Abstract
Stochastic fluctuations, termed “noise,” in the level of biological molecules can greatly impact cellular functions. While biological noise can sometimes be detrimental, recent studies have provided an increasing number of examples in which biological noise can be functionally beneficial. Rather than provide an exhaustive review of the growing literature in this field, in this review we focus on single cell studies based on quantitative microscopy that have generated a deeper understanding of the sources, characteristics, limitations, and benefits of biological noise. Specifically, we highlight studies showing how noise can help coordinate the expression of multiple downstream target genes, impact the channel capacity of signaling networks, and interact synergistically with oscillatory dynamics to enhance the sensitivity of signal processing. We conclude with a discussion of current challenges and future opportunities.
Graphical Abstract
Introduction
At any given moment, cells perform a multitude of complex functions based on diverse biochemical reactions that can be highly susceptible to stochastic fluctuations in molecular reactant levels [1–3]. Such fluctuations in either a single or many molecular reactants across multiple reactions is referred to as molecular noise [4]. Biological noise can arise from multiple sources, including: molecular reactants being present in small quantities [1, 2]; variability in internal states between cells due to differences in cell cycle progression [5]; differences in levels of cellular components due to uneven segregation during cell division [6]; stochasticity in gene expression [7, 8]; or, differences in local microenvironment [4]. In some cases, variations from many sources of noise accumulate and produce cell-to-cell variability in observable phenotypes, while in other cases even a single varying source can produce distinct cellular outcomes for populations that are genetically identical [7, 9, 10].
In many fields, such as electrical engineering, noise is often viewed as a detriment, causing a reduction in the transmission of information. However, from even the early days of molecular biology, biological noise was observed to confer benefits for some biological systems. For example, stochasticity has long been appreciated as serving a key role in regulating the lysis/lysogeny state switch of bacteriophage lambda in which the decision between infection and dormancy states is determined by biochemical fluctuations [11–14]; however, even in this relatively well-characterized system the exact mechanisms and extent to which stochastic biochemical reactions impact the state switch are still being investigated (for a discussion see [15]). As another classic example, early studies of microbial communities showed that cell-to-cell variability caused by biological noise can result in cellular bet-hedging, a process where a fraction of cells randomly enters a state of reduced fitness to increase the likelihood of a subpopulation surviving environmental stresses. Bet-hedging in microbial populations was first observed during studies in the 1940s, in which populations of genetically identical bacterial cells were not completely killed when treated with antibiotics [16]. A subpopulation of cells was able to survive, or “persist,” in the presence of antibiotic; however, the persistence phenotype was nonheritable. Bacterial cultures derived from the persistent cells spontaneously reverted to the normal, non-persistent state, regaining sensitivity to antibiotics [17]. Variations in genetically identical cell populations were also observed in bacterial competence studies, in which only 10–20% of cells entered into a specialized competent state at the expense of cell growth to uptake and incorporate environmental DNA into their chromosome [18, 19]. Although these population-level studies demonstrated the favorable role of noise in cellular bet-hedging strategies, the advent of technologies to study individual cells have illuminated the complex, and often conflicting role of noise in multiple regulatory networks, particularly those involved in signal transduction.
In recent years, the investigation of non-genetic cell-to-cell variability has been propelled by the development of sophisticated tools to analyze individual living cells [20, 21]. Single cell methods deliver invaluable insights into mechanisms of regulation and function that are concealed in bulk, population-level studies measuring ensemble-averaged cellular responses [22–25]. Single cell studies have not only furthered our understanding of the process of bet-hedging in single-celled organisms [26], but have also revealed new mechanisms in which signaling networks have evolved to either circumvent or exploit noise to process inputs. Technologies based on fluorescent protein reporters have been particularly useful in challenging the notion that noisy expression is detrimental to signal processing.
The number of groundbreaking studies over recent years in the area of biological noise precludes an exhaustive analysis of this important topic. Therefore, in this review we choose to focus on recent work in which microscopy-based single cell approaches have generated new insights into the regulation and function of biological noise in well-studied regulatory networks. We first review select studies that enabled better characterization of general features of biological noise, focusing on both live-cell fluorescence microscopy and fixed cell single molecule RNA fluorescence in situ hybridization (smRNA FISH) studies. We then describe select experimental and computational studies showing how variability in transcription factor dynamics may provide benefits of coordination in target gene expression. We next describe recent work analyzing the effects of noise in biological signal transduction systems, revealing how noise generates specific limitations for the transmission of information and how some cells circumvent such limitations. In addition, we discuss specific examples of how cells use biological noise to generate functional benefits for information processing tasks in well-studied signaling networks. Finally, we conclude with a discussion of future research challenges and opportunities.
Characterization of Noise in Gene Expression
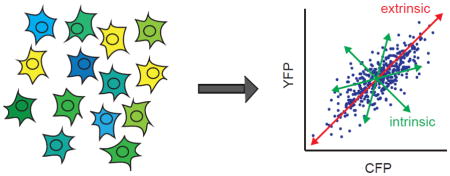
In a broad sense, biological noise can be categorized as being comprised of intrinsic and extrinsic components. Intrinsic noise refers to fluctuations caused by stochastic events in biochemical reactions within any given cell, e.g., the random binding of a transcription factor to a promoter or the number of transcripts produced in a given unit of time. Extrinsic noise refers to variations among different cells in a shared environment, e.g., variation due to cells having different numbers of ribosomes. Extrinsic noise can be further specified as either global extrinsic noise (i.e., fluctuations in molecular components that are the basis for essential biochemical reactions that widely affect gene expression) or pathway-specific extrinsic noise (i.e., fluctuations in the level of a key transcription factor involved in a specific signal transduction pathway) [8, 27].
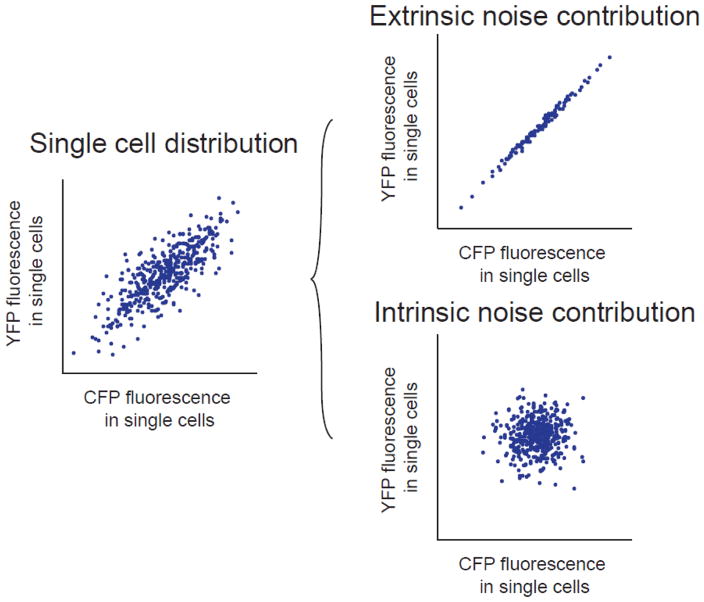
Fluorescence microscopy-based analysis of single cells provided a method to determine the extent to which varied gene expression in a cell population is generated by extrinsic or intrinsic noise sources. Elowitz and colleagues developed a method that employs two distinguishable fluorescent reporter genes from the same origin, i.e., the cyan and yellow fluorescent protein variants derived from the green fluorescent protein [9]. To differentiate between intrinsic and extrinsic noise contributions, the reporter genes are expressed from identical promoters but at distinct integration sites in the genome, such as distinct loci in the E. coli chromosome, as in the work by Elowitz and colleagues [9], or two different alleles of the same gene for a diploid organism, as applied in follow-up studies [8, 28]. Intrinsic noise sources produce differential expression between the reporter genes in a single cell. Extrinsic noise leads to equivalent reporter gene expression within a single cell, but not between two cells (Figure 1). The dual reporter method enabled determination of the contributions of intrinsic and extrinsic noise sources to non-genetic variation in cell populations and identified general mechanisms by which the sources of noise can influence global gene expression and regulation [8, 9, 28, 29].
