces cerevisiae (SI Appendix, Fig. S1B). HPLC-MS analysis of the fermentation broth showed that all four S. lividans exconjugants (individually harboring cosmids 26D9, 5E10, 25G11, and 18C6) produced 1 (SI Appendix, Fig. S2). This finding confirms that these cosmids contain the complete BGC for 1. To further verify involvement of the genes harbored by the cosmids in the biosynthesis of 1, we constructed a large fragment deletion mutation from S. hygrospinosus var. beijingensis, ZXQ01. We found that ZXQ01 completely lost the ability to produce 1, which could be restored when ZXQ01 was complemented with 26D9 cosmid (SI Appendix, Fig. S3). This confirms that the region corresponding to 26D9 indeed contains the 1 BGC (herein referred as the ani cluster).
To narrow down the genes involved in the biosynthesis of 1, we performed terminal sequencing of the four cosmid inserts. Alignment with the S. hygrospinosus var. beijingensis genome sequences led to the identification of an overlapping 28.67-kb region among 26D9, 5E10, 25G11, and 18C6 cosmids that contains 25 ORFs (SI Appendix, Fig. S1B and Table S3). To identify the genes responsible for biosynthesis of 1, we individually deleted the 25 ORFs on the 26D9 cosmid and transferred the mutant cosmids to S. lividans TK24. A metabolite profile analysis of the S. lividans TK24 strains harboring the mutant 26D9 cosmids suggested that eight of the genes (aniF, aniI, aniK, aniL, aniN, aniO, aniP, and aniQ) are essential for the biosynthesis of 1, because strains lacking these individual genes lost the ability to produce 1. However, the production of the pathway intermediates is very low in S. lividans TK24, which hinders further biosynthetic pathway elucidation. For this reason, the expression of the 26D9 mutant cosmids was repeated in ZXQ01, which lacks the genomic region corresponding to the 26D9 insert (SI Appendix, Fig. S3).
Like the S. lividans TK24, ZXQ01 strains harboring 26D9 mutant cosmids lacking the foregoing eight genes (ZXQ02–07, ZXQ09, and ZXQ10) lost the ability to produce 1; several of these mutants accumulated various intermediates (Fig. 1C and SI Appendix, Fig. S4). Further bioinformatics analysis predicted that aniF, aniI, aniK, and aniL are involved in transcriptional regulation, tailoring modifications (O-acetylation and O-methylation), and transport, whereas aniN, aniO, aniP, and aniQ may be responsible for biosynthesis of the 1 core scaffold (Fig. 1B and Table 1).
Table 1.
Deduced functions of ORFs in the anisomycin biosynthetic gene cluster
| Gene | Size, aa | Predicted function |
| aniF | 208 | LuxR family transcriptional regulator |
| aniG | 577 | α-glucosidase |
| aniI | 222 | O-acetyltransferase |
| aniK | 331 | SAM-dependent O-methyltransferase |
| aniL | 393 | Major facilitator superfamily transporter |
| aniN | 275 | NAD(P)-dependent short-chain dehydrogenase |
| aniO | 462 | Glycosyltransferase |
| aniP | 603 | Transketolase |
| aniQ | 441 | PLP-dependent aminotransferase |
Characterization of the Tailoring Genes in Anisomycin Biosynthesis Revealed Unexpected Glycosylated Intermediates.
We first sought to characterize the function of the tailoring genes aniI and aniK encoding an O-acetyltransferase and an O-methyltransferase, respectively. The ZXQ01 26D9∆aniI mutant (ZXQ04) accumulated a new intermediate, 5, with an m/z of 224.1287 [M+H]+, which we structurally characterized as deacetylanisomycin (Fig. 1 and SI Appendix, Fig. S4). Thus, as expected, the 3-O-acetylation by AniI is not required for pyrrolidine core biosynthesis. Interestingly, ZXQ03 lacking the O-methyltransferase gene aniK accumulated several intermediates (3, 3a, 4, and 4a), which exhibited a UV spectrum very similar to that of 1 but with a different retention time on HPLC (SI Appendix, Fig. S4B). Quadrupole time-of-flight (Q-TOF) analysis revealed that 3, 3a, 4, and 4a yielded [M+H]+ ions at m/z of 372.1788, 210.1180, 414.1713, and 252.1400, respectively (SI Appendix, Fig. S4C). The ZXQ03 culture was upscaled, and 3–4a were obtained for structural elucidation. Comprehensive NMR analysis identified the compounds 3 and 3a, 4 and 4a as two pairs (SI Appendix, Fig. S16 and Table S4). Compared with 3a and 4a, which were identified as desmethyl-deacetylanisomycin and desmethyl-anisomycin, respectively, compounds 3 and 4 contained an additional glucose unit not found in the final product 1. No glycosylated derivative of 1 had been reported or detected previously; thus, this highly unexpected finding suggests that a cryptic glycosylation reaction occurred during the biosynthesis of 1.