Figure 1.
Components of gene expression noise. Gene expression noise can be decomposed into intrinsic and extrinsic contributions in single cells using two distinguishable fluorescent reporter genes, CFP and YFP, expressed from identical promoters and integrated at distinct genomic loci. Extrinsic noise produces proportional CFP and YFP expression in each cell. Intrinsic noise produces independent CFP and YFP expression in each cell. Adapted from [21].
To identify more specific molecular mechanisms contributing to extrinsic and intrinsic noise sources in individual cells, several groups have made use of a variety of single cell microscopy-based approaches. While technically challenging due to the often low copy numbers of biomolecular species being analyzed, such studies have enabled precise quantification of mRNA and proteins with single molecule sensitivity. One particularly noteworthy phenomenon identified and characterized through such approaches is transcriptional bursting, a source of noise in gene expression for a variety of organisms [7, 30, 31].
Golding and colleagues monitored transcriptional bursting in real time by measuring mRNA transcript levels with single molecule resolution in individual living E. coli cells [7]. To attain this level of resolution, they used an in vivo mRNA tagging method based on a translational fusion of GFP to the RNA-binding protein MS2 [32, 33], and previously shown to be capable of tracking mRNA movement in living cells [34, 35]. Visualization of MS2-GFP enabled observation of the synthesis of target mRNA transcripts with genetically encoded MS2 binding sites as they were transcribed. Using this approach, Golding and colleagues demonstrated that transcription in E. coli is consistent with a gene activation/inactivation model in that it occurs in quantal bursts, has geometrically distributed burst sizes, and has exponentially distributed time intervals. In addition to transcriptional bursting, studies based on single cell fluorescence microscopy in E. coli have identified sources of cell-to-cell variability in gene activation related to the cellular response to external stimuli, such as the kinetics of cellular uptake mechanisms for inducer molecules [36, 37] Specifically, studies of the arabinose utilization system in individual, living E. coli cells demonstrated that cell-to-cell heterogeneity in gene activation was influenced by the kinetics of arabinose uptake, which may be attributed to varying levels of uptake proteins in cells prior to arabinose induction [36, 37]. Single-cell bioluminescence measurements enabled Suter and colleagues to observe transcriptional bursting in real time in mammalian cells [30]. Using cell lines expressing a short-lived nuclear-localized luciferase encoded by an unstable mRNA, they quantified bursting kinetics of the reporter at high temporal resolution. Through this method, they determined that bursts are gene-specific, which was primarily attributed to cis-acting regulatory elements.
In addition to approaches using live-cell microscopy, several recent studies have highlighted the power of smRNA FISH[38, 39] of fixed cells for characterizing the sources of noise in gene expression. For example, Chong and colleagues used a high-throughput, in vitro method based on smRNA FISH to monitor transcription in real time on individual DNA templates to identify mechanisms generating transcriptional bursting [31]. From this analysis, they determined that the build-up and release of DNA supercoiling during transcription is a physical cause of bursting in E. coli [31]. Jones and colleagues [40] used smRNA FISH to determine the extent to which gene-specific characteristics impact variation in expression. Taking a synthetic biology approach, they generated a suite of synthetic genes in E. coli with distinct promoter strengths, transcription factor binding strengths, and transcription factor copy numbers. Through this systematic approach, they determined that different promoter architectures are associated with different degrees of variability in gene expression. This result suggests that gene expression noise is a tunable property that may be subject to evolutionary selection pressures.
Given that different genes may inherently have distinct levels of expression noise, cells may have a potential difficulty in coordinating the expression of multiple genes that code for multi-protein complexes in which the subunits must be expressed in a proper stoichiometry. Recent work by Gandhi and colleagues using smRNA FISH explored the extent to which transcriptional noise impacts multi-protein complex formation [41]. They implemented two-color smRNA FISH in S. cerevisiae to determine whether mRNA levels of functionally related genes that are constitutively expressed (e.g., essential genes for the stoichiometric formation of a multi-protein complex) are more correlated than those of constitutively expressed genes that are functionally unrelated. Strikingly, their study indicated that mRNA levels of functionally related genes, or even two alleles of the same gene with identical promoters, were as weakly correlated as functionally unrelated genes. This finding suggests that the stochastic nature of transcription initiation may preclude cells from coordinating constitutively expressed genes at the mRNA level, and that the proper stoichiometric formation of multi-protein complexes is more dependent on post-transcriptional or post-translational regulatory mechanisms.
In addition to fluctuations in gene expression and mRNA stability, stochastic events can affect translation and protein stability to alter protein concentrations and generate phenotypic variations in individual cells. Determining the extent to which mRNA levels correlate with protein abundances has been a recent challenge, and several studies highlight the importance of post-transcriptional regulation in determining the concentration and variation of protein abundances [42]. For example, to better characterize fluctuations in not only transcript levels but also protein levels in individual cells, Taniguchi and colleagues carried out a relatively global quantitative analysis of mRNA and protein expression with single molecule sensitivity by conducting smRNA FISH and single molecule imaging of a novel yellow fluorescent protein library in individual E. coli cells [43]. They quantified protein and mRNA levels for over 1000 genes in individual E. coli cells and determined that mRNA and protein levels for any given gene are uncorrelated within a single cell. They reported that this observation was likely caused by the large discrepancy in half-lives between mRNA and proteins; however, this interpretation was recently challenged by a theoretical study conducted by Hilfinger and colleagues [44]. Additionally, through global profiling of the proteome, Taniguchi and colleagues determined that intrinsic noise characterized by transcription rate and burst size could only account for large fluctuations in low-abundance proteins, indicating that variation in high-abundance proteins is dominated by a global extrinsic noise source. One caveat when interpreting such results is that they are based on correlations of mRNA and protein abundances at the same point in time. A more appropriate comparison, which is the next challenge for the field, is to measure the correlation between mRNA levels at one time and protein levels at a slightly later time to take into account the inherent time required for events such as translation and protein maturation.
In addition to characterizing and identifying sources of noise in gene expression, innovative smRNA FISH approaches have advanced investigations of mechanisms for noise suppression. Battich and colleagues [45] showed that cells can employ a buffering mechanism to diminish stochastic fluctuations triggered by transcriptional bursting. Using a high-throughput smRNA FISH technique based on branched DNA technology they measured single cell transcript abundances for nearly 1000 genes and simultaneously quantified 183 cell state features (e.g., cell or nucleus area and shape) and microenvironment (e.g., local cell crowding and number of cell neighbors). In acquiring this large-scale dataset, they determined that cytoplasmic mRNA transcript abundances are minimally stochastic and correlate with features related to cellular state and microenvironment, while nuclear mRNA transcript abundances were highly variable. The disparity between the stochasticity of cytoplasmic and nuclear transcript abundances suggests that cells leverage nuclear retention times of transcripts to filter out stochastic fluctuations arising from transcriptional bursting. In a related study, Padovan-Merhar and colleagues [46] implemented a smRNA FISH approach to demonstrate that mRNA transcript abundance strongly correlates with cellular volume in single mammalian cells, despite having the same absolute number of DNA molecules. This study identified two independent, global mechanisms that enable cells to maintain appropriate mRNA concentrations by compensating for variations in cell size or DNA content during the cell cycle: a trans mechanism to adjust transcriptional burst size for larger cells to produce higher mRNA levels from low DNA copy numbers, and a cis mechanism to regulate burst frequency throughout the cell cycle, preserving the appropriate RNA concentrations even with varying DNA content.
A limitation of early smRNA FISH studies, whether in microbes [31, 41], tissue culture [44], or in vivo [45], is that they could reliably detect transcripts for up to only a few distinct genes in any given cell due to limitations in spectrally distinct fluorophores. However, recent developments in this field have included methods to quantify dozens of distinct transcript species simultaneously. For example, Lubeck and colleagues used super resolution microscopy to quantify spatially barcoded transcripts to simultaneously detect mRNA abundances for 32 genes in single S. cerevisiae cells [46]. They later developed a temporal barcoding scheme in which sequential rounds of mRNA labeling with up to four different fluorescent probes was followed by enzymatic digestion of the probes and labeling with new sets of probes [46]. A benefit of such a technique is that it makes use of traditional fluorescence microscopy instead of the more technically challenging and costly super-resolution microscopy. Using such a scheme with four distinct fluorophores, only two rounds of hybridization were required to label 16 different mRNA species in yeast cells. Theoretically, only eight rounds of hybridization would be required to probe the entire transcriptome. Advances such as these will likely continue to provide powerful methods to probe the impact and mechanisms of noise in gene expression.
Potential Functional Benefits of Noise in Coordinating Gene Expression
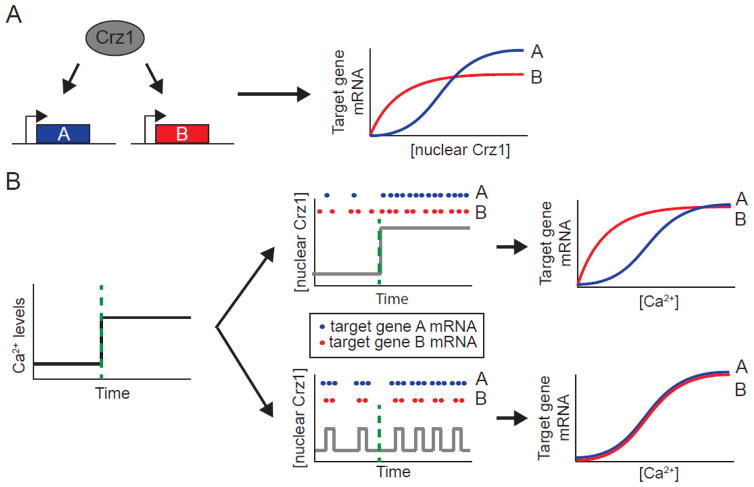
Several recent studies point to a potential role for noise in the coordination of target gene expression in cellular stress responses. Cai and colleagues determined that in the yeast calcium stress response, a wide array of target genes are co-expressed by a single transcription factor undergoing noisy frequency-modulated pulses of nuclear localization [50]. They focused on the transcription factor Crz1, which becomes upregulated in response to calcium (Figure 2A). Using time-lapse fluorescence microscopy, Cai and colleagues showed that Crz1 localizes to the nucleus in short, stochastic bursts with an average duration of 2 minutes. Increasing the calcium concentration affects the number of stochastic bursts per unit of time; however, the average amplitude and duration of the individual bursts are unaltered. Upon translocation to the nucleus, Crz1 regulates the expression of a large number of calcium-response genes. The noisy, short duration nuclear translocations of Crz1 enabled the promoters of different target genes to be activated at a fixed ratio, regardless of the concentration of calcium to which the cells were responding [50] (Figure 2B). This coordination of target gene expression would be difficult to achieve in an amplitude-modulated system where an increase in calcium leads to a proportional increase in nuclear Crz1 concentration, generating different ratios of target gene expression for each concentration of calcium (Figure 2B). A subsequent fluorescence microscopy-based screen by Dalal and colleagues showed that at least twelve different yeast transcription factors undergo similarly noisy short duration, frequency-modulated bursts of nuclear translocation [51]. These results suggest that such noisy transcription factor dynamics may be evolutionarily beneficial for a wide range of cellular responses. While deterministic fixed frequency pulses of transcription factor activity could theoretically confer such gene expression coordination, the fixed-frequency pulses of the mammalian transcription factor p53 were recently shown to reduce coordination of target gene expression in response to DNA damage [52]. Taken together, these results suggest that the noise in the yeast pulsatile transcription factor dynamics may aid in generating the gene coordination observed in yeast systems.
Figure 2.
Frequency-modulation of stochastic nuclear bursts facilitates gene expression coordination in yeast. A) In response to Ca2+, the Crz1 transcription factor localizes to the nucleus in stochastic bursts to activate target genes A and B. Target gene promoters have differential responses to activation by nuclear Crz1 due to variations in affinities or cooperativities (blue and red curves). B) For amplitude-modulation regulation (top panels), an increase in Ca2+ leads to a proportional increase in nuclear Crz1 levels (gray line), generating different ratios of expression of target gene A (blue dots, blue curve normalized to maximal expression) relative to target gene B (red dots, red curve normalized to maximal expression) for different Ca2+ concentrations. For frequency-modulation regulation (bottom panels), an increase in Ca2+ generates stochastic nuclear localization of Crz1 (gray line) that produces a fixed ratio of expression of target gene A (blue dots, blue curve normalized to maximal expression) relative to target gene B (red dots, red curve normalized to maximal expression) regardless of the Ca2+ concentration. Adapted from [3].
Similar to the yeast response networks characterized by Dalal and colleagues, many regulatory networks require multiple target genes to be coordinately expressed. To identify general principles by which noise could impact target gene coordination, Garcia-Bernardo and Dunlop used a stochastic computational approach to characterize the potential impact of noise and transcription factor dynamics in the coordination of target genes [53]. The stochastic model was based on a general “single input regulatory module” in which a single transcription factor regulates several downstream target genes. Target gene expression showed the highest temporal coordination when activated by fluctuating inputs of the transcription factor, such as that generated by pulsatile dynamics or a high level of noise. Additionally, fluctuating inputs enabled the coordination of multiple downstream target genes without an overall increase in the cost imposed on cells when activating stress response genes. In support of these predictions, Meouche and colleagues used time-lapse fluorescence microscopy to show that stochastic expression of MarA, a key transcription factor regulating numerous downstream targets required for multidrug resistance in E. coli, correlated with transient resistance of a subpopulation of cells in the presence of antibiotics [54].