To further corroborate the function of AniK, the protein was expressed and purified from Escherichia coli BL21(DE3) (SI Appendix, Fig. S5). Here 3a or 4a was incubated with AniK along with S-adenosylmethionine (SAM), and the reaction mixtures were analyzed by LC-MS. Conversion of 3a and 4a to 5 and 1, respectively, were detected along with SAM to S-adenosylhomocysteine (SAH) (Fig. 2). This demonstrates that AniK is responsible for methylation of the phenyl moiety in 1.
Fig. 2.
Reconstitution of methylation catalyzed by AniK in vitro. (A) HPLC profiles of (i) standard 5, (ii) standard 3a, (iii) boiled AniK incubated with 3a as a negative control, and (iv) AniK incubated with 3a. (B) HPLC profiles of (i) standard 1, (ii) standard 4a, (iii) boiled AniK incubated with 4a as a negative control, and (iv) AniK incubated with 4a.
An Unusual Short-Chain Dehydrogenase Is Involved in the Synthesis of the Pyrrolidine Core.
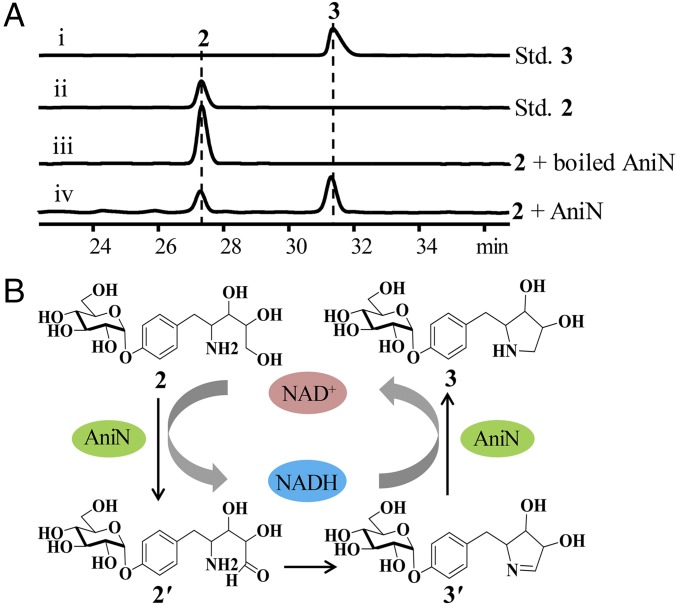
ZXQ02, another mutant strain (26D9∆aniN), yielded a pathway intermediate 2 with a UV spectrum similar to 1 and [M+H]+ ions at an m/z of 390.1726 (SI Appendix, Fig. S4). We managed to purify 2 from a larger-scale culture of ZXQ02. Based on NMR analysis, 2 was found to be an open ring intermediate of the 1 pathway containing a glycosyl moiety like 3 and 4 (SI Appendix, Fig. S17 and Table S5). This suggests that AniN, which exhibits high sequence homology to a family of NAD(P)-dependent short chain dehydrogenases, is involved in the synthesis of the five-membered pyrrolidine ring and likely acts on glycosylated intermediate 2. We hypothesized that AniN catalyzes the dehydrogenation of hydroxyl in 2 to form an aldehyde, which can be spontaneously cyclized by condensation with the amine to form an imine before eventually reducing to 3. To validate this hypothesis, we incubated purified AniN with NAD+ and 2, and then subjected the reaction mixtures to LC-MS analysis. Surprisingly, AniN was able to convert 2 to a single product with an [M+H]+ ion at m/z 372.1788 corresponding to 3 (Fig. 3A). These results demonstrate that AniN can catalyze a multistep reaction converting 2 directly to 3 without the involvement of another enzyme.
Fig. 3.
Reconstitution of pyrrolidine assembly catalyzed by dehydrogenase AniN in vitro. (A) HPLC profiles of (i) standard 3, (ii) standard 2, (iii) boiled AniN incubated with 2 as a negative control, and (iv) AniN incubated with 2. (B) The hypothetical mechanism of the AniN reaction.