The potential benefits of noise in coordinating multiple target genes within a pathway was further explored by Stewart-Ornstein and colleagues [55]. They developed a noise-based method to identify clusters of genes with a common upstream regulator in yeast cells. Using single-cell correlation analysis, they identified gene regulatory networks based on the concept that noise generated by an upstream regulator can be transmitted to its downstream target genes. Based on pair-wise correlations of yeast proteins tagged with the fluorescent proteins mCherry or GFP, the authors inferred gene regulatory networks by identifying “noise regulons,” i.e., groups of proteins with strongly correlated stochastic fluctuations in their expression. Noise regulons were identified in multiple well-known pathways, including stress response, amino acid biosynthesis, and mitochondrial regulation, suggesting that gene coordination through stochastic fluctuations may be important for coordinating multiple target genes required for a variety of cell physiological needs. Future work will be required to more directly determine the extent to which noise confers functional benefits in the individual pathways.
Effects of Noise on the Channel Capacity of Signal Transduction Networks
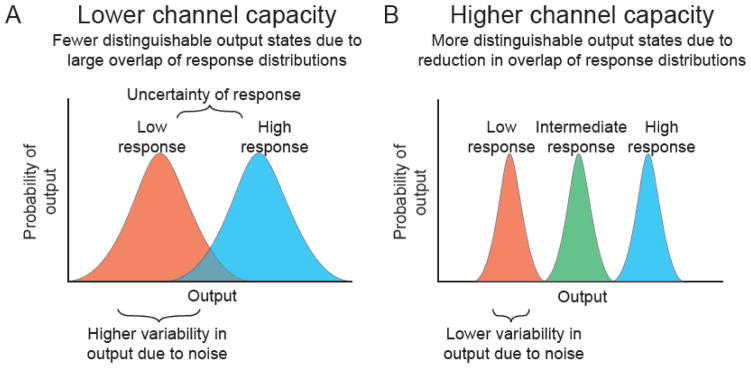
In addition to characterizing the potential sources of noise and their impact on gene expression, single cell approaches have more recently been applied upstream of gene regulation to study the integral role noise plays in signal transduction systems. One particularly interesting feature recently analyzed is the channel capacity of a signal transduction pathway. Channel capacity is a general characteristic of signaling systems, whether they are electronic circuits or kinase cascades, that defines the maximal amount of information that can be transduced along a pathway given the amount of noise present in the pathway components [56–58]. The amount of information is given in units of bits, with n bits corresponding to 2n possible output states that can be distinguished for a given range of inputs. For a greater amount of noise in a system, one generally finds a reduced channel capacity owing to the inability to distinguish between distinct possible states of the signal being transduced (Figure 3).
Figure 3.
Limitation of the channel capacity of a signal transduction system by noise. A) Biochemical noise increases the variability in biological response distributions (red and blue output distributions), increasing the region of overlap between output distributions and resulting in a reduction of the possible number of distinguishable output states. B) For a signal transduction pathway with lower noise, the variability in an output response distribution is smaller, generating a higher number of distinguishable output states. Adapted from [56].
Recent studies have quantified the channel capacity of a variety of biological systems. Building on previous work applying information theory approaches to neural signaling [59, 60] and transcription factor activity in Drosophila embryo patterning [61], several groups quantified the channel capacity of kinase networks. For the quorum sensing systems in the prokaryote V. harveyi, Mehta and coworkers calculated that the channel capacity for each of the kinases responding to the presence of auto-inducer stimuli is in the range of 0.6–1.5 bits [62]. Thus, for these signaling systems operating with a channel capacity on the order of 1 bit, only two possible output states can reliably be encoded over the physiological range of stimuli. One could interpret these two states as an “off” state and an “on” state for the absence or presence of an input signal, respectively.
Information theoretic approaches have also determined the channel capacity of several mammalian signaling systems. Using immunocytochemistry of individual cells, Cheong and colleagues quantified the concentrations of the transcription factor nuclear factor-kappa B (NF-κB) in response to TNF stimulation [63]. They determined that the channel capacity for the TNF response in any given cell was about 1 bit, similar to that for the V. harveyi quorum sensing pathways [63]. Analysis of several other pathways including signaling by platelet-derived growth factor, epidermal growth factor, and G protein-coupled receptor signaling also revealed channel capacities on the order of 1 bit [56], indicating a potentially common limitation generated by noise for multiple signaling systems in both prokaryotes and eukaryotes.
How then might cells distinguish a greater variety of signaling states beyond a binary system? One possible method is to reduce the noise in the system. Negative feedback is a common mechanism for reducing noise levels in electrical circuits, and previous studies have shown that negative feedback loops are one potential mechanism for attenuating noise in biological circuits [64, 65]. However, noise reduction via negative feedback may come at the cost of reduced signal sensitivity [66], and there are fundamental limits for the extent to which noise suppression can occur [67]. In addition, at least for the NF-κB response to TNF stimulation, negative feedback has experimentally been found to increase channel capacity by only a modest amount [56]. Another method for cells to increase the possible number of output states for a signaling network is to integrate information from several parallel pathways. Analogous to a string of zeroes and ones being used in digital circuits to represent a larger number of output states, separate signaling molecules individually distinguishable as only a high or a low level could be combinatorially arranged to generate a larger number of cellular output states. It has been speculated that such a system could have evolved in early Drosophila embryo patterning as a mechanism for generating multiple states of positional information from the combinatorial activation of four different transcription factor genes (hunchback, krüppel, giant, and knirps)[61]. Another potential mechanism for increasing information in a signaling network is for a group of cells to act as an ensemble in responding to a given signal, relying on cell-to-cell communication via diffusible signals to minimize the effects of information loss from biochemical noise. Cheong and coworkers calculated that for the NF-κB response to TNF, even a modestly sized ensemble of 14 cells could theoretically increase the channel capacity of the signaling pathway to nearly double the number of bits [56].
Another particularly intriguing method by which biological systems can circumvent signaling bottlenecks generated by biochemical noise is to make use of temporal dynamics. As an illustrative example of the power of temporal sampling, consider Morse code, in which one of two states – a “dot” or a “dash” – is possible at any given time for a transmitted signal. Even though the signal is represented by two possible states, by considering a string of these two states over time – a chain of dots and dashes—the number of possible messages (and thus the information that can be transmitted via Morse code) is limitless. In biological contexts, it has been well-established that neural circuits make use of temporally encoded information via the frequency of neural spikes [68]. More recently, fluorescence microscopy approaches have shown that several intracellular signaling systems including the ERK, Ca+2, and NF-κB pathways can make use of dynamics to reduce information loss due to extrinsic noise [69]. Complex temporal regulation has been identified in several eukaryotic signaling networks through the use of time-lapsed fluorescence microscopy. For example, oscillatory dynamics have been observed in a variety of systems including the Ca+2 [70–73], NF-κB[23, 74, 75], p53 [22, 76–78], and ERK [79–81] signaling pathways. With greater use of single-cell approaches to track the dynamics of important signaling molecules, it is likely that we may yet identify additional mechanisms by which cells have evolved dynamic-encoding schemes to increase the information content of signaling pathways to circumvent limitations of biological noise.