A time course analysis of the reaction revealed that, as expected, the production of 3 and consumption of 2 proceeded in a time-dependent manner, but no reaction intermediate was detected (SI Appendix, Fig. S6A). In addition, we detected an initial increase in NADH, followed by a gradual decrement during the course of the reaction, and found that the reaction could be inhibited by adding the aldehyde dehydrogenase inhibitor methylene blue (SI Appendix, Fig. S6 B and C). These results demonstrate that both oxidation and reduction occur during the conversion of 2 to 3, likely via an initial oxidative dehydrogenation of 2 to the corresponding aldehyde 2′. A spontaneous redox-neutral condensation between the amine and aldehyde resulted in the imine 3′, which was further reduced by AniN to form 3 (Fig. 3B).
AniQ Is a Tyrosine Aminotransferase Catalyzing the First Step in the Anisomycin Pathway.
Earlier isotope-labeled precursor feeding experiments have established that 1 biosynthesis likely is initiated by utilization of tyrosine and glycine or acetate as starting substrates (12); however, the origin of the nitrogen atom in 1 has remained obscure. We performed feeding experiments using 15N-glycine and LD-15N-tyrosine, which demonstrated that the nitrogen does not come from either glycine or tyrosine (SI Appendix, Fig. S7). Based on the presence of an aminotransferase gene aniQ in the ani cluster, we hypothesized that the nitrogen from tyrosine might have been exchanged at some stage during the biosynthesis of 1, perhaps via transamination. The 26D9ΔaniQ strain (ZXQ07) completely lost the ability to produce 1, and no intermediate was accumulated in the culture (SI Appendix, Fig. S4B). The production of 1 can be partially restored in ZXQ07 when complemented with aniQ (SI Appendix, Fig. S8A). Thus, AniQ may act at the beginning of the pathway, perhaps using tyrosine as a substrate.
To test the foregoing hypothesis, we expressed AniQ in E. coli BL21 (DE3). The purified recombinant protein was yellowish with a characteristic λmax at 417 nm (SI Appendix, Fig. S9A), indicating that the enzyme was bound to the pyridoxal 5′-phosphate (PLP) cofactor. AniQ was assayed using l-tyrosine or d-tyrosine as substrate in the presence of PLP as the cofactor and α-ketoglutarate (α-KG) as the amine acceptor. We used heat-inactivated AniQ as a negative control and the purified TyrB, which had been confirmed to catalyze the transamination of l-tyrosine (16), as a positive control. Both AniQ and TyrB could convert l-tyrosine to a distinct product, 6, which shares the same retention time and Q-TOF data with the synthesized authentic 4-hydroxyphenylpyruvic acid (Fig. 4A and SI Appendix, Fig. S9C); however, AniQ cannot accept d-tyrosine as a substrate (SI Appendix, Fig. S9B).
Fig. 4.
Reconstitution of two transamination reactions catalyzed by AniQ and glycosylation by AniO in vitro. (A) HPLC profiles of (i) standard 6 in 50 mM Hepes buffer (pH 8.0), (ii) boiled AniQ incubated with l-tyrosine as a negative control, (iii) TyrB incubated with l-tyrosine as a positive control, and (iv) AniQ incubated with l-tyrosine. l-Tyr, l-tyrosine. (B) HPLC profiles of (i) standard 2, (ii) standard 8, (iii) boiled AniQ incubated with 8 as a negative control, and (iv) AniQ incubated with 8. (C) HPLC profiles of (i) standard 8, (ii) boiled AniO incubated with 7 as a negative control, and (iii) AniO incubated with 7.
The foregoing in vitro results also suggest that the α-keto acid 6 is the first intermediate in the 1 pathway. This resonates with the presence of aniP, which encodes a protein with a conserved transketolase domain similar to the 1-deoxy-d-xylulose-5-phosphate (DXP) synthase in the nonmevalonate isoprenoid pathway (17, 18). DXP synthase is a thiamine diphosphate-dependent enzyme that generates DXP from pyruvate and glyceraldehyde-3-phosphate (G3P). Likewise, AniP may convert the α-keto acid 6 and G3P to the putative intermediate 7 via a transketolase-type condensation reaction (SI Appendix, Fig. S10A). Like the ΔaniQ, the 26D9ΔaniP strain (ZXQ06) completely lost the ability to produce 1 and did not accumulate any intermediate in the culture (SI Appendix, Fig. S4B).
We attempted to characterize the enzyme activity of AniP, but unfortunately, the recombinant AniP protein has poor solubility, and we failed to reconstitute the activity in vitro. Given that glycerol can be converted to G3P via a glycerol utilization pathway in Streptomyces (19), we fed 1,3-13C2-labeled glycerol into S. hygrospinosus var. beijingensis culture. Indeed, Q-TOF analysis revealed significant enrichment of the +2-Da isotope peak (m/z 268.1295 [M+H]+) in the mass spectrum of 1 compared with the negative control (SI Appendix, Fig. S10B), supporting the proposed function for AniP.
The Second Transamination by AniQ Is Dependent on a Cryptic Glycosylation by AniO.
The implication of the aforementioned AniQ and AniP function is that an amino group needs to be reintroduced into the pathway to afford 2 as the substrate for AniN to form the pyrrolidine moiety. Given that AniQ is the only aminotransferase encoded in the ani cluster, we postulated that it also might be capable of converting 7 to 2a or 8 to 2. Because substrates 7 and 8 were unavailable, and because several aminotransferases have been shown to catalyze the reverse reaction (20), we first tested whether AniQ can convert 2 to 8 by incubating AniQ with 2 in the presence of PLP and α-KG. LC-MS analysis showed that AniQ converted 2 to a single product 8 with the [M+NH4]+ ion at m/z 406.1644, 1 Da less than that for 2 (SI Appendix, Fig. S11A), suggesting successful transamination of 2. We then scaled up the enzymatic reaction and purified 2.5 mg of 8 for NMR analysis and confirmed its structure (SI Appendix, Fig. S18 and Table S6). Equipped with 8, we next tested the forward reaction of AniQ in this second transamination. We assayed AniQ with 8 and PLP along with various l-amino acids as the amine donor. HPLC analysis of the enzymatic reactions revealed the conversion of 8 to 2 (Fig. 4B). Surprisingly, although AniQ uses α-KG as amine acceptor, l-glutamine was identified as the best amine donor. Approximately 92% of the substrate 8 could be converted to 2 within 20 min with 20 μM AniQ (SI Appendix, Fig. S11 B and C).
We next sought to investigate the effect of the glycosyl group on the second transamination. The presence of a glucose unit among the pathway intermediates (2, 3, and 4) was puzzling, because the final product 1 does not contain any glucosyl moiety. Because the ani cluster encodes an α-glucosidase AniG, we tested whether AniG could deglycosylate 2 to 2a, which would provide the substrate required to assay AniQ. LC-MS analysis showed that the purified AniG could efficiently convert 2 to a new product with mass ion m/z of 228.1221 (C11H17NO4; calcd. 228.1230 [M+H]+), 162 Da less than that of 2 (C17H27NO9; calcd. 390.1759 [M+H]+) (SI Appendix, Fig. S12A). This indicates that the glycosyl unit has been removed from 2. By performing this enzymatic hydrolysis reaction on a larger scale, we purified 10 mg of 2a and confirmed its structure by NMR analysis (SI Appendix, Fig. S17 and Table S5). We also demonstrated that AniG can efficiently deglycosylate 3 to 3a and 4 to 4a (SI Appendix, Fig. S12 B and C).
Having obtained a good amount of 2a, we next assayed AniQ with 2a using α-KG as amine acceptor. We were able to observe the production of a new peak with a mass ion at m/z 249.4 ([M+Na]+), which was 1 Da less than that of 2a, suggesting successful transamination of 2a. The molecular formula of this new product with m/z 249.0734 [M+Na]+ (calcd. 249.0733) as determined by Q-TOF is C11H14O5, which corresponds to 7 (SI Appendix, Fig. S11D). We scaled up the AniQ enzymatic reaction and purified this new compound. The full NMR analysis revealed that it was indeed 7 (SI Appendix, Fig. S18 and Table S6). Accordingly, we assayed the forward reaction of AniQ with 7 using l-glutamine as the amine donor. The results show that the glucosyl unit plays an important role in the biosynthesis of 1, because AniQ transaminates the glycosylated substrates at a 10-fold faster rate than the nonglycosylated equivalents (SI Appendix, Fig. S11E). Taken together, our results demonstrate that AniQ is responsible for two different transamination reactions during the biosynthesis of 1.
The selectivity of AniQ toward glycosylated substrates prompted us to reinvestigate AniN. Using 2a obtained above, we found that, besides 2, AniN can accept the nonglycosylated 2a and form the pyrrolidone ring to afford 3a. Comparing the initial conversion rate at the same enzyme concentration, AniN is able to convert 2 to 3 at an approximately threefold-faster rate than it can convert 2a to 3a (SI Appendix, Fig. S11F). Therefore, like AniQ, AniN prefers glycosylated substrate over the nonglycosylated substrate.
AniO Is Responsible for the Cryptic Glycosylation and the Subsequent Deglycosylation Is Important for the Bioactivity of 1.