Noise Can Enhance Dynamic Signal Processing
Novel single cell fluorescence microscopy-based studies have shown that cells can potentially harness noise not only to promote the coordination of target gene expression but also to improve dynamic signal processing. For example, recent studies have shown that biological noise in nonlinear, oscillatory systems can facilitate signal amplification and induce cellular heterogeneity. Experimental and computational work has shown that noise can improve signal detection of dynamic inputs using either stochastic resonance, i.e., where noise fluctuations promote the detection of weak signals,[82, 83] or stochastic focusing, i.e., where noise fluctuations enable a system with a gradual response mechanism to function as if it has a threshold response mechanism [84, 85]. Stochastic resonance, in particular, has been shown to be beneficial for increasing the sensitivity of biological signaling networks by modulating the periodic input to entrain, or synchronize, oscillators in a cell to generate an amplified response [86–90]. Entrainment occurs when an oscillator becomes phase-locked with a driving stimulus, i.e., when the natural frequency of the biological oscillator matches that of a periodic input. Such a condition enables a system to have increased sensitivity to a weak input signal. Entrainment has been observed in circadian clocks [91] as well as sensory neuron processing [92].
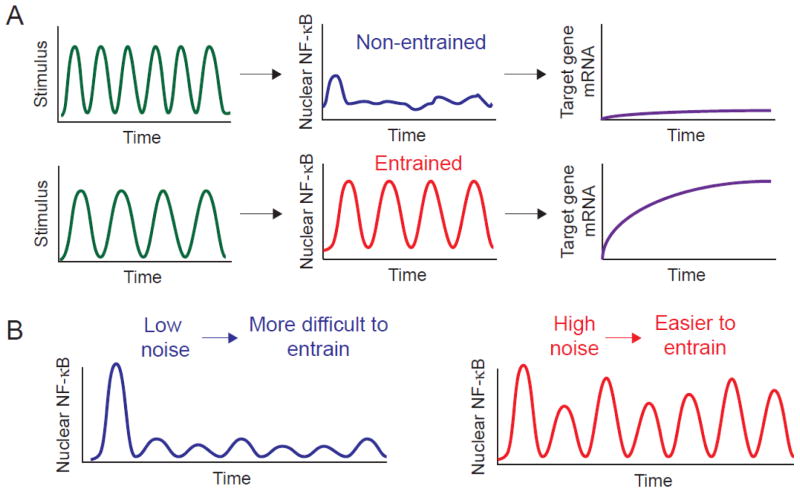
Recent single cell fluorescence microscopy studies have shown the importance of entrainment in dynamic signal processing. Kellogg and Tay demonstrated how noise and oscillations positively impact the extensively studied NF-κB signaling network [93]. Using a microfluidic-based approach to deliver sustained or dynamic TNF cytokine signals to live cells, they tracked fluorescently-tagged NF-κB in individual cells and assayed NF-κB target gene expression (Figure 4A). They determined that the NF-κB system achieves entrainment and synchronizes oscillations at specific TNF input frequencies, leading to improved gene expression efficiency and reduced cell-to-cell variability [93].
Figure 4.
Enhancing the sensitivity of biological oscillators through entrainment. A) Entrainment occurs when biological oscillators become phase-locked with a driving stimulus such that the natural frequency of the oscillator matches the frequency of the input signal. Oscillators such as the transcription factor NF-κB can generate a greater output when entrained, such as more highly activating downstream target gene expression. B) Both intrinsic and extrinsic noise can facilitate entrainment. Adapted from ref. [93].
Intrinsic and extrinsic noise impact entrainment differently, enabling signaling pathways to carry out distinct functions. Kellogg and Tay used a stochastic model based on the nonlinear binding of NF-κB to DNA to demonstrate that entrainment in the NF-κB pathway was enhanced by intrinsic noise (Figure 4B), which directly amplified the NF-κB oscillations to strengthen the transcriptional output and improve gene expression efficiency [93]. Conversely, intrinsic noise can be leveraged to disrupt entrainment and maintain robust single cell oscillations in the presence of cellular perturbations, and is predicted to be important for the overall coordination and stability of population-level responses. Paszek and colleagues showed that the NF-κB pathway implements a dual delayed feedback motif, which involves the optimally timed stochastic transcription of the NF-κB inhibitors IκBα and IκBε, to generate robust oscillations in individual cells [94, 95]. The importance of noise for maintaining robust oscillations in the presence of a fluctuating environment has also been observed in Ca+2 signaling [96]. While intrinsic noise can facilitate or disrupt entrainment to evoke specific functions, extrinsic noise can enhance entrainment by providing population diversity, which promotes bet-hedging mechanisms in unpredictable environments [97, 98]. Kellogg and Tay used stochastic modeling to demonstrate that extrinsic noise facilitates bet-hedging mechanisms by inducing variability in the natural period of NF-κB, ensuring that a subpopulation of cells can entrain to a wide range of TNF input frequencies [93].
Entrainment of individual oscillators may also prove useful for the development of robust large-scale synthetic oscillators. Mondragón-Palomino and colleagues applied single cell time-lapse fluorescence microscopy and computational modeling to monitor hundreds of bacterial synthetic oscillators. They identified extrinsic noise as the source of stochastic variability, enabling the oscillators to entrain to a wide range of frequencies [99].
Outlook and Opportunities
We have discussed numerous studies demonstrating the complex nature of noise in gene regulation and signal transduction. In certain conditions, noise presents a challenge to cells, as was shown in studies addressing channel capacity; in other conditions, cells may leverage noise, as was shown in examples where noise enhanced dynamic signal processing. There is much work needed to fully understand the sources and impact of biological noise, and this effort will likely require the development of novel single cell technologies based on fluorescence microscopy. For example, single-cell correlation analysis is an important tool in the study of noise propagation; however, it provides a limited view when only a single time point is considered, as cellular networks are dynamic [55, 100, 101]. Another pressing challenge in studies probing the contributions of noise in signal transduction is the ability to study the coordination of multiple network nodes simultaneously and dynamically in cells. To address these questions, new methods and technologies that enable combinatorial labeling and multiplex detection of mRNAs or proteins with single molecule sensitivity will likely need to be developed. In addition, the study of noise propagation in signaling networks remains a difficult task in multicellular tissues and will require technologies that can detect mRNAs or proteins with single molecule sensitivity in dense, opaque matter [102, 103]. With the development of advanced fluorescence imaging methods and technologies, as well as integration of knowledge obtained through the emerging fields of single cell “omics” approaches, we expect future studies to continue to unravel the intricate role of noise in dynamic gene regulation and signal processing.
Research Highlights.
fluorescence-based approaches have advanced our understanding of biological noise
recent studies highlight the importance of noise in gene regulation and signaling
cells have evolved methods to circumvent limitations generated by noise
in some situations, mechanisms have evolved to generate benefits from noise
Acknowledgments
We thank the members of the Batchelor lab for useful discussions during the writing of this review. M.D.H. and E.B. are supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. The authors declare no competing financial interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Van Kampen NG. Stochastic Processes in Physics and Chemistry. 3. Amsterdam: Elsevier; 2007. Chapter III - Stochastic Processes; pp. 52–72. [Google Scholar]
- 2.McAdams HH, Arkin A. It’s a noisy business! Genetic regulation at the nanomolar scale. Trends in Genetics. 1999;15:65–9. doi: 10.1016/s0168-9525(98)01659-x. [DOI] [PubMed] [Google Scholar]
- 3.Eldar A, Elowitz MB. Functional roles for noise in genetic circuits. Nature. 2010;467:167–73. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCullagh E, Farlow J, Fuller C, Girard J, Lipinski-Kruszka J, Lu D, et al. Not all quiet on the noise front. Nat Chem Biol. 2009;5:699–704. doi: 10.1038/nchembio.222. [DOI] [PubMed] [Google Scholar]
- 5.Raser JM, O’Shea EK. Noise in Gene Expression: Origins, Consequences, and Control. Science. 2005;309:2010–3. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huh D, Paulsson J. Non-genetic heterogeneity from stochastic partitioning at cell division. Nat Genet. 2011;43:95–100. doi: 10.1038/ng.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Golding I, Paulsson J, Zawilski SM, Cox EC. Real-Time Kinetics of Gene Activity in Individual Bacteria. Cell. 2005;123:1025–36. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 8.Raser JM, O’Shea EK. Control of Stochasticity in Eukaryotic Gene Expression. Science. 2004;304:1811–4. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic Gene Expression in a Single Cell. Science. 2002;297:1183–6. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 10.Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet. 2005;6:451–64. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- 11.Lwoff A. Lysogeny. Bacteriological Reviews. 1953;17:269–337. doi: 10.1128/br.17.4.269-337.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neubauer Z, Calef E. Immunity phase-shift in defective lysogens: Non-mutational hereditary change of early regulation of λ prophage. Journal of Molecular Biology. 1970;51:1–13. doi: 10.1016/0022-2836(70)90265-2. [DOI] [PubMed] [Google Scholar]
- 13.Arkin A, Ross J, McAdams HH. Stochastic Kinetic Analysis of Developmental Pathway Bifurcation in Phage λ-Infected Escherichia coli Cells. Genetics. 1998;149:1633–48. doi: 10.1093/genetics/149.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ptashne M. A Genetic Switch: Phage Lambda Revisited. 3. Cold Spring Harbor Laboratory Press; 2004. [Google Scholar]
- 15.Golding I. Decision Making in Living Cells: Lessons from a Simple System. Annual Review of Biophysics. 2011;40:63–80. doi: 10.1146/annurev-biophys-042910-155227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bigger J. Treatment Of Staphylococcal Infections With Penicillin By Intermittent Sterilisation. The Lancet. 1944;244:497–500. [Google Scholar]
- 17.Moyed HS, Broderick SH. Molecular cloning and expression of hipA, a gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. Journal of Bacteriology. 1986;166:399–403. doi: 10.1128/jb.166.2.399-403.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hadden C, Nester EW. Purification of Competent Cells in the Bacillus subtilis Transformation System. Journal of Bacteriology. 1968;95:876–85. doi: 10.1128/jb.95.3.876-885.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cahn FH, Fox MS. Fractionation of Transformable Bacteria from Competent Cultures of Bacillus subtilis on Renografin Gradients. Journal of Bacteriology. 1968;95:867–75. doi: 10.1128/jb.95.3.867-875.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaudet S, Miller-Jensen K. Redefining Signaling Pathways with an Expanding Single-Cell Toolbox. Trends in Biotechnology. 2016;34:458–69. doi: 10.1016/j.tibtech.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raj A, Oudenaarden Av. Single-Molecule Approaches to Stochastic Gene Expression. Annual Review of Biophysics. 2009;38:255–70. doi: 10.1146/annurev.biophys.37.032807.125928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB, et al. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet. 2004;36:147–50. doi: 10.1038/ng1293. [DOI] [PubMed] [Google Scholar]
- 23.Nelson DE, Ihekwaba AEC, Elliott M, Johnson JR, Gibney CA, Foreman BE, et al. Oscillations in NF-κB Signaling Control the Dynamics of Gene Expression. Science. 2004;306:704–8. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- 24.Aoki K, Kumagai Y, Sakurai A, Komatsu N, Fujita Y, Shionyu C, et al. Stochastic ERK Activation Induced by Noise and Cell-to-Cell Propagation Regulates Cell Density-Dependent Proliferation. Molecular Cell. 2013;52:529–40. doi: 10.1016/j.molcel.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 25.Locke JCW, Elowitz MB. Using Movies to Analyse Gene Circuit Dynamics in Single Cells. Nature reviews Microbiology. 2009;7:383–92. doi: 10.1038/nrmicro2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maamar H, Dubnau D. Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Molecular Microbiology. 2005;56:615–24. doi: 10.1111/j.1365-2958.2005.04592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pedraza JM, van Oudenaarden A. Noise Propagation in Gene Networks. Science. 2005;307:1965–9. doi: 10.1126/science.1109090. [DOI] [PubMed] [Google Scholar]
- 28.Bar-Even A, Paulsson J, Maheshri N, Carmi M, O’Shea E, Pilpel Y, et al. Noise in protein expression scales with natural protein abundance. Nat Genet. 2006;38:636–43. doi: 10.1038/ng1807. [DOI] [PubMed] [Google Scholar]
- 29.Hansen AS, O’Shea EK. Promoter decoding of transcription factor dynamics involves a trade-off between noise and control of gene expression. Molecular Systems Biology. 2013;9:704. doi: 10.1038/msb.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suter DM, Molina N, Gatfield D, Schneider K, Schibler U, Naef F. Mammalian Genes Are Transcribed with Widely Different Bursting Kinetics. Science. 2011;332:472–4. doi: 10.1126/science.1198817. [DOI] [PubMed] [Google Scholar]
- 31.Chong S, Chen C, Ge H, Xie XS. Mechanism of Transcriptional Bursting in Bacteria. Cell. 158:314–26. doi: 10.1016/j.cell.2014.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peabody DS. The RNA binding site of bacteriophage MS2 coat protein. The EMBO Journal. 1993;12:595–600. doi: 10.1002/j.1460-2075.1993.tb05691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA Particles in Living Yeast. Molecular Cell. 1998;2:437–45. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 34.Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard J-M, Singer RH, et al. Single mRNA Molecules Demonstrate Probabilistic Movement in Living Mammalian Cells. Current Biology. 2003;13:161–7. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Golding I, Cox EC. RNA dynamics in live Escherichia coli cells. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:11310–5. doi: 10.1073/pnas.0404443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mäkelä J, Kandhavelu M, Oliveira SMD, Chandraseelan JG, Lloyd-Price J, Peltonen J, et al. In vivo single-molecule kinetics of activation and subsequent activity of the arabinose promoter. Nucleic Acids Research. 2013;41:6544–52. doi: 10.1093/nar/gkt350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Megerle JA, Fritz G, Gerland U, Jung K, Rädler JO. Timing and Dynamics of Single Cell Gene Expression in the Arabinose Utilization System. Biophysical Journal. 2008;95:2103–15. doi: 10.1529/biophysj.107.127191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of Single RNA Transcripts in Situ. Science. 1998;280:585–90. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 39.Raj A, Tyagi S. Chapter 17 - Detection of Individual Endogenous RNA Transcripts In Situ Using Multiple Singly Labeled Probes. In: Nils GW, editor. Methods in Enzymology. Academic Press; 2010. pp. 365–86. [DOI] [PubMed] [Google Scholar]