We next turned our attention to the cryptic but crucial glycosylation reaction in the biosynthesis of 1. Indeed, the ani cluster encodes a putative UDP-glucose–dependent glycosyltransferase AniO. The accumulation of 2 in the ZXQ02 strain lacking aniN suggests that the glycosyl transfer occurred before the pyrrolidine formation catalyzed by AniN. Inactivation of aniO on 26D9 (ZXQ05) led to the loss of 1 production, but not to accumulation of the expected nonglycosylated 2a or of any other intermediates (SI Appendix, Fig. S4B). The production of 1 in ZXQ05 can be partially restored when complemented with aniO (SI Appendix, Fig. S8B). We suspected that AniO is responsible for glycosylating 7 to 8, which in turn can be reaminated by AniQ to 2. Fortuitously, 7 was made available earlier from 2 via the two-step enzymatic transformation using AniG and AniQ.
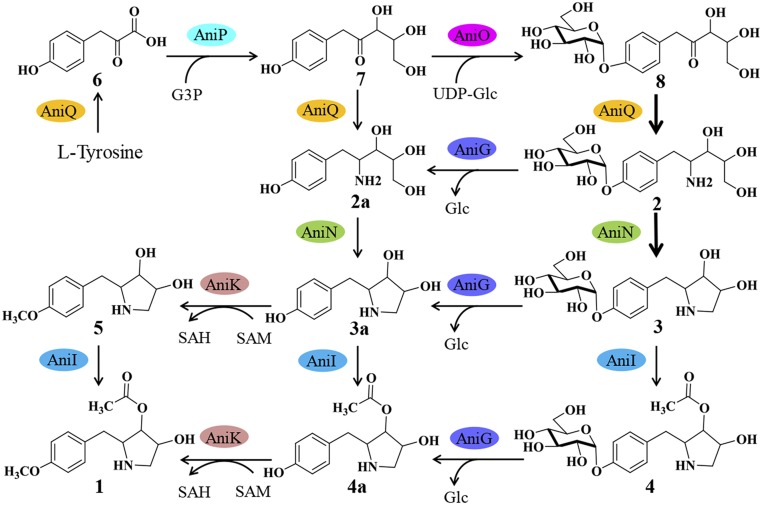
Unable to express AniO as a soluble His-tagged protein, we expressed AniO as an N-terminal SUMO-tagged protein in E. coli BL21(DE3). In the presence of UDP-glucose, we assayed AniO with l-tyrosine, 6, and 7, respectively, and also tested compounds 2a, 3a, and 4a as potential substrates for AniO. The results show that AniO could only convert 7 to a new product, 8, but could not recognize l-tyrosine, 6, 2a, 3a, and 4a as substrates (Fig. 4C and SI Appendix, Fig. S13). Thus, AniO is responsible for the unexpected glycosyltransfer reaction in the 1 pathway by converting the intermediate 7 to 8, which acts as the main substrate for AniQ to form 2 (Fig. 5).
Fig. 5.
Proposed anisomycin biosynthetic pathway.
The accumulation of various derivatives of 1 in this study provided us with a good opportunity to test the structure-activity relationship and investigate the effect of various tailoring modifications on the bioactivity of 1. Interestingly, among the compounds tested (3, 3a, 4, 4a, and 1 as a positive control), only 4a exhibited growth-inhibiting activity on an agar plate assay, similar to that of 1; the other compounds showed no activity (SI Appendix, Fig. S14A). These results demonstrate that glycosylation at the phenol ring significantly reduces the bioactivity of 1, while the acetylation at C3 is also important for bioactivity.
Discussion
Significant advances have been made in the ability to search for and predict BGCs from microbial genomes. BGCs with well-known backbone biosynthetic enzymes, such as polyketide synthases (PKSs), NRPSs, and terpene synthases, can be readily identified with the aid of bioinformatics software, such as antiSMASH 3.0 (21). Nonetheless, challenges remain in identifying those biosynthetic gene clusters lacking the obvious or well-known backbone biosynthetic genes. 1 is an example of such natural products that are difficult to categorize into the well-known natural product classes, and identifying its BGC was challenging. The sole isotope feeding study reported in the 1960s provided some insight into the biosynthetic origin of 1 (12). The involvement of tyrosine and potentially glycine or acetate prompted us to focus initially on the NRPS-containing gene clusters in S. hygrospinosus var. beijingensis; however, the deletion of multiple candidate NRPS gene clusters did not affect the production of 1. We also attempted to identify candidate gene clusters from the cosmid library with phenol O-methyltransferase as a gene probe, but without success. Eventually, we successfully identified the ani cluster using a bioactivity-guided high-throughput library screening approach (Fig. 1A) (15). This study has finally revealed the molecular genetic basis for biosynthesis of 1, which has been unknown since it was isolated more than 60 y ago (1).