- 40.Jones DL, Brewster RC, Phillips R. Promoter architecture dictates cell-to-cell variability in gene expression. Science. 2014;346:1533–6. doi: 10.1126/science.1255301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gandhi SJ, Zenklusen D, Lionnet T, Singer RH. Transcription of functionally related constitutive genes is not coordinated. Nat Struct Mol Biol. 2011;18:27–34. doi: 10.1038/nsmb.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–32. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taniguchi Y, Choi PJ, Li G-W, Chen H, Babu M, Hearn J, et al. Quantifying E. coli Proteome and Transcriptome with Single-Molecule Sensitivity in Single Cells. Science. 2010;329:533–8. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hilfinger A, Norman Thomas M, Paulsson J. Exploiting Natural Fluctuations to Identify Kinetic Mechanisms in Sparsely Characterized Systems. Cell Systems. 2016;2:251–9. doi: 10.1016/j.cels.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 45.Battich N, Stoeger T, Pelkmans L. Control of Transcript Variability in Single Mammalian Cells. Cell. 2015;163:1596–610. doi: 10.1016/j.cell.2015.11.018. [DOI] [PubMed] [Google Scholar]
- 46.Padovan-Merhar O, Nair Gautham P, Biaesch Andrew G, Mayer A, Scarfone S, Foley Shawn W, et al. Single Mammalian Cells Compensate for Differences in Cellular Volume and DNA Copy Number through Independent Global Transcriptional Mechanisms. Molecular Cell. 2015;58:339–52. doi: 10.1016/j.molcel.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singer Zakary S, Yong J, Tischler J, Hackett Jamie A, Altinok A, Surani MA, et al. Dynamic Heterogeneity and DNA Methylation in Embryonic Stem Cells. Molecular Cell. 55:319–31. doi: 10.1016/j.molcel.2014.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Meth. 2008;5:877–9. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lubeck E, Cai L. Single-cell systems biology by super-resolution imaging and combinatorial labeling. Nat Meth. 2012;9:743–8. doi: 10.1038/nmeth.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cai L, Dalal CK, Elowitz MB. Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature. 2008;455:485–90. doi: 10.1038/nature07292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dalal Chiraj K, Cai L, Lin Y, Rahbar K, Elowitz Michael B. Pulsatile Dynamics in the Yeast Proteome. Current Biology. 2014;24:2189–94. doi: 10.1016/j.cub.2014.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Porter Joshua R, Fisher Brian E, Batchelor E. p53 Pulses Diversify Target Gene Expression Dynamics in an mRNA Half-Life-Dependent Manner and Delineate Co-regulated Target Gene Subnetworks. Cell Systems. 2:272–82. doi: 10.1016/j.cels.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garcia-Bernardo J, Dunlop Mary J. Noise and Low-Level Dynamics Can Coordinate Multicomponent Bet Hedging Mechanisms. Biophysical Journal. 2015;108:184–93. doi: 10.1016/j.bpj.2014.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.El Meouche I, Siu Y, Dunlop MJ. Stochastic expression of a multiple antibiotic resistance activator confers transient resistance in single cells. Scientific Reports. 2016;6:19538. doi: 10.1038/srep19538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stewart-Ornstein J, Weissman Jonathan S, El-Samad H. Cellular Noise Regulons Underlie Fluctuations in Saccharomyces cerevisiae. Molecular Cell. 45:483–93. doi: 10.1016/j.molcel.2011.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheong R, Rhee A, Wang CJ, Nemenman I, Levchenko A. Information Transduction Capacity of Noisy Biochemical Signaling Networks. Science. 2011;334:354–8. doi: 10.1126/science.1204553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alex R, Raymond C, Andre L. The application of information theory to biochemical signaling systems. Physical Biology. 2012;9:045011. doi: 10.1088/1478-3975/9/4/045011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hansen AS, O’Shea EK. Limits on information transduction through amplitude and frequency regulation of transcription factor activity. eLife. 2015;4:e06559. doi: 10.7554/eLife.06559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Ruyter van Steveninck RR, Lewen GD, Strong SP, Koberle R, Bialek W. Reproducibility and Variability in Neural Spike Trains. Science. 1997;275:1805–8. doi: 10.1126/science.275.5307.1805. [DOI] [PubMed] [Google Scholar]
- 60.Borst A, Theunissen FE. Information theory and neural coding. Nat Neurosci. 1999;2:947–57. doi: 10.1038/14731. [DOI] [PubMed] [Google Scholar]
- 61.Tkačik G, Callan CG, Bialek W. Information flow and optimization in transcriptional regulation. Proceedings of the National Academy of Sciences. 2008;105:12265–70. doi: 10.1073/pnas.0806077105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mehta P, Goyal S, Long T, Bassler BL, Wingreen NS. Information processing and signal integration in bacterial quorum sensing. Molecular Systems Biology. 2009:5. doi: 10.1038/msb.2009.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rhee A, Cheong R, Levchenko A. Noise decomposition of intracellular biochemical signaling networks using nonequivalent reporters. Proceedings of the National Academy of Sciences. 2014;111:17330–5. doi: 10.1073/pnas.1411932111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Becskei A, Serrano L. Engineering stability in gene networks by autoregulation. Nature. 2000;405:590–3. doi: 10.1038/35014651. [DOI] [PubMed] [Google Scholar]
- 65.Dublanche Y, Michalodimitrakis K, Kümmerer N, Foglierini M, Serrano L. Noise in transcription negative feedback loops: simulation and experimental analysis. Molecular Systems Biology. 2006;2:41. doi: 10.1038/msb4100081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hornung G, Barkai N. Noise Propagation and Signaling Sensitivity in Biological Networks: A Role for Positive Feedback. PLoS Comput Biol. 2008;4:e8. doi: 10.1371/journal.pcbi.0040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lestas I, Vinnicombe G, Paulsson J. Fundamental limits on the suppression of molecular fluctuations. Nature. 2010;467:174–8. doi: 10.1038/nature09333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rieke F, Warland David, Bialek William. Spikes: exploring the neural code. MIT Press; 1999. [Google Scholar]
- 69.Selimkhanov J, Taylor B, Yao J, Pilko A, Albeck J, Hoffmann A, et al. Accurate information transmission through dynamic biochemical signaling networks. Science. 2014;346:1370–3. doi: 10.1126/science.1254933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dupont G, Combettes L, Bird GS, Putney JW. Calcium Oscillations. Cold Spring Harbor Perspectives in Biology. 2011;3:a004226. doi: 10.1101/cshperspect.a004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dolmetsch RE, Xu K, Lewis RS. Calcium oscillations increase the efficiency and specificity of gene expression. Nature. 1998;392:933–6. doi: 10.1038/31960. [DOI] [PubMed] [Google Scholar]