The identity of the ani cluster was further masked by the involvement of several unexpected cryptic enzymatic steps in the biosynthesis of 1. The presence of the aminotransferase gene aniQ and the glycosyltransferase gene aniO was unexpected, because tyrosine readily provides the nitrogen in the pyrrolidine ring, whereas glycosylated anisomycin derivatives were never reported or observed. Through construction of 24 gene inactivation strains, elucidation of the structures for eight pathway intermediates, and isotope feeding and biochemical characterization of five biosynthetic enzymes (AniN, O, Q, G, and K), we can now confidently propose a pathway for biosynthesis of 1 (Fig. 5).
The precursor l-tyrosine is first deaminated by AniQ to yield 4-hydroxyphenylpyruvic acid 6. This sets the stage for the transketolase-type condensation reaction between 6 and G3P catalyzed by AniP to yield 7. This is followed by the cryptic and seemingly redundant glycosylation step catalyzed by AniO. Several cryptic reactions in other natural product biosynthetic pathways have been reported recently. Cryptic methyl esterification (22), chlorination (23), acylation (24), and demethoxylation (25, 26) have been shown to be indispensable in the biosynthesis of streptonigrin, coronatine, saframycin, and xantholipin, respectively. However, to the best of our knowledge, glucose, which is the main carbon source, is used to mediate the biosynthesis of natural products, has not been previously reported in microbial natural product pathways. We first suspected that glycosylation protects the host from toxic intermediates, and assayed the inhibitory activity of 7 and 8 on S. hygrospinosus var. beijingensis and the mutant ZXQ01 (∆26D9) strain, but found that they had no influence on WT or ZXQ01 growth at concentrations of up to 250 μg (SI Appendix, Fig. S14B).
Biochemical characterization showed that the enzymes immediately downstream of AniO (AniQ and AniN) have strong selectivity toward glycosylated substrates. Thus, the cryptic glycosylation step is crucial for enzyme substrate recognition and for the biosynthesis of 1 to proceed smoothly. Glycosylation also may protect the intermediates from being degraded or consumed in the host cell. This is supported by the fact that no unglycosylated intermediates were observed in the ΔaniO strain.
After formation of the glycosylated 8, the amino group was reintroduced into the pathway by AniQ to form 2. As mentioned earlier, this second transamination step of AniQ is highly selective for the glycosylated 8 compared with the unglycosylated counterpart 7 with a 10-fold difference in the conversion rate. The next step catalyzed by AniN has strong selectivity for glycosylated substrate as well. AniN is a unique alcohol dehydrogenase that mediates pyrrolidine formation, likely via an initial alcohol dehydrogenation step, followed by spontaneous annulation and then imine reduction. Although the involvement of an alcohol dehydrogenase in pyrrolidine formation has not been reported previously, the mechanism of pyrrolidine formation in 1 is similar to that of the proline biosynthesis and 2-methyl-3-n-amyl-dihydropyrrole formation in prodigiosin biosynthesis (27, 28).
On formation of the pyrrolidine system, 3 is deglycosylated by AniG, acetylated by AniI, and methylated by AniK. The three tailoring reactions need not follow any particular sequence, except that deglycosylation must precede O-methylation on the phenol moiety. All three tailoring enzymes are relatively flexible and can tolerate different substrates (Fig. 2 and SI Appendix, Fig. S12). This likely accounts for the observation that the 1 producer strain is very “clean” (i.e., no accumulation of pathway intermediates in the WT strain), as the flexible enzymes can redirect shunt intermediates back into the pathway to 1.
Interestingly, deletion of the α-glucosidase gene aniG had no effect on the production of 1; however, in in vitro assays, AniG could efficiently convert 3 to 3a and 4 to 4a (SI Appendix, Fig. S12), confirming that AniG is indeed involved in the biosynthesis of 1. Three other putative α-glucosidases, ORF 5229 (52%), ORF 5233 (54%), and ORF 0931 (52%), were found encoded elsewhere in the S. hygrospinosus var. beijingensis genome, and one putative α-glucosidase (70%) was found in the S. lividans TK24 genome. All exhibited high sequence homology to AniG, and it is possible that they could complement the function of AniG in vivo.