- 72.Hannanta-anan P, Chow Brian Y. Optogenetic Control of Calcium Oscillation Waveform Defines NFAT as an Integrator of Calcium Load. Cell Systems. 2016;2:283–8. doi: 10.1016/j.cels.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kupzig S, Walker SA, Cullen PJ. The frequencies of calcium oscillations are optimized for efficient calcium-mediated activation of Ras and the ERK/MAPK cascade. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:7577–82. doi: 10.1073/pnas.0409611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Y, Paszek P, Horton CA, Kell DB, White MR, Broomhead DS, et al. Interactions among oscillatory pathways in NF-kappa B signaling. BMC Systems Biology. 2011;5:23. doi: 10.1186/1752-0509-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ashall L, Horton CA, Nelson DE, Paszek P, Harper CV, Sillitoe K, et al. Pulsatile stimulation determines timing and specificity of NF-kappaB-dependent transcription. Science. 2009;324:242–6. doi: 10.1126/science.1164860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Purvis JE, Karhohs KW, Mock C, Batchelor E, Loewer A, Lahav G. p53 Dynamics Control Cell Fate. Science. 2012;336:1440–4. doi: 10.1126/science.1218351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Batchelor E, Loewer A, Lahav G. The ups and downs of p53: understanding protein dynamics in single cells. Nat Rev Cancer. 2009;9:371–7. doi: 10.1038/nrc2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Batchelor E, Loewer A, Mock C, Lahav G. Stimulus-dependent dynamics of p53 in single cells. Molecular Systems Biology. 2011;7:488. doi: 10.1038/msb.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shankaran H, Ippolito DL, Chrisler WB, Resat H, Bollinger N, Opresko LK, et al. Rapid and sustained nuclear–cytoplasmic ERK oscillations induced by epidermal growth factor. Molecular Systems Biology. 2009;5:332. doi: 10.1038/msb.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Aoki K, Kumagai Y, Sakurai A, Komatsu N, Fujita Y, Shionyu C, et al. Stochastic ERK Activation Induced by Noise and Cell-to-Cell Propagation Regulates Cell Density-Dependent Proliferation. Molecular Cell. 52:529–40. doi: 10.1016/j.molcel.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 81.Albeck John G, Mills Gordon B, Brugge Joan S. Frequency-Modulated Pulses of ERK Activity Transmit Quantitative Proliferation Signals. Molecular Cell. 2013;49:249–61. doi: 10.1016/j.molcel.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Collins JJ, Imhoff TT, Grigg P. Noise-enhanced information transmission in rat SA1 cutaneous mechanoreceptors via aperiodic stochastic resonance. Journal of Neurophysiology. 1996;76:642–5. doi: 10.1152/jn.1996.76.1.642. [DOI] [PubMed] [Google Scholar]
- 83.Douglass JK, Wilkens L, Pantazelou E, Moss F. Noise enhancement of information transfer in crayfish mechanoreceptors by stochastic resonance. Nature. 1993;365:337–40. doi: 10.1038/365337a0. [DOI] [PubMed] [Google Scholar]
- 84.Paulsson J, Berg OG, Ehrenberg M. Stochastic focusing: Fluctuation-enhanced sensitivity of intracellular regulation. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:7148–53. doi: 10.1073/pnas.110057697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Berg OG, Paulsson J, Ehrenberg M. Fluctuations in Repressor Control: Thermodynamic Constraints on Stochastic Focusing. Biophysical Journal. 2000;79:2944–53. doi: 10.1016/S0006-3495(00)76531-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Abraham U, Granada AE, Westermark PO, Heine M, Kramer A, Herzel H. Coupling governs entrainment range of circadian clocks. Molecular Systems Biology. 2010;6:438. doi: 10.1038/msb.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mori T, Kai S. Noise-Induced Entrainment and Stochastic Resonance in Human Brain Waves. Physical Review Letters. 2002;88:218101. doi: 10.1103/PhysRevLett.88.218101. [DOI] [PubMed] [Google Scholar]
- 88.Jensen MH, Krishna S. Inducing phase-locking and chaos in cellular oscillators by modulating the driving stimuli. FEBS Letters. 2012;586:1664–8. doi: 10.1016/j.febslet.2012.04.044. [DOI] [PubMed] [Google Scholar]
- 89.Zhou C, Kurths J, Kiss IZ, Hudson JL. Noise-Enhanced Phase Synchronization of Chaotic Oscillators. Physical Review Letters. 2002;89:014101. doi: 10.1103/PhysRevLett.89.014101. [DOI] [PubMed] [Google Scholar]
- 90.Läer L, Kloppstech M, Schöfl C, Sejnowski TJ, Brabant G, Prank K. Noise enhanced hormonal signal transduction through intracellular calcium oscillations. Biophysical Chemistry. 2001;91:157–66. doi: 10.1016/s0301-4622(01)00167-3. [DOI] [PubMed] [Google Scholar]
- 91.Gérard C, Goldbeter A. Entrainment of the Mammalian Cell Cycle by the Circadian Clock: Modeling Two Coupled Cellular Rhythms. PLoS Comput Biol. 2012;8:e1002516. doi: 10.1371/journal.pcbi.1002516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McDonnell MD, Ward LM. The benefits of noise in neural systems: bridging theory and experiment. Nat Rev Neurosci. 2011;12:415–26. doi: 10.1038/nrn3061. [DOI] [PubMed] [Google Scholar]
- 93.Kellogg Ryan A, Tay S. Noise Facilitates Transcriptional Control under Dynamic Inputs. Cell. 2015;160:381–92. doi: 10.1016/j.cell.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 94.Paszek P, Ryan S, Ashall L, Sillitoe K, Harper CV, Spiller DG, et al. Population robustness arising from cellular heterogeneity. Proceedings of the National Academy of Sciences. 2010;107:11644–9. doi: 10.1073/pnas.0913798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ashall L, Horton CA, Nelson DE, Paszek P, Harper CV, Sillitoe K, et al. Pulsatile Stimulation Determines Timing and Specificity of NF-κB-Dependent Transcription. Science. 2009;324:242–6. doi: 10.1126/science.1164860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Perc M, Marhl M. Noise enhances robustness of intracellular Ca2+ oscillations. Physics Letters A. 2003;316:304–10. [Google Scholar]
- 97.Wakamoto Y, Dhar N, Chait R, Schneider K, Signorino-Gelo F, Leibler S, et al. Dynamic Persistence of Antibiotic-Stressed Mycobacteria. Science. 2013;339:91–5. doi: 10.1126/science.1229858. [DOI] [PubMed] [Google Scholar]
- 98.Süel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB. An excitable gene regulatory circuit induces transient cellular differentiation. Nature. 2006;440:545–50. doi: 10.1038/nature04588. [DOI] [PubMed] [Google Scholar]
- 99.Mondragón-Palomino O, Danino T, Selimkhanov J, Tsimring L, Hasty J. Entrainment of a Population of Synthetic Genetic Oscillators. Science. 2011;333:1315–9. doi: 10.1126/science.1205369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Munsky B, Neuert G, van Oudenaarden A. Using Gene Expression Noise to Understand Gene Regulation. Science. 2012;336:183–7. doi: 10.1126/science.1216379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dunlop MJ, Cox RS, Levine JH, Murray RM, Elowitz MB. Regulatory activity revealed by dynamic correlations in gene expression noise. Nat Genet. 2008;40:1493–8. doi: 10.1038/ng.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang B, Treweek Jennifer B, Kulkarni Rajan P, Deverman Benjamin E, Chen C-K, Lubeck E, et al. Single-Cell Phenotyping within Transparent Intact Tissue through Whole-Body Clearing. Cell. 2014;158:945–58. doi: 10.1016/j.cell.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shah S, Lubeck E, Schwarzkopf M, He T-F, Greenbaum A, Sohn CH, et al. Single-molecule RNA detection at depth by hybridization chain reaction and tissue hydrogel embedding and clearing. Development. 2016;143:2862–7. doi: 10.1242/dev.138560. [DOI] [PMC free article] [PubMed] [Google Scholar]