Together, AniQ, AniP, AniO, and AniN compose an enzyme ensemble capable of forming pyrrolidine ring systems. A genomic search using MultiGeneBlast (29) showed that homologs of such biosynthetic gene ensembles can be found in several Streptomyces species as well as in two Proteobacteria, Pseudomonas protegens and Chromobacterium violaceum (SI Appendix, Fig. S15). As such, the distribution of this class of BGCs is not restricted to the Actinobacteria. Interestingly, these Proteobacteria lack the homolog for the glycosyltransferase gene aniO, suggesting that the corresponding pathways have diverged and do not involved any glycosylated intermediates. It is tempting to speculate that these homologous BGCs may encode novel pyrrolidine antibiotics.
In summary, this study reveals a previously undescribed BGC encoding the important protein synthesis inhibitor 1, and provides valuable insight into the biosynthesis of pyrrolidine alkaloids. The findings provide opportunities for the genome mining of novel pyrrolidine natural products and set the foundation for engineering the biosynthesis of 1 to advance future drug development.
Materials and Methods
Bacterial strains, plasmids, and primers are listed SI Appendix, Tables S1 and S2. Methods for genomic library construction, generation of Streptomyces mutants, structure elucidation, in vitro biochemical assays, and isotope feeding are included in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Prof. Meifeng Tao for supporting our high-throughput screening experiment. This work was supported by grants from the National Natural Science Foundation of China (31630002, 21661140002, 31470183, and 31400029), the Ministry of Science and Technology, the Shanghai Pujiang Program from the Shanghai Municipal Council of Science and Technology, and the Australian Research Council (FT160100233).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. KY014292).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1701361114/-/DCSupplemental.
References
- 1.Sobin BA, Tanner FW. Anisomycin, a new anti-protozoan antibiotic. J Am Chem Soc. 1954;76:4053. [Google Scholar]
- 2.Wang X, Ren Q, Tong Z, Gu H, Zhang Z. Studies on the effective components of agricultural antibiotic 120. Zhongguo Shengwu Fangzhi Xuebao. 1994;10:131–134. [Google Scholar]
- 3.Butler K. Anisomycin, III: Conformational studies of the pyrrolidine ring. J Org Chem. 1968;33:2136–2141. [Google Scholar]
- 4.Grollman AP. Inhibitors of protein biosynthesis, II: Mode of action of anisomycin. J Biol Chem. 1967;242:3226–3233. [PubMed] [Google Scholar]
- 5.Flood JF, Bennett EL, Orme AE, Rosenzweig MR, Jarvik ME. Memory: Modification of anisomycin-induced amnesia by stimulants and depressants. Science. 1978;199:324–326. doi: 10.1126/science.619461. [DOI] [PubMed] [Google Scholar]
- 6.Nader K, Schafe GE, Le Doux JE. Fear memories require protein synthesis in the amygdala for reconsolidation after retrieval. Nature. 2000;406:722–726. doi: 10.1038/35021052. [DOI] [PubMed] [Google Scholar]
- 7.Hosoya Y, Kameyama T, Naganawa H, Okami Y, Takeuchi T. Anisomycin and new congeners active against human tumor cell lines. J Antibiot (Tokyo) 1993;46:1300–1302. doi: 10.7164/antibiotics.46.1300. [DOI] [PubMed] [Google Scholar]
- 8.Mawji IA, et al. A chemical screen identifies anisomycin as an anoikis sensitizer that functions by decreasing FLIP protein synthesis. Cancer Res. 2007;67:8307–8315. doi: 10.1158/0008-5472.CAN-07-1687. [DOI] [PubMed] [Google Scholar]
- 9.Cano E, Doza YN, Ben-Levy R, Cohen P, Mahadevan LC. Identification of anisomycin-activated kinases p45 and p55 in murine cells as MAPKAP kinase-2. Oncogene. 1996;12:805–812. [PubMed] [Google Scholar]
- 10.Ballini R, Marcantoni E, Petrini M. A nitrone-based approach to the enantioselective total synthesis of (-)-anisomycin. J Org Chem. 1992;57:1316–1318. [Google Scholar]
- 11.Joo JE, Lee KY, Pham VT, Tian YS, Ham WH. Application of Pd(0)-catalyzed intramolecular oxazine formation to the efficient total synthesis of (-)-anisomycin. Org Lett. 2007;9:3627–3630. doi: 10.1021/ol701519b. [DOI] [PubMed] [Google Scholar]
- 12.Butler K. Anisomycin, II: Biosynthesis of anisomycin. J Org Chem. 1966;31:317–320. doi: 10.1021/jo01339a503. [DOI] [PubMed] [Google Scholar]
- 13.Walsh CT, Garneau-Tsodikova S, Howard-Jones AR. Biological formation of pyrroles: Nature’s logic and enzymatic machinery. Nat Prod Rep. 2006;23:517–531. doi: 10.1039/b605245m. [DOI] [PubMed] [Google Scholar]
- 14.Cao B, et al. Genome mining of the biosynthetic gene cluster of the polyene macrolide antibiotic tetramycin and characterization of a P450 monooxygenase involved in the hydroxylation of the tetramycin B polyol segment. ChemBioChem. 2012;13:2234–2242. doi: 10.1002/cbic.201200402. [DOI] [PubMed] [Google Scholar]
- 15.Xu M, et al. Functional genome mining for metabolites encoded by large gene clusters through heterologous expression of a whole-genome bacterial artificial chromosome library in Streptomyces spp. Appl Environ Microbiol. 2016;82:5795–5805. doi: 10.1128/AEM.01383-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayashi H, Inoue K, Nagata T, Kuramitsu S, Kagamiyama H. Escherichia coli aromatic amino acid aminotransferase: Characterization and comparison with aspartate aminotransferase. Biochemistry. 1993;32:12229–12239. doi: 10.1021/bi00096a036. [DOI] [PubMed] [Google Scholar]
- 17.Rohmer M, Knani M, Simonin P, Sutter B, Sahm H. Isoprenoid biosynthesis in bacteria: A novel pathway for the early steps leading to isopentenyl diphosphate. Biochem J. 1993;295:517–524. doi: 10.1042/bj2950517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lange BM, Wildung MR, McCaskill D, Croteau R. A family of transketolases that directs isoprenoid biosynthesis via a mevalonate-independent pathway. Proc Natl Acad Sci USA. 1998;95:2100–2104. doi: 10.1073/pnas.95.5.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baños S, Pérez-Redondo R, Koekman B, Liras P. Glycerol utilization gene cluster in Streptomyces clavuligerus. Appl Environ Microbiol. 2009;75:2991–2995. doi: 10.1128/AEM.00181-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maeda H, Yoo H, Dudareva N. Prephenate aminotransferase directs plant phenylalanine biosynthesis via arogenate. Nat Chem Biol. 2011;7:19–21. doi: 10.1038/nchembio.485. [DOI] [PubMed] [Google Scholar]
- 21.Weber T, et al. antiSMASH 3.0: A comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43:W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu F, et al. Characterization of streptonigrin biosynthesis reveals a cryptic carboxyl methylation and an unusual oxidative cleavage of a N-C bond. J Am Chem Soc. 2013;135:1739–1748. doi: 10.1021/ja3069243. [DOI] [PubMed] [Google Scholar]
- 23.Vaillancourt FH, Yeh E, Vosburg DA, O’Connor SE, Walsh CT. Cryptic chlorination by a non-haem iron enzyme during cyclopropyl amino acid biosynthesis. Nature. 2005;436:1191–1194. doi: 10.1038/nature03797. [DOI] [PubMed] [Google Scholar]
- 24.Koketsu K, Watanabe K, Suda H, Oguri H, Oikawa H. Reconstruction of the saframycin core scaffold defines dual Pictet-Spengler mechanisms. Nat Chem Biol. 2010;6:408–410. doi: 10.1038/nchembio.365. [DOI] [PubMed] [Google Scholar]
- 25.Zhang W, et al. Unveiling the post-PKS redox tailoring steps in biosynthesis of the type II polyketide antitumor antibiotic xantholipin. Chem Biol. 2012;19:422–432. doi: 10.1016/j.chembiol.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 26.Kong L, et al. A multifunctional monooxygenase XanO4 catalyzes xanthone formation in xantholipin biosynthesis via a cryptic demethoxylation. Cell Chem Biol. 2016;23:508–516. doi: 10.1016/j.chembiol.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 27.Williamson NR, et al. Biosynthesis of the red antibiotic, prodigiosin, in Serratia: Identification of a novel 2-methyl-3-n-amyl-pyrrole (MAP) assembly pathway, definition of the terminal condensing enzyme, and implications for undecylprodigiosin biosynthesis in Streptomyces. Mol Microbiol. 2005;56:971–989. doi: 10.1111/j.1365-2958.2005.04602.x. [DOI] [PubMed] [Google Scholar]
- 28.Adams E, Frank L. Metabolism of proline and the hydroxyprolines. Annu Rev Biochem. 1980;49:1005–1061. doi: 10.1146/annurev.bi.49.070180.005041. [DOI] [PubMed] [Google Scholar]
- 29.Medema MH, Takano E, Breitling R. Detecting sequence homology at the gene cluster level with MultiGeneBlast. Mol Biol Evol. 2013;30:1218–1223. doi: 10.1093/molbev/mst025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